Abstract
A modification of spoligotyping with primers derived from the direct repeat (DR) and IS6110 sequences was used to identify IS6110 insertions in the DR locus of Mycobacterium tuberculosis clinical strains from the St. Petersburg area of Russia. Novel IS6110 insertions were identified: (i) in two epidemiologically unlinked Beijing family strains, an asymmetrical direct insertion in DR37; (ii) in a non-Beijing strain, an asymmetrical insertion in the opposite orientation in DR38; (iii) in another non-Beijing strain, a direct insertion in DR38 and one in the opposite orientation in DR14 (DR numbering is according to standard spoligotyping). Our results strengthen an observation that the DR locus structure is extremely conserved in the Beijing genotype. Asymmetrical insertions prevented detection of the adjacent spacer by standard spoligotyping. This, therefore, should be taken into consideration when similar spoligoprofiles that differ only in signals 37 and 38 are interpreted.
The direct repeat (DR) locus is characteristic of the Mycobacterium tuberculosis complex (9). No significant homology between the M. tuberculosis DR sequence and the rest of its genome and DNA of other bacterial genera was found (18). The DR locus consists of multiple tandem 36-bp repeats (virtually exact) interspersed with variable spacers of about equal size. The DR and the adjacent variable sequence form a direct variant repeat (DVR). Polymorphism of the DR locus (absence or presence of single DVRs) has been exploited widely for distinguishing among clinical isolates of the M. tuberculosis complex by using spacer oligonucleotide typing (spoligotyping) (10, 18, 19). Initially, 43 distinct DVRs were described (10), but recently it has been shown that up to 68 copies may be present in the genome of M. tuberculosis and that their order is strongly conserved in the locus (2, 18).
Though less discriminative than IS6110 RFLP typing (17), spoligotyping has been used successfully in many cases for strain differentiation (10, 15, 16). At the same time, the strains of the Beijing genetic family within M. tuberculosis could not be differentiated by spoligotyping alone, since DR spacers 35 to 43 were present in virtually all isolates (13, 15, 19). This notorious genotype, originally found to be endemic in East Asia (19), currently shows worldwide distribution (16). The well-defined strain W belongs to this family, too (5). Previously, it was also shown to be predominant in the northwestern region of Russia, and suspected ongoing transmission of the multidrug-resistant strains of the Beijing family was shown to be the principal cause of the epidemic spread of multidrug-resistant tuberculosis in this region (12a, 13).
The DR locus presents one of the IS6110 insertion preferential loci (ipl) in the M. tuberculosis genome (9). It was shown that the IS6110 insertion can disrupt it asymmetrically. This led to a loss of target for any of the DR primers used for spoligotyping (3, 7, 16) and nonamplification of the spacer otherwise present in the locus (7, 11). These findings prompted us to take a closer look at the structure of the DR locus in M. tuberculosis clinical strains, circulating in the St. Petersburg area of Russia, with special attention to missing single signals in spoligoprofiles and, consequently, to eventual IS6110 insertions into particular DRs.
Conventional bacteriological procedures were used for isolation and culturing of M. tuberculosis strains from patients with chronic and newly diagnosed pulmonary tuberculosis. These patients were randomly selected for a population-based study; they were from St. Petersburg and three neighboring provinces of northwestern Russia (Leningrad Oblast, Novgorod, and Pskov) and were admitted to the hospital of the St. Petersburg Institute of Phthisiopulmonology and City Anti-Tuberculosis Dispensary of St. Petersburg between 1997 and 2001. DNA was isolated as described previously (17). Standard spoligotyping with primers DRb (forward) and DRa (reverse, 5′ biotinylated) was performed as described by Kamerbeek et al. (10), and IS6110 RFLP typing was done as described by van Embden et al. (17). The profiles obtained were processed by the GelCompar 4.1 package (BVBI Applied Maths, Kortrijk, Belgium) and constituted our local database. For left-right (LR) spoligotyping (7) we used primers Ris1 and Ris2 (8), located outwardly at the IS6110 right and left ends, respectively, and DRa. The PCR products amplified with Ris1/DRa and Ris2/DRa primer pairs were subjected to the standard spoligotyping procedure. This allowed amplification and detection of the particular DR spacers to the left and right of the IS6110 copy, depending on its orientation towards and within the DR locus.
The 198 epidemiologically unlinked Beijing strains of 381 studied were identified in our laboratory in 1997 to 2000 by IS6110 RFLP and standard spoligotyping (primers DRb/DRa). We searched our spoligoprofiles database for particular strains of the Beijing family missing any of the nine signature spacers, spacers 35 to 43 (5 strains out of 198), and reexamined them together with a selection of 20 randomly chosen typical Beijing strains by spoligotyping with three different primer combinations (Fig. 1).
FIG. 1.
Schematic presentation of M. tuberculosis spoligotyping profiles generated by three combinations of IS6110 and DR primers.
A schematic presentation of different spoligoprofiles is given in Fig. 1. Combined analysis of all three kinds of profiles for each strain permitted us to deduce the DR locus composition and the positions of IS6110 insertions, as well as their orientation with respect to the DR locus. We therefore assumed that 20 of the 25 Beijing strains retested had a single IS6110 copy in the opposite orientation upstream the DR locus, which itself consisted of nine DVRs from 35 to 43 (Fig. 1 and 2). This insertion position has already been described as common for the Beijing/W family genotype (2, 18). However, incomplete spoligoprofiles were identified in 5 of the 25 Beijing strains. Three atypical Beijing strains were missing spacers 37 and 38 (strains 6104 and 71043) and spacer 40 (strain 2563; shared spoligotype 190, also includes strains from Cuba and the United States [16]; W14 subgroup [4]), although further analysis by LR spoligotyping revealed no additional IS6110 copy in their DR loci (Fig. 1). Bifani et al. (4) also showed that spoligoprofile disruption in spacer 40 in the W14 subgroup strains was not due to an IS6110 insertion. According to the profiles presented in Fig. 1, two Beijing strains missing spacer 37 only harbored two copies of IS6110 (strains 4387 and 74512). One IS6110 copy had a location typical of the Beijing type, as described above. Another IS6110 copy was located forwardly either in DR37 (more likely) or in the adjacent spacer 37, in both cases rendering spacer 37 unamplified and consequently unrevealed by standard spoligotyping (Fig. 2). As both of these epidemiologically unlinked strains showed such a genotype, it did not seem to be coincidental.
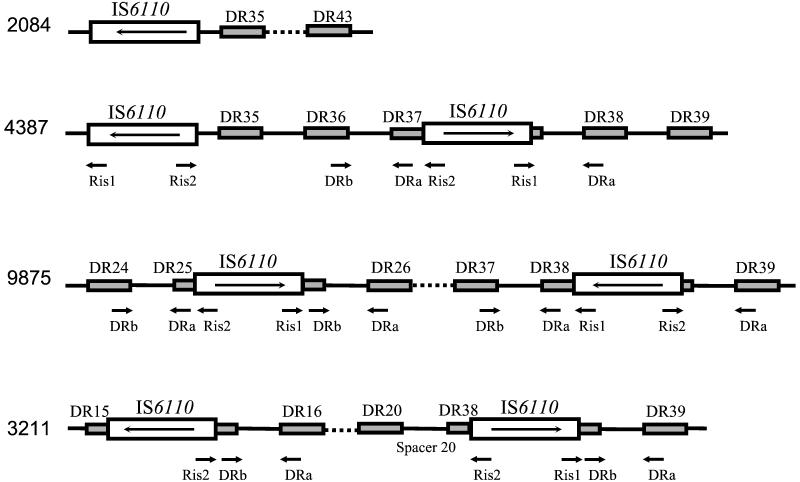
FIG. 2.
Schematic illustration of IS6110 insertions in the DR locus of M. tuberculosis strains inferred from spoligoprofiles (Fig. 1). The DVRs of standard spoligotyping (10) are included. Strain 2084 represents the typical Beijing family pattern found in 20 strains. Strain 4387 represents the atypical Beijing profile missing spacer 37 by standard spoligotyping.
We applied the same strategy to the spoligoprofiles of non-Beijing strains missing any of the spacers discussed above (Fig. 1). In strain 9875, an IS6110 insertion site was found in the center of DR25, not disrupting the targets for the DR primers (Fig. 2), as observed previously in both high- and low-IS6110-copy-number strains (7, 11, 14, 18). In addition, a second insertion of IS6110 in the opposite orientation was apparently present in DVR38 of this strain (Fig. 2), resulting in nonamplification of spacer 38 and subsequent loss of a signal in the spoligoprofile (Fig. 1). In strain 3211, two previously undescribed IS6110 insertions were found: an inverted one in DR15 and a direct one in DR38, both located in the middle of these DRs and not resulting in false disappearance of the adjacent spacers in spoligoprofile (Fig. 2).
We speculate that the occurrence of IS6110 insertions in different directions at the same site (DVRs 37 and 38) in unlinked and genotypically (IS6110 restriction fragment length polymorphism) distinct strains is likely to be independent. Similarly, the independent occurrence of IS6110 insertions in the IS1547/ipl sequences was described by Fang et al. (6). We therefore hypothesize that DVR37/DVR38 could present a “hot spot” for IS6110 insertion in the DR locus, though further study on more strains with such particular spoligoprofiles from different geographic locations is undoubtedly required.
It is noteworthy that for all the Beijing strains studied the strength of the LR-spoligotyping hybridization signals steadily decreased slightly from spacers 35 and 37 to spacer 43. This can be explained by their increasing distance from the IS6110 copy that, itself, served as the only target for a forward primer (Fig. 2). In general, the performance of LR spoligotyping is also sensitive to the “separation effect” of an additional IS6110 copy insertion that prevents effective primer extension and amplification of the outlying spacers. For example, in strain 9875, the DR spacers starting from spacer 38 could not be amplified by Ris1/DRa due to IS6110 located in the opposite orientation in DR38 (Fig. 1 and 2).
To sum up, four novel insertion sites of IS6110 were identified in the DR locus of M. tuberculosis. We have demonstrated that certain DR spacers (37 and 38) may be misleadingly absent from a profile generated by the routine spoligotyping scheme due to specific asymmetrical IS6110 insertion. We suggest using caution in epidemiological interpretation of similar profiles that differ only in these particular signals (37 and 38). Our data strengthen an observation that the Beijing genotype of M. tuberculosis is extremely conserved in its DR locus composition, as certain spacers, though unrevealed by spoligotyping, may be not effectively missing but “hidden” due to particular IS6110 insertions. One can hypothesize that such a constant structure of the DR locus in this family could have been inherited since an evolutionarily distant time and may reflect better adaptation to the host and better ability to propagate in the human population. The DR locus does not have any proven function in the M. tuberculosis genome, and its evolution appears to be neutral, as a rule (18). A case involving the Beijing genotype could present an exception. Once its specific DVR arrangement, including the 4.4-kb upstream region deletion (1, 18), was achieved, this state could have been advantageous for the genome and been subsequently maintained and inherited. van Embden et al. (18) have hypothesized that the DR region may be involved in the regulation of replication similar to a locus of short tandem repeats in Haloferax spp. (12), but for M. tuberculosis a definite biological significance of the particular arrangements of the DR locus is yet to be discovered.
Acknowledgments
We thank Lidia Steklova for providing us with some clinical isolates. We are thankful to Alessandra Riva and Daniel Velton for critical reading of the manuscript and English language corrections. I.M. is grateful to Christophe Sola for inspiring discussions on the DR locus structure and evolution.
This study was partly supported by International Atomic Energy Agency Research contract no. 9924 and by “Reseau International des Instituts Pasteur et Instituts Associes,” Institut Pasteur, Paris, France.
REFERENCES
- 1.Beggs, M. L., K. D. Eisenach, and M. D. Cave. 2000. Mapping of IS6110 insertion sites in two epidemic strains of Mycobacterium tuberculosis. J. Clin. Microbiol. 38:2923-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beggs, M. L., M. D. Cave, C. Marlowe, L. Cloney, P. Duck, and K. D. Eisenach. 1996. Characterization of Mycobacterium tuberculosis direct repeat sequence for use in cycling probe reaction. J. Clin. Microbiol. 34:2985-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benjamin, W. H., Jr., K. H. Lok, R. Harris, N. Brook, L. Bond, D. Mulcahy, N. Robinson, V. Pruitt, D. Y. P. Kirkpatrick, M. E. Kimerling, and N. E. Dunlap. 2001. Identification of a contaminating Mycobacterium tuberculosis strain with a transposition of an IS6110 insertion element resulting in an altered spoligotype. J. Clin. Microbiol. 37:1092-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bifani, P. J., B. Mathema, M. Campo, S. Moghazeh, B. Nivin, E. Shashkina, J. Driscoll, S. S. Munsiff, R. Frothingham, and B. N. Kreiswirth. 2001. Molecular identification of streptomycin monoresistant Mycobacterium tuberculosis related to multidrug-resistant W strain. Emerg. Infect. Dis. 7:842-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bifani, P. J., B. Mathema, Z. Liu, S. L. Moghazeh, B. Shopsin, B. Tempalski, J. Driscol, R. Frothingham, J. M. Musser, P. Alcabes, B. N. Kreiswirth. 1999. Identification of a W variant outbreak of Mycobacterium tuberculosis via population-based molecular epidemiology. JAMA 282:2321-2327. [DOI] [PubMed] [Google Scholar]
- 6.Fang, Z., D. T. Kenna, C. Doig, D. N. Smittipat, P. Palittapongarnpim, B. Watt, and K. J. Forbes. 2001. Molecular evidence for independent occurrence of IS6110 insertions at the same sites of the genome of Mycobacterium tuberculosis in different clinical isolates. J. Bacteriol. 183:5279-5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filliol, I., C. Sola, and N. Rastogi. 2000. Detection of a previously unamplified spacer within the DR locus of Mycobacterium tuberculosis: epidemiological implications. J. Clin. Microbiol. 38:1231-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedman, C. R., M. Stoeckle, W. D. Johnson, Jr., and L. W. Riley. 1995. Double-repetitive-element PCR method for subtyping Mycobacterium tuberculosis clinical isolates. J. Clin. Microbiol. 33:1383-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hermans, P. W. M., D. van Soolingen, E. M. Bik, P. E. W. de Haas, J. W. Dale, and J. D. A. van Embden. 1991. The insertion element IS987 from Mycobacterium bovis BCG is located in a hot spot integration region for insertion elements in Mycobacterium tuberculosis complex strains. Infect. Immun. 59:2695-2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. D. A. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Legrand, E., I. Filliol, C. Sola, and N. Rastogi. 2001. Use of spoligotyping to study the evolution of the direct repeat locus by IS6110 transposition in Mycobacterium tuberculosis. J. Clin. Microbiol. 39:1595-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mojica, F. J. M., C. Ferrer, G. Juez, and F. Rodriguez-Valera. 1995. Long stretches of short tandem repeats are present in the largest replicons of the Archaea Haloferax mediterranei and Haloferax volcanii and could be involved in replicon partitioning. Mol. Microbiol. 17:85-93. [DOI] [PubMed] [Google Scholar]
- 12a.Narvskaya, O., I. Mokrousov, E.Limeschenko, T. Otten, L. Steklova, O. Graschenkova, and B. Vishnevskiy. 2000. Molecular characterisation of Mycobacterium tuberculosis strains from the northwest region of Russia. Epinorth 1:22-24. [Online.] [Google Scholar]
- 13.Narvskaya, O., I. Mokrousov, T. F. Otten, and B. I. Vyshnevskiy. 1999. Genetic marking of polyresistant Mycobacterium tuberculosis strains isolated in the north-west of Russia. Probl. Tuberk. 3:39-41. (In Russian.) [PubMed] [Google Scholar]
- 14.Plikaytis, B. B., N. Kurepina, C. L. Woodley, R. Fleischmann, B. Kreiswirth, and, T. M. Shinnick. 1999. Multiplex PCR assay to aid in the identification of the highly transmissible Mycobacterium tuberculosis strain CDC1551. Tuber. Lung Dis. 79:273-278. [DOI] [PubMed] [Google Scholar]
- 15.Soini, H., X. Pan, A. Amin, E. A. Graviss, A. Siddiqui, and J. M. Musser. 2000. Characterization of Mycobacterium tuberculosis isolates from patients in Houston, Texas, by spoligotyping. J. Clin. Microbiol. 38:669-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sola, C., I. Filliol, M. C. Guttieres, I. Mokrousov, V. Vincent, and N. Rastogi. 2001. Spoligotype database of Mycobacterium tuberculosis: biogeographic distribution of shared types and epidemiologic and phylogenetic perspectives. Emerg. Infect. Dis. 7:390-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Embden, J. D. A., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnik, and P. M. Small. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Embden, J. D. A., T. van Gorkom, K. Kremer, T. Jansen, B. A. M. van der Zeijst, and L. M. Schouls. 2000. Genetic variation and evolutionary origin of the direct repeat locus of Mycobacterium tuberculosis complex bacteria. J. Bacteriol. 182:2393-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Soolingen, D., L. Qian, P. E. W. de Haas, J. T. Douglas, H. Traore, F. Portaels, H. Z. Quing, D. Enkhasaikan, P. Nymadawa, and J. D. A. van Embden. 1995. Predominance of a single genotype of Mycobacterium tuberculosis in countries of East Asia. J. Clin. Microbiol. 33:3234-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]