Abstract
Eleven strains of a rapidly growing mycobacterium were isolated from patient specimens originating from various regions of the province of Ontario, Canada, over a 2-year period. Unique high-performance liquid chromatography (HPLC) and PCR-restriction enzyme pattern analysis (PRA) profiles initially suggested a new Mycobacterium species, while sequencing of the 16S rRNA gene revealed a sequence match with Mycobacterium sp. strain MCRO 17 (GenBank accession no. X93028), an isolate determined to be unique which is to date uncharacterized, and also a close similarity to M. elephantis (GenBank accession no. AJ010747), with six base pair variations. A complete biochemical profile of these isolates revealed a species of mycobacteria with phenotypic characteristics similar to those of M. flavescens. HPLC, PRA, and 16S rRNA sequencing of strain M. elephantis DSM 44368T and result comparisons with the clinical isolates revealed that these strains were in fact M. elephantis, a newly described species isolated from an elephant. All strains were isolated from human samples, 10 from sputum and 1 from an axillary lymph node.
Since 1997 alone, 17 new species of mycobacteria have been established. With the exponential discovery of new species, phenotypic profiles determined by conventional testing are less reliable as a means of accurately identifying the 95 mycobacterial species currently established (J. P. Euzéby, http://www.bacterio.cict.fr/m/mycobacterium.html). The increased use of sequence-based identification techniques, as well as high-performance liquid chromatography (HPLC) analysis of mycolic acids, has become a necessity in identifying less common species of mycobacteria.
In this study we examined 11 mycobacterial strains, which gave unique and identical profiles when analyzed by HPLC and PCR-restriction pattern enzyme analysis (PRA). Sequencing of the 16S rRNA gene revealed a perfect identity with an uncharacterized Mycobacterium strain from the GenBank database designated MCRO 17 (GenBank accession no. X93028) and a close relationship to M. elephantis DSM 44368T (GenBank accession no. AJ010747). Ten strains were isolated from sputa of different patients, and the 11th strain was isolated from an axillary lymph node from a 27-year-old male. This patient presented with lymphadenopathy, and after surgical excision, histopathology of the node showed caseating granulomas. Subsequent mycobacterial culture yielded a lightly pigmented mycobacterium that corresponded to the above cluster. The patients resided in various regions of the province of Ontario, Canada.
This paper describes a cluster of mycobacterial species isolated on 11 separate occasions from human specimens, subsequently identified as M. elephantis based on further characterization of the type strain of the species in our laboratories. New characteristics of this species are included.
MATERIALS AND METHODS
Mycobacterial strains.
Seven of 10 clinical strains analyzed derived from sputum cultures obtained at the Mycobacteriology Laboratory, Central Public Health Laboratory, Toronto, Ontario, Canada, which isolates approximately 2,000 nontuberculous mycobacteria per year. Three other strains were received at the laboratory for culture identification after isolation from sputa at three separate regional Public Health Laboratories in Ontario. The 11th strain was isolated from the culture of an excised axillary lymph node, grown at the London, Ontario, Public Health Laboratory and sent to the Toronto laboratory for identification. Eight of the 11 strains were subsequently sent to the National Reference Centre for Mycobacteriology, Health Canada, Winnipeg, Manitoba, Canada, for further characterization. The specimen source, sex, and age of the patient and a brief clinical history are found in Table 1. Reference strains used for the in-house National Reference Centre for Mycobacteriology mycobacterial 16S rRNA gene sequence database (23) were obtained from the American Type Culture Collection and Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH. Strain 01-17 has been deposited in the American Type Culture Collection as ATCC BAA-324.
TABLE 1.
Case histories of patients associated with the isolated clinical strains of M. elephantisa
| Location (Ontario, Canada) | Strain identifier | Sex | Age (yr) | Source of specimen | History |
|---|---|---|---|---|---|
| Toronto | 00-160 | M | 49 | Sputum | 3-yr follow-up post-TB infection |
| Whitefish | 00-252 | M | 60 | Sputum | Sore side and back; occasional morning hemoptysis; no treatment |
| Ottawa | 72189 | M | 60 | Sputum | Smoker; chronic cough; from Somalia |
| Thunder Bay | 01-19 | F | 64 | Sputum | Heavy smoker; COPD; chronic bronchitis |
| Mississauga | 00-254 | M | 73 | Sputum | Pneumothorax |
| St. Catherine's | 00-253 | F | 73 | Sputum | COPD; chronic bronchitis |
| Toronto | 71901 | M | 79 | Sputum | COPD |
| Owen Sound | 01-17 | M | 27 | Left axillary node | Enlarged left axillary node; see text |
| Toronto | 01-81 | F | 74 | Sputum | Chronic cough; from Korea. |
| Brampton | 01-101 | F | 70 | Sputum | Renal patient on dialysis with coughing and hemoptysis, under observation due to TB patient in the same unit |
| Mississauga | 01-111 | F | 89 | Sputum | Refugee from Kosovo with hypertension |
Abbreviations: M, male; F, female; TB, tuberculosis; COPD, chronic obstructive pulmonary disease.
Phenotypic characterization.
Seven of the 11 clinical strains and M. elephantis DSM 44368T were subcultured on Löwenstein-Jensen (L-J) medium at 25, 30, 37, 42, and 52°C and observed for growth at 3, 5, 7, and 14 days. Photochromogenicity was determined at 37°C on L-J medium. Cultures were maintained at 37°C on Middlebrook 7H10 agar for subsequent testing and determination of colony characteristics. Auramine O fluorescence and Kinyoun acid-fast stainings were both performed as previously described (11) on fresh cultures from both solid and 12B broth media. Biochemical tests performed included those for niacin, nitrate reduction, semiquantitative (>45-mm foam) and heat-stable (68°C) catalase tests, Tween 80 hydrolysis, tellurite reduction, tolerance of 5% NaCl, iron uptake, arylsulfatase activity at 3 and 14 days, growth on MacConkey agar without crystal violet, urease (Wayne method), pyrazinamidase, and sodium citrate as sole carbon source, as previously described (11). Additional tests included the acid phosphatase and β-glucosidase tests (5) and those for the following amidases: acetamidase, benzamidase, nicotinamidase, and succinamidase (1).
HPLC.
Mycolic acids were extracted and analyzed according to the standardized method (2). Briefly, the fatty acids were saponified with 25% KOH solution in methanol-water (1:1) for 1 h in an autoclave at 121°C. After acidification with 18.5% HCl, the mycolic acids were extracted with chloroform and derivatized to their UV-absorbing p-bromophenacyl esters. A high-molecular-weight internal standard (RIBI ImmunoChem Research Inc., Hamilton, Mont.) was added to each sample before testing. The HPLC hardware consisted of a Waters Corp. (Milford, Mass.) system, including a 717 auto sampler, a 600 E pump, a 486 UV detector, and a Beckman Ultrasphere XL-octyldecyl silane column. For chromatographic data analysis, the HPLC chromatograms were imported into Millennium 32 software (Waters Corp.). For species identification, the chromatographic data were compared with an extensive in-house library of mycobacterial species HPLC profiles as well as with published data (3). As M. elephantis was a very recently established species, its HPLC profile was not available publicly and was determined in-house with M. elephantis DSM 44368T.
PRA.
PRA of the hsp65 gene was performed as described previously (19). Briefly, a 439-bp fragment of the 65-kDa heat shock protein gene (hsp65) was amplified by PCR with primers which are conserved throughout the Mycobacterium genus. The fragment produced was digested by two enzymes, BstEII and HaeIII. The restriction fragments were separated by agarose gel electrophoresis and visualized by ethidium bromide. The pattern of restriction was compared to the published algorithm (19) and to the pattern determined in-house for M. elephantis DSM 44368T.
PCR of the nearly complete 16S rRNA gene.
Organisms were heat killed by being boiled at 100°C for 10 min and mechanically lysed using the Mini Bead-Beater (Biospec Products, Bartlesville, Okla.) for 2 min. The lysate was centrifuged to precipitate cellular debris, and the supernatant was used for PCR. Genomic DNA was quantitated using the PicoGreen double-stranded DNA quantitation kit (Molecular Probes, Inc., Eugene, Oreg.) with a TD-700 laboratory fluorometer (Turner Designs, Sunnyvale, Calif.). Each reaction mixture contained approximately 10 ng of DNA; 2.5 mM MgCl2; 1× PCR buffer (Amersham Pharmacia Biotech, Baie d'Urfé, Quebec, Canada); 200 μM (each) dCTP, dGTP, dATP, and dTTP; 1,000 pmol of each forward and reverse primer; and 1.25 U of Taq DNA polymerase (Amersham Pharmacia Biotech) in a final volume of 50 μl. Primers used were pA (5′ AGA GTT TGA TCC TGG CTC AG 3′) (6) and primer 1492 (5′ GGT TAC CTT GTT ACG ACT T 3′) (14). The PCR was performed using the Perkin-Elmer GeneAmp PCR System 2400 with a cycle of 94°C for 5 min; 30 cycles of 94, 60, and 72°C for 1 min each; and final extension at 72°C for 10 min, and the mixture was held at 4°C. The PCR product was purified using a MicroCon centrifugal filter device (Millipore Corporation, Nepean, Ontario, Canada) and quantified using UV absorbance at 260 nm.
16S rRNA sequencing and phylogenetic analysis.
The ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems, Foster City, Calif.) was used for the sequencing of the PCR product. A combination of universal primers was chosen to sequence the nearly complete gene (6). The sequencing reaction and template preparation were performed in accordance with the directions of the manufacturer (Applied Biosystems). The sequencing product was purified using the recommended Centricep columns (Princeton Separations, Adelphia, N.J.). Samples were run on an ABI PRISM 310 Genetic Analyzer (Applied Biosystems). The sequencing output was analyzed using the accompanying DNA Sequence Analyzer computer software (Applied Biosystems). The Lasergene program (version 4.01; DNASTAR, Inc., Madison, Wis.) was used for sequence assembly, sequence alignment, and phylogenetic analysis. Multiple sequence alignments were determined using the Clustal method algorithm. Analysis was performed by comparing sequences with the sequences of reference strains determined in our laboratory and select sequences obtained from GenBank.
Antimicrobial susceptibility testing.
Bactec 12B radiometric broth macrodilution sensitivity testing was performed on isolate 01-17 according to the method used for M. avium complex (9, 17). The following drugs were tested, with their MICs indicated in Table : amikacin, capreomycin, clarithromycin, clofazamine, ciprofloxacin, ethambutol, ethionamide, isoniazid, kanamycin, ofloxacin, rifabutin, rifampin, and streptomycin (Table 3).
TABLE 3.
Antimicrobial sensitivity results for strain 01-17 determined by the Bactec 12B radiometric broth macrodilution method
| Antibiotic | Concns tested (μg/ml) | MIC (μg/ml) |
|---|---|---|
| Amikacin | 0.5, 1.0, 2.0, 4.0, 8.0 | ≤0.5 |
| Capreomycin | 1.25, 2.5, 5.0, 10.0, 20.0 | ≤1.25 |
| Clarithromycin | 2.0, 4.0, 8.0, 16.0, 32.0, 64.0 | ≤2.0 |
| Ciprofloxacin | 1.0, 2.0, 4.0, 8.0, 16.0 | ≤1.0 |
| Ethambutol | 1.0, 2.0, 4.0, 8.0, 16.0 | ≤1.0 |
| Ethionamide | 1.25, 2.5, 5.0, 10.0, 20.0 | 5.0 |
| Isoniazid | 0.025, 0.05, 0.1, 0.2, 0.4 | 0.1 |
| Kanamycin | 1.25, 2.5, 5.0, 10.0, 20.0 | ≤1.25 |
| Levofloxacin | 0.5, 1.0, 2.0, 4.0, 8.0 | ≤0.5 |
| Ofloxacin | 0.5, 1.0, 2.0, 4.0, 8.0 | ≤0.5 |
| Rifabutin | 0.12, 0.25, 0.5, 1.0, 2.0 | >2.0 |
| Rifampin | 0.5, 1.0, 2.0, 4.0, 8.0 | >8.0 |
| Streptomycin | 0.5, 1.0, 2.0, 4.0, 8.0 | ≤0.5 |
Nucleotide sequence accession number.
The nucleotide sequence of strain 01-17 has been deposited in the GenBank database under accession no. AF385898.
RESULTS
Microscopy of the clinical strains from 12B broth and L-J solid medium revealed small, dispersed, beaded, coccobacillary acid-fast organisms. Acid-fast properties became weak in older cultures. No cords, spores, or hyphae were seen. On L-J medium, subcultured strains first appeared at 42°C in 3 days, at 37°C in 5 days, and at 25 and 30°C within 7 days. No growth occurred at 52°C. The species is a rapid grower, defined as growing in less than 7 days; however, growth is slower than that for the majority of rapidly growing species, such as M. smegmatis and M. fortuitum, as it begins to appear on solid media only after 3 days under the best conditions, attaining full growth by day 7. Mature colonies (i.e., 7 days) on Middlebrook 7H10 and L-J media appeared smooth and domed, with a pale yellow pigment becoming brighter with age, in both light and dark conditions. One strain (00-160) appeared somewhat mucoid, this difference being more evident on L-J medium. M. elephantis DSM 44368T revealed two colony types, designated 1 and 2. On Middlebrook 7H10 agar, colony 1 appeared cream colored, domed, and matte, acquiring a lobular center and yellow pigmentation with age. Colony 2 resembled those of the 11 clinical strains, domed and smooth, with a pale yellow pigmentation. On L-J medium, both colony types were wet in appearance, with colony 1 appearing a brighter yellow than colony 2.
Phenotypic and biochemical profiles were determined for 7 of the 11 clinical strains and are indicated in Table 2, along with the results of M. elephantis per the work of Shojaei et al. (16) and those determined for M. elephantis DSM 44368T in our laboratory. Characteristics of M. elephantis were also compared with those of other species having phenotypic and molecular similarities. Test results were identical for the two colony types of M. elephantis DSM 44368T and correlated well with those for the clinical strains.
TABLE 2.
Phenotypic characteristics of the M. elephantis cluster in comparison with the type strain of the species and closely related speciesa
| Test | Result for strain(s):
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Clinical strains | M. elephantis DSM 44368b (our laboratory) | M. elephantis DSM 44368b (Shojaei et al. [16]) | MCRO 17c | MCRO 20d | M. flavescens | M. scrofulaceum | M. pulveris | |
| Growth ratee | M | M | R | M | S | M | ||
| Growth temp (°C) | ||||||||
| 25 | + | + | + | + | ||||
| 31 | + | + | + (28°C) | |||||
| 37 | + | + | + | + | + | + | ||
| 42 | + (best) | + (best) | + | Vi | − | + | ||
| 52 | − | − | − | − | ||||
| Pigmentationf | Sg | Sg | N | S | S | N | ||
| Morphologyh | S, Y | S, Y | S, Y | S, Y | S | |||
| Niacin | − | − | − | − | − | |||
| Nitrate reductase | + | + | + | + | + | + | − | + |
| Semiquantitative catalase (mm) | >45 | >45 | >45 | >45 | >45 | >45 | >45 | |
| 68°C catalase | + | + | + | + | ||||
| Tween 80 hydrolysis | + | + | + | + | + | + | − | 88% (7 days) |
| Tellurite reduction | − | − | − (5 days) | + | − | −/+ (44%) | +/− (64%) | |
| 5% NaCl tolerance | + | + | − | − | − | + (62%) | − | + |
| Iron uptake | − | − | − | − | ||||
| Arylsulfatase activity | ||||||||
| 3 days | − | − | − (84%) | − (99%) | − | |||
| 14 days | − | − | − | + (78%) | − (64%) | + | ||
| Growth on MACj | − | − | − | − | ||||
| Urease activity | + | + | + | + | + | + (72%) | + | 25% |
| Pyrazinamidase | + | + | + | V | + | |||
| Acetamidase | − | − | − | − | ||||
| Benzamidase | − | − | − | |||||
| Nicotinamidase | + | + | + | + | ||||
| Succinamidase | − | − | − | |||||
| Acid phosphatase | + | + | + | − | + | |||
| β-Glucosidase | + | + | ||||||
| Sodium citrate | − | − | ||||||
| Acid production | ||||||||
| Fructose | − | − | ||||||
| Mannitol | − | − | V | |||||
| Sorbitol | − | − | V | |||||
| Inositol | − | − | ||||||
Species closely related to the M. elephantis cluster on the basis of the 16S rRNA gene were Mycobacterium sp. strains MCRO 17 (18) and MCRO 20 (18), M. elephantis (16), and M. pulveris (22). Those closely related to the cluster on the basis of phenotypic characteristics were M. flavescens and M. scrofulaceum (7, 13). Seven of the 11 strains (00-160, 00-253, 00-254, 01-17, 01-19, 01-81, and 01-101) were tested by biochemical methods, and results were identical for all strains.
Testing was performed in our laboratory for each colony type (1 and 2). Results were identical, with the exception of the morphology.
Previously identified as M. scrofulaceum.
Previously identified as M. flavescens.
M, moderate; S, slow; R, rapid.
S, scotochromogen; N, nonchromogen.
Pigmentation was difficult to establish, as isolates were generally a pale yellow, intensifying to a bright yellow within a few days.
S, smooth; Y, yellow.
V, variable.
MAC, MacConkey agar without crystal violet.
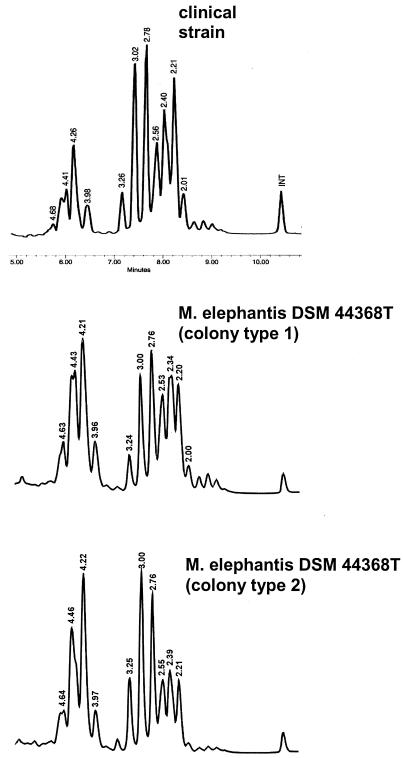
HPLC of mycolic acids was performed on all clinical isolates, as well as the two colony types of M. elephantis. The HPLC chromatogram of the clinical isolates as well as for both colonies of M. elephantis showed multiple peaks in a double cluster pattern (Fig. 1). A small cluster of five early peaks is followed by a larger cluster of multiple peaks. The profile did not correspond to any other in the library of patterns or in published data. Isolate 00-254 was sent to the Centers for Disease Control and Prevention in Atlanta, Ga., and was identified as NCP (Not Common Pattern) 201A.
FIG. 1.
Mycolic acid pattern comparison of M. elephantis strain 01-19 with both colony types of M. elephantis DSM 44368T, obtained by HPLC analysis. The relative retention time is indicated for each peak. INT, high-molecular-weight internal standard.
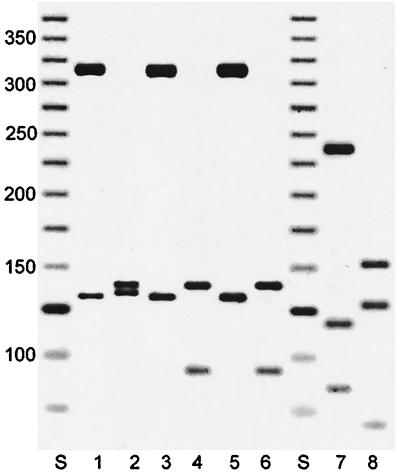
The PRA showed a pattern that was distinct from previously published patterns as well as from those in the in-house library of PRA patterns. BstEII digestion showed two fragments, 325 and 130 bp, and HaeIII digestion showed three fragments, 140, 100, and 60 bp (Fig. 2). The PRA pattern of M. elephantis DSM 44368T colony 2 was identical to that of the clinical strains, whereas that of colony 1 presented with a band at 135 bp and no band at 60 bp with the HaeIII digestion.
FIG. 2.
PRA patterns obtained from digestion of the amplified hsp65 gene. Lanes 1 and 2, M. elephantis DSM 44368T colony 1; lanes 3 and 4, M. elephantis DSM 44368T colony 2; lanes 5 and 6, clinical strain 00-254; lanes 7 and 8, M. tuberculosis; lanes S, DNA ladder, 25 bp; lanes 1, 3, 5, and 7, BstEII digestion; lanes 2, 4, 6, and 8, HaeIII digestion. Numbers at left are molecular sizes in base pairs.
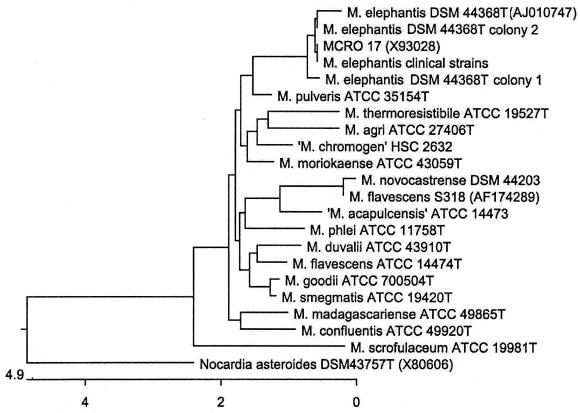
Analysis of 1,463 bases of the clinical strains of mycobacteria, from position 28 to 1506 on the Escherichia coli 16S rRNA gene, revealed 99.8% similarity with and 0% divergence from Mycobacterium sp. strain MCRO 17 (GenBank accession no. X93028). Three N′s in the GenBank sequence contributed to a result of less than 100% similarity; otherwise, the sequences are identical. Its closest established species relative was M. elephantis DSM 44368T (GenBank accession no. AJ010747), with 99.5% similarity and 0.4% divergence, differing in the 16S rRNA gene by six bases, all beyond the first 500 bases of the gene. Another close relative is M. pulveris ATCC 35154T (Fig. 3), the sequence of which was determined in our laboratory, with 98.6% similarity and 1.1% divergence, differing by 17 bases. Upon receipt of the strain M. elephantis DSM 44368T, 16S rRNA gene sequencing was determined for both colony types. Sequence differences were detected between the two colony types as well as from the sequence of M. elephantis from GenBank and the clinical strains (Fig. 4).
FIG. 3.
Phylogenetic relationships of M. elephantis, clinical and type strains, with its closest genotypic and phenotypic relatives, based on the 16S rRNA gene. M. scrofulaceum is included due to its reported similarity with MCRO 17 (18). Multiple sequence alignments were determined from bp 54 to 1470 of the E. coli 16S rRNA gene by using the Clustal method algorithm in the Megalign component of the Lasergene program, version 4.01. The tree was rooted using Nocardia asteroides as the outgroup sequence. Sequences were determined in our laboratory unless indicated by a GenBank accession number.
FIG. 4.
Alignment of variable regions within the 16S rRNA gene. (A) Regions of variability among strains of M. elephantis; (B) other regions of variability between M. elephantis and its closest genotypic relative, M. pulveris. Numbers in parentheses indicate the base position on the E. coli 16S rRNA gene.
Bactec 12B radiometric broth macrodilution sensitivity testing results are indicated in Table 3.
DISCUSSION
M. elephantis is a recently described new species of mycobacteria, published in May 2000 (16). The described strain was obtained from an elephant, and apart from limited biochemical characteristics, only the availability of the 16S rRNA gene sequence in GenBank would allow another laboratory to detect this particular species in its own specimen collections. However, it is well known that analyses resulting from BLAST searches against GenBank submissions are too often misleading due to errors that they might contain (23; S. Dostal, E. Richter, A. Roth, S. Niemann, J. Rothgänger, J. Albert, M. Frosch, and D. Harmsen, Abstr. 101st Gen. Meet. Am. Soc. Microbiol., abstr. C-240, 2001). The sequence of events leading to the identification of these isolates as M. elephantis was not direct and was confirmed only after obtaining the type strain from the Deutsche Sammlung von Mikroorganismen und Zellkulturen collection. Prior to this, we were faced with a cluster of mycobacterium strains with unique HPLC and PRA patterns. 16S rRNA sequencing analysis indicated an identical match with the sequence of MCRO 17 (GenBank accession no. X93028), followed by a close match with M. elephantis (GenBank accession no. AJ010747), with six base pair variations. Other factors lead us to believe that this cluster of strains was likely not M. elephantis, as the variations in the 16S rRNA gene would suggest. First, we found that recently deposited 16S rRNA gene sequences of mycobacterial species generally contain few errors in comparison to submissions in the early 1990s. Second, our clinical strains grew well on 5% NaCl and were considered scotochromogenic due to their yellow pigment on solid media and their similarity in characteristics to M. flavescens. M. elephantis is described as a nonchromogen and was reported to be negative for growth on 5% NaCl.
The MCRO strains were described by Springer et al. as part of an evaluation of difficult-to-identify clinical isolates of mycobacteria the 16S ribosomal DNA (rDNA) sequences of which were determined to help clarify their identity (18). MCRO 17 was described as most closely resembling M. scrofulaceum based on biochemical test results. Another strain, MCRO 20, described as most closely resembling M. flavescens, was found to have a sequence identical to that of MCRO 17 in regions A and B, corresponding to E. coli 16S rDNA positions 129 to 267 and 430 to 500, respectively. It is unknown whether the full gene was sequenced and found to be identical to that in MCRO 17. The sequence of MCRO 20 is not present in GenBank. Regions A and B of both MCRO 17 and MCRO 20 were determined to be “unique and identical” and “related to a group of thermotolerant rapid-growers which show an insertion of 2 nucleotides in helix 10” (18).
According to their biochemical profiles, the clinical isolates studied most closely resembled M. flavescens, as both are scotochromogenic, nitrate positive, and tolerant of 5% NaCl. Furthermore, they were both urease and pyrazinamidase positive, which would classify this organism as M. flavescens based on the work-flow chart for scotochromogens as previously described (10), which include the five most common species, M. gordonae, M. scrofulaceum, M. xenopi, M. flavescens, and M. szulgai. The growth pattern, of intermediate-rate growth, contributes further to the phenotypic similarities with M. flavescens. However, sequence-based data confirm that the strains were not M. flavescens, even in light of the questionable integrity of this species (15).
Several observations were made once the type strain of M. elephantis was obtained and studied in our laboratories. First, two colony types were equally present. This phenomenon has been observed in our laboratory with several other reference strains of mycobacteria, such as M. branderi, M. gilvum, M. obuense, and M. neoaurum, among others. We have also determined the 16S rRNA gene sequences of their various colony types and found them to be identical within the same species. While we anticipated that the same would occur with M. elephantis, this was not the case. M. elephantis DSM 44368T designated colony 1 appeared nonchromogenic on 7H10 medium, domed, and matte and acquiring a lobular center and a pale yellow pigmentation with age. The 16S rDNA sequence of this colony type showed nine base pair variations and two deletions in comparison with GenBank accession no. AJ010747. M. elephantis DSM 44368T designated colony 2 was pale yellow, smooth, and domed, like the clinical strains. The 16S rDNA sequence of this colony type showed five variations in comparison with the GenBank sequence. Shojaei et al. included cloning of the PCR product in their protocol (4), which may explain the variations between the GenBank submission of the M. elephantis type strain and that determined in our laboratory for the same strain. All sequencing performed in our laboratory was determined directly from the PCR product, thereby averaging out random Taq errors. The clinical strains showed only one variation from the sequence determined for M. elephantis DSM 44368T colony type 2. All clinical strains were sequenced once, and all presented the same sequence, while the two colony types of M. elephantis DSM 44368T were individually sequenced twice and with the same results as described above.
Based on biochemical testing, the clinical strains are phenotypically similar to the species M. flavescens. However, the strains are more distantly related to this species based on 16S rRNA gene sequence analysis, having a 96.8% similarity. Sequence analysis of the 16S rRNA gene reveals that M. elephantis is most closely related to M. pulveris. There are 17 base differences between M. pulveris and the clinical strains of M. elephantis, 3 of them situated in the first 500 bases of the gene, as observed when using RIDOM (http://www.ridom.de/) for sequence comparisons. Phenotypically, clinical strains of M. elephantis differ in characteristics from the type strain described in reference 16 by presenting pale yellow colonies and tolerating 5% NaCl. Repeat biochemical testing performed in our laboratory showed that the type strain did tolerate 5% NaCl.
All strains, with the exception of 01-17 from the lymph node, were isolated from sputum specimens. None of the specimens were positive for acid-fast bacilli on the initial smear. The patients who yielded these respiratory isolates were generally elderly with preexisting nonmycobacterial disease. Since none of these patients had repeat isolation of the strain, the clinical relevance is difficult to ascertain. The lymph node isolate, however, is significant. The isolation of a nontuberculous mycobacterium from a sterile site is generally considered to be clinically relevant (24). Lymphadenitis caused by nontuberculous mycobacteria is not uncommon, with M. avium complex being the most common etiologic agent (12). Recently, there have been reports of novel species of mycobacteria such as M. tusciae, M. heidelbergense, and M. bohemicum causing lymphadenitis (8, 20, 21). The patient with a mycobacterium isolated from the cervical lymph node in this study was a 27-year-old immunocompetent male with extensive, recent tattoos on his skin. It is possible that this procedure could have caused intradermal inoculation of the organism. The histopathology findings of granulomatous changes in the node tissue and the subsequent isolation of a mycobacterium species concur with the diagnosis of mycobacterial lymphadenitis.
Pigmented rapidly growing mycobacteria are normally not known to cause disease. However, M. neoaurum, a pigmented rapid grower, has been associated previously with several cases of catheter-related bacteremia and has been best identified using 16S rRNA gene sequencing (25). Definitive identification of these species is difficult without advanced methods such as HPLC or sequencing. New or less commonly isolated species of mycobacteria are likely to be misidentified by conventional methods, and laboratories lacking advanced technology should consider submitting to a reference laboratory nontuberculous mycobacterial strains of clinical significance that are not identified with commercial DNA probes.
The increased use of molecular methods in mycobacteriology laboratories has contributed to the discovery of many new species in this genus in recent years. It is also not unreasonable to predict that sequence-based identification, with the 16S rRNA gene being the most likely target, will establish itself in routine microbiology in the near future. The commercial kit MicroSeq (Applied Biosystems) and the publicly available RIDOM (http://www.ridom.de), both quality-controlled 16S rRNA gene sequence databases, have been developed in anticipation of this event.
REFERENCES
- 1.Bönicke, R. 1962. L'identification des mycobactéries a l'aide de méthodes biochimiques. Bull. Int. Union Tuberc. 32:13-76. [Google Scholar]
- 2.Butler, W. R., M. M. Floyd, V. A. Silcox, G. D. Cage, E. Desmond, P. S. Duffey, L. Guthertz, W. Gross, K. C. Jost, Jr., L. S. Ramos, L. Thibert, and N. G. Warren. 1996. Standardized method for HPLC identification of mycobacteria. Centers for Disease Control and Prevention, U.S. Department of Health and Human Services, Atlanta, Ga.
- 3.Butler, W. R., M. M. Floyd, V. A. Silcox, G. D. Cage, E. Desmond, P. S. Duffey, L. Guthertz, W. Gross, K. C. Jost, Jr., L. S. Ramos, L. Thibert, and N. G. Warren. 1999. Mycolic acid pattern standard for HPLC identification of mycobacteria. Centers for Disease Control and Prevention, U.S. Department of Health and Human Services, Atlanta, Ga.
- 4.Chun, J., and M. Goodfellow. 1995. A phylogenetic analysis of the genus Nocardia with 16S rRNA gene sequences. Int. J. Syst. Bacteriol. 45:240-245. [DOI] [PubMed] [Google Scholar]
- 5.David, H. L., and M. T. Jahan. 1977. β-Glucosidase activity in mycobacteria. J. Clin. Microbiol. 5:383-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edwards, U., T. Rogall, H. Blocker, M. Emde, and E. C. Böttger. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodfellow, M., and J. G. Magee. 1998. Taxonomy of mycobacteria, p. 1-71. In P. R. Gangadharam and P. A. Jenkin (ed.), Mycobacteria I: basic aspects. International Thomson Publishing, New York, N.Y.
- 8.Haas, W. H., W. R. Butler, P. Kirschner, B. B. Plikaytis, M. B. Coyle, B. Amthor, A. G. Steigerwalt, D. J. Brenner, M. Salfinger, J. T. Crawford, E. C. Bottger, and H. J. Bremer. 1997. A new agent of mycobacterial lymphadenitis in children: Mycobacterium heidelbergense sp. nov. J. Clin. Microbiol. 35:3203-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heifets, L., P. Lindholm-Levy, J. Libonati, N. Hooper, A. Laszlo, M. Cynamon, and S. Siddiqi. 1993. Radiometric broth macrodilution method for the determination of minimal inhibitory concentrations (MIC) with Mycobacterium avium complex isolates. National Jewish Center for Immunology and Respiratory Medicine. Denver, Colo.
- 10.Heifets, L. B., and P. A. Jenkins. 1998. Speciation of mycobacteria in clinical laboratories, p. 308-349. In P. R. Gangadharam and P. A. Jenkin (ed.), Mycobacteria I: basic aspects. International Thomson Publishing, New York, N.Y.
- 11.Kent, P. T., and G. P. Kubica. 1985. Public health mycobacteriology: a guide for the level III laboratory. Centers for Disease Control, U.S. Department of Health and Human Services, Atlanta, Ga.
- 12.Lai, K. K., K. D. Stottmeier, I. H. Sherman, and W. R. McCabe. 1984. Mycobacterial cervical lymphadenopathy. Relation of etiologic agents to age. JAMA 251:1286-1288. [DOI] [PubMed] [Google Scholar]
- 13.Metchock, B., F. S. Nolte, and R. J. Wallace, Jr. 1999. Mycobacterium, p. 399-437. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. ASM Press, Washington, D.C.
- 14.Relman, D. A. 1993. Universal bacterial 16S rRNA amplification and sequencing, p. 489-495. In D. H. Persing, T. F. Smith, F. C. Tenover, and T. J. White (ed.), Diagnostic molecular microbiology: principles and applications. American Society for Microbiology, Washington, D.C.
- 15.Roth, A., U. Reischl, A. Streubel, L. Naumann, R. M. Kroppenstedt, M. Habicht, M. Fischer, and H. Mauch. 2000. Novel diagnostic algorithm for identification of mycobacteria using genus-specific amplification of the 16S-23S rRNA gene spacer and restriction endonucleases. J. Clin. Microbiol. 38:1094-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shojaei, H., J. G. Magee, R. Freeman, M. D. Yates, N. U. Horadagoda, and M. Goodfellow. 2000. Mycobacterium elephantis sp. nov., a rapidly growing non-chromogenic mycobacterium isolated from an elephant. Int. J. Syst. E vol. Microbiol. 50:1817-1820. [DOI] [PubMed] [Google Scholar]
- 17.Siddiqi, S. H., L. B. Heifets, M. H. Cynamon, N. M. Hooper, A. Laszlo, J. P. Libonati, L. P. Lindholm, and N. Pearson. 1993. Rapid broth macrodilution method for determination of MICs for Mycobacterium avium isolates. J. Clin. Microbiol. 31:2332-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Springer, B., L. Stockman, K. Teschner, G. D. Roberts, and E. C. Böttger. 1996. Two-laboratory collaborative study on identification of mycobacteria: molecular versus phenotypic methods. J. Clin. Microbiol. 34:296-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Telenti, A., F. Marchesi, M. Balz, F. Bally, E. C. Bottger, and T. Bodmer. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31:175-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tortoli, E., A. Bartoloni, V. Manfrin, A. Mantella, C. Scarparo, and E. Bottger. 2000. Cervical lymphadenitis due to Mycobacterium bohemicum. Clin. Infect. Dis. 30:210-211. [DOI] [PubMed] [Google Scholar]
- 21.Tortoli, E., R. M. Kroppenstedt, A. Bartoloni, G. Caroli, I. Jan, J. Pawlowski, and S. Emler. 1999. Mycobacterium tusciae sp. nov. Int. J. Syst. Bacteriol. 49:1839-1844. [DOI] [PubMed] [Google Scholar]
- 22.Tsukamura, M., S. Mizuno, and H. Toyama. 1983. Mycobacterium pulveris sp. nov., a nonphotochromogenic mycobacterium with an intermediate growth rate. Int. J. Syst. Bacteriol. 33:811-815. [Google Scholar]
- 23.Turenne, C. Y., L. Tschetter, J. Wolfe, and A. Kabani. 2001. Necessity of quality-controlled 16S rRNA gene sequence databases: identifying nontuberculous Mycobacterium species. J. Clin. Microbiol. 39:3637-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wallace, R. J., Jr., R. O'Brien, J. Glassroth, J. Raleigh, and A. Dutt. 1990. Diagnosis and treatment of disease caused by nontuberculous mycobacteria. Am. Rev. Respir. Dis. 142:940-953. [DOI] [PubMed] [Google Scholar]
- 25.Woo, P. C. Y., H. W. Tsoi, K. W. Leung, P. N. L. Lum, A. S. P. Leung, C. H. Ma, K. M. Kam, and K. Y. Yuen. 2000. Identification of Mycobacterium neoaurum isolated from a neutropenic patient with catheter-related bacteremia by 16S rRNA sequencing. J. Clin. Microbiol. 38:3515-3517. [DOI] [PMC free article] [PubMed] [Google Scholar]