Abstract
Variability of disease manifestations has been noted in patients with Lyme disease. A contributing factor to this variation may be the number of spirochetes present in infected patients. We evaluated clinical and laboratory findings for patients with erythema migrans with regard to the number of Borrelia burgdorferi organisms detected by quantitative PCR (qPCR) in 2-mm skin biopsy specimens. B. burgdorferi was detected in 80% (40 of 50) of the specimens tested; the mean number of spirochetes in these specimens ranged over 3 orders of magnitude (10 to 11,000 spirochetes per 2-mm biopsy specimen). Larger numbers of spirochetes were significantly associated with a shorter duration of the erythema migrans skin lesion (P = 0.020), smaller skin lesions (P = 0.020), and infection with a specific genotype of B. burgdorferi (P = 0.008) but not with the number or severity of symptoms. Skin culture positivity was significantly associated with skin lesions containing larger numbers of spirochetes (P = 0.019).
Lyme borreliosis, the most common vector-borne disease of the Northern Hemisphere, is caused by a group of spirochetes referred to as Borrelia burgdorferi sensu lato. Humans become infected with these organisms during the feeding of certain Ixodes ticks (2, 20, 21). In North America, all known cases of Lyme disease are caused by infection with B. burgdorferi sensu stricto (8, 10). Variations in the clinical presentation of patients with Lyme borreliosis in the United States have been noted previously (15, 22). Reasons for these differences for which there is supportive evidence include coinfection with other tick-borne pathogens such as Ehrlichia phagocytophila or Babesia microti (15) and genetic heterogeneity of the infecting B. burgdorferi sensu stricto strain (6-8, 10, 26). Whether differences in the number of spirochetes in infected patients is a contributing factor is unknown. However, in a murine model of B. burgdorferi infection, the number of B. burgdorferi organisms detected by quantitative PCR (qPCR) in animal tissues closely correlated with development of joint swelling and inflammation (23).
Adaptation of a real-time qPCR method for enumeration of B. burgdorferi organisms in mouse tissues (12) to human skin samples allowed us to address the impact of the quantity of spirochetes on the clinical and laboratory features of Lyme disease patients with erythema migrans (EM).
MATERIALS AND METHODS
Patient population.
Fifty untreated adult patients presenting with EM to the Lyme Disease Diagnostic Centers of the Westchester Medical Center during the spring and summer of 2000 were included in the study. All cases of Lyme disease satisfied the surveillance case definition of the Centers for Disease Control and Prevention (5). Written informed consent was obtained from all participants, and the study was approved by the Institutional Review Board of New York Medical College. A comparative study of different modalities for diagnosis of early Lyme disease for 47 of these patients was reported previously (16).
Signs and symptoms were recorded as present or absent. If present, the severities of the symptoms were characterized by the patients by use of an 8-cm-long visual analogue scale, as described previously (9). For symptomatic patients, a symptom severity index was calculated by using the cumulative symptom score divided by the number of symptoms present.
Skin biopsy and culture.
Skin biopsy specimens (diameters, 2 mm) were obtained from the advancing border of primary EM lesions as described elsewhere (19). The biopsy specimens were placed in incomplete Barbour-Stoenner-Kelly (BSK) medium (this preparation of BSK medium lacks rabbit serum and bovine serum albumin but contains rifampin [40 μg/ml]) for later laboratory processing. Tissues were transferred to 0.5 ml of incomplete BSK medium and were ground in a microtissue grinder (Spectrum Medical Industries, Los Angeles, Calif.). One-half of this suspension was added to a 7-ml screw-cap tube containing 6 ml of complete BSK medium (with rabbit serum and 35% bovine serum albumin solution but without antibiotics). The tube was tightly capped and incubated at 33°C for the duration of culture. Cultures were first examined by dark-field microscopy at 2 weeks and were incubated for at least 8 weeks. The remaining suspension plus the skin fragment itself was processed for PCR amplification.
DNA extraction and standard PCR conditions.
DNA was extracted separately from both skin tissue and its suspension medium by use of a commercial kit (IsoQuick; ORCA Research, Bothell, Wash.), as reported previously (8). Each extracted sample was resuspended in 50 μl of water, and corresponding skin and supernatant samples were combined prior to PCR amplification (a total of 100 μl per biopsy specimen). Five microliters of DNA suspension was used for conventional PCR, and 2 μl of DNA suspension was used for each qPCR amplification.
A 353-bp region of the B. burgdorferi flagellin gene (flaB) was amplified by a nested PCR protocol previously described by Barbour et al. (3). First-round PCR amplification was performed in 25-μl reaction mixtures containing 5 μl of DNA and 100 μM (each) deoxynucleoside triphosphates, 1.25 U of Taq DNA polymerase (Boehringer Mannheim), and 20 pmol of each primer in a DNA engine thermocycler (MJ Research, Watertown, Mass.). For second-round PCR, 1 μl of the first-round reaction mixture was added to 24 μl of the second-round master mixture. The thermal cycling profile of both the first- and the second-round PCRs consisted of one 3-min cycle at 94°C, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s. To avoid cross-contamination and sample carryover, pre- and post-PCR sample processing and PCR amplification were performed in separate rooms and all fluid transfers were carried out with plugged pipette tips to eliminate aerosols. DNA from isolate B31-MI was used as a positive control, and sterile water was used as the template for negative control amplifications, which were included with each PCR run. Amplified DNA products were detected by agarose gel electrophoresis in Tris-borate-EDTA buffer.
Quantification of B. burgdorferi DNA in skin biopsy samples by qPCR.
A B. burgdorferi-specific 222-bp fragment of the recA gene from patient skin biopsy specimens was amplified and quantified on a LightCycler real-time PCR instrument (Roche Diagnostics, Mannheim, Germany), as described previously (12, 23). PCR was performed in glass capillaries in a final volume of 10 or 20 μl containing 1× LightCycler Master Mixture (Roche), 3 mM MgCl2, 1 μM each primer, and 2 μl of DNA template. The forward and reverse primers were nTM17.F (5′-GTG GAT CTA TTG TAT TAG ATG AGG CTC TCG-3′) and nTM17.R (5′-GCC AAA GTT CTG CAA CAT TAA CAC CTA AAG-3′), respectively (12). The amplification program consisted of heating at 95°C for 30 s, followed by 45 cycles of heating at 20°C/s to 95°C with a 1-s hold, cooling at 20°C/s to 60°C with a 4-s hold, and heating at 20°C/s to 72°C with 10-s hold. The fluorescent product was collected at 80°C at the last step of each cycle to minimize signals from nonspecific products. A melting curve was acquired by heating the product at 20°C/s to 95°C, cooling it at 20°C/s to 60°C, and slowly heating it at 0.2°C/s to 95°C, with fluorescence collection at 0.2°C intervals. Specific and nonspecific PCR products were distinguished by melting curve analysis, since the melting temperature (Tm) of the nonspecific products was below 80°C, whereas the Tm of recA-specific amplicons was approximately 84°C. Data were analyzed with the LightCycler software provided by the manufacturer. Only data from the log-linear portion of the amplification were chosen for analysis. Since 1/2 of the total skin biopsy material was processed for DNA extraction and 1/50 (2 μl out of 100 μl) of the DNA prepared from each skin biopsy specimen was used as the template for qPCR, the spirochete number obtained for each qPCR was multiplied by 100 to obtain the total number of spirochetes in the entire 2-mm skin biopsy specimen. All qPCR-negative samples were diluted (1:5 or 1:10) and/or spiked with the DNA equivalent of 100 spirochetes and were subjected to another PCR amplification to determine whether potential PCR inhibitors were present.
An external standard set for B. burgdorferi-specific recA was developed as follows. Genomic DNA was prepared from a cultured B. burgdorferi clinical isolate, strain B356. The concentration of recA (number of copies per microliter) in purified genomic DNA was estimated as the DNA concentration determined by measurement of the optical density at 260 nm, assuming that the genome size is 1.5 Mbp and that there is one copy of recA per genome. This was further confirmed by a PCR-based limited-dilution assay. DNA templates containing 10 to 105 copies of B. burgdorferi-specific recA were included in each qPCR in order to generate a standard curve.
Determination of B. burgdorferi genotype.
Genotyping of the B. burgdorferi isolates was accomplished by a nested PCR protocol as described previously (8). Briefly, first-round PCR amplification of the 16S-23S ribosomal DNA spacer was carried out with primers PA and P95. One microliter of the first-round amplification mixture was used for a second round of PCR with primers PB and P97. Ten microliters of the nested PCR product was digested overnight at 65°C with TruI (MBI Fermentas, Hanover, Md.). The resultant digestion products were analyzed by electrophoresis on 2.4% agarose gels.
Serology.
Acute- and convalescent-phase serum specimens were tested by polyvalent (immunoglobulin M and immunoglobulin G) enzyme-linked immunosorbent assay (Wampole Laboratories, Cranbury, N.J.), in accordance with the instructions of the manufacturer.
Statistical analysis.
For categorical variables, P values were determined by the chi-square test or Fisher's exact test. For continuous variables that were normally distributed, Student's t test was used. If data were nonnormally distributed, the Mann-Whitney U test was used. A one-way analysis of variance or a Kruskal-Wallis test was used to compare three groups. The Spearman rank correlation was used to assess associations between skewed or ordinal variables. Multiple linear regression analyses were performed to test whether candidate variables were predictive of the number of spirochetes from EM lesions. All tests were two tailed. Analysis was performed with several statistical packages (Minitab [version 12.1] and True Epistat [version 5.1] from Epistat Services, Richardson, Tex., and SPSS [version 10.0] from SPSS, Chicago, Ill.). P values ≤0.05 were considered significant.
RESULTS
Clinical data.
The clinical characteristics of 50 untreated adult patients with EM evaluated during the spring and summer of 2000 are summarized in Table 1.
TABLE 1.
Clinical characteristics of 50 adult patients with EM
| Characteristic | Value | Range | Median |
|---|---|---|---|
| Age (yr [mean ± SD]) | 46.6 ± 11.7 | 21-72 | 46.0 |
| Duration of EM (days [mean ± SD]) | 8.6 ± 8.8 | 1-34 | 4.0 |
| No. (%) of patients who: | |||
| Were female | 27 (54) | ||
| Had symptoms on presentation | 39 (78) | ||
| Had a single EM lesion | 37 (74) |
Comparison of culture, nested PCR, and qPCR.
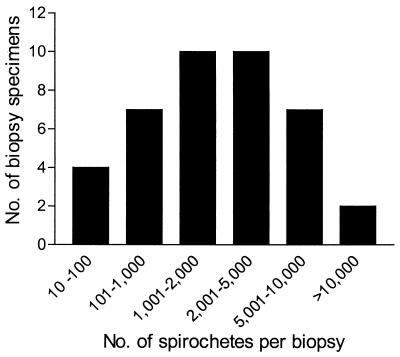
Skin biopsy specimens (diameter, 2 mm) obtained from the leading edge of the primary EM lesion (if known) were analyzed for the presence of B. burgdorferi by culture of the skin biopsy specimen, conventional nested PCR, and real-time qPCR. Comparison of the results obtained by these different methods is presented in Table 2. Among these diagnostic methods, qPCR was the most sensitive, with 40 of 50 (80%) biopsy specimens being positive, followed by conventional nested PCR (32 of 50 specimens [64%] were positive) and culture of skin biopsy specimen (27 of 50 specimens [54%] were positive). Of the culture-positive biopsy specimens, 7 of 27 (25.9%) were negative by conventional PCR and 3 of 27 (11.1%) were negative by qPCR. The rate of concordance between all three methods was 54%. For those specimens in which B. burgdorferi was detectable by qPCR, the mean ± standard deviation number of spirochetes present in a 2-mm skin biopsy specimen was 2,462 ± 2,942, the median number of organisms was 1,450, and the number of spirochetes in positive specimens varied over 3 orders of magnitude (Fig. 1). The mean number of spirochetes detected in culture-positive specimens was more than double the number detected in culture-negative specimens (3,940 versus 1,642 spirochetes [P = 0.019]).
TABLE 2.
Comparison of laboratory methods for detection of B. burgdorferi in EM lesions of 50 patients
| Result by the following detection method: | No. (%) positive | ||
|---|---|---|---|
| Culture | Nested PCR | qPCR | |
| + | + | + | 20 (40) |
| + | + | − | 0 (0) |
| + | − | + | 4 (8) |
| + | − | − | 3 (6) |
| − | + | + | 12 (24) |
| − | + | − | 0 (0) |
| − | − | + | 4 (8) |
| − | − | − | 7 (14) |
FIG. 1.
Spirochete burden in individual 2-mm EM lesion biopsy specimens. The number of B. burgdorferi organisms present in a 2-mm skin biopsy specimen from individual patients was determined by qPCR. Data are presented for 40 specimens which yielded positive values by qPCR.
Clinical findings and spirochete number.
In addition to EM, 39 patients (78%) had another symptom or sign on presentation (Table 1). To evaluate whether high or low spirochete numbers were associated with clinical symptomatology, analyses were performed by comparison of patients with more than the median number of B. burgdorferi organisms in the 2-mm skin biopsy sample versus those with less than this number. No significant association between clinical symptoms and spirochete number was observed (Table 3). There was also no significant association between the symptom score for each individual symptom and the number of spirochetes detected (data not shown).
TABLE 3.
Comparison of patient signs and symptoms with the median spirochete number
| Sign, symptom, or score | % of patients or score for patients with the following no. of spirochetes:
|
P value | |
|---|---|---|---|
| ≤1,450 (n = 25) | >1,450 (n = 25) | ||
| Loss of appetite | 28 | 28 | 1.000a |
| Dizziness | 16 | 28 | 0.496a |
| Tiredness or lack of energy | 56 | 48 | 0.778a |
| Headache | 32 | 44 | 0.561a |
| Muscle pain | 52 | 40 | 0.571a |
| Joint pain | 36 | 40 | 1.000a |
| Tingling or abnormal sensation | 40 | 16 | 0.114a |
| Stiff neck | 60 | 40 | 0.258a |
| Concentration or memory problems | 32 | 40 | 0.769a |
| Fever or chills | 36 | 32 | 1.000a |
| Cumulative symptom score (median) | 6.2 (1.7-21.6)b | 5.1 (0-25.4) | 0.884c |
| No. of symptoms (median) | 3.0 (1.5-7.5) | 3.0 (0-6) | 0.487c |
| Symptom severity index (median) | 2.6 (1.2-3.3) | 2.9 (0-4.4) | 0.668c |
| Largest EM diameter (cm [median]) | 19 (14-23) | 14 (9-17) | 0.005c |
| Baseline temp (°C, [mean ± SD]) | 36.68 ± 0.6 | 36.86 ± 0.6 | 0.294c |
| Regional lymphadenopathy | 7 | 12 | 0.244d |
Fisher's exact test.
Values in parentheses are interquartile range (i.e., 25th and 75th percentiles).
Mann-Whitney U test.
Chi-square test.
In contrast, the number of B. burgdorferi organisms detected in the 2-mm biopsy specimen was significantly inversely associated both with the duration of EM (rs = −0.329; P = 0.020) and with the size of the largest EM lesion (rs = −0.329; P = 0.020). Since the largest EM lesion was not necessarily the one that was biopsied in patients with multiple EM lesions, an additional analysis was performed by comparing the lesion size with the number of spirochetes only for those patients with a single EM lesion. A similarly significant inverse correlation between the largest EM lesion diameter and the number of spirochetes was observed (rs = −0.345; P = 0.037).
Serology.
Twenty-six patients (52%) were seropositive by enzyme-linked immunosorbent assay on presentation (Table 4). Seropositivity was not significantly associated with the number of spirochetes in the 2-mm sample of the EM lesion (P = 0.16).
TABLE 4.
Seropositivity and skin culture positivity as a function of median spirochete number
| Test result | No. (%) of patients with the following no. of spirochetes:
|
P value | |
|---|---|---|---|
| ≤1,450 (n = 25) | >1,450 (n = 25) | ||
| Seropositivity on presentation | 16 (64) | 10 (40) | 0.156a |
| Skin culture positivity | 8 (32) | 18 (72) | 0.010a |
Fisher's exact test.
B. burgdorferi genotype and spirochete load.
Genotyping of B. burgdorferi isolates from the 27 culture-positive biopsy specimens was carried out by a previously described PCR-restriction fragment length polymorphism (RFLP) typing method directed at the 16S-23S ribosomal DNA spacer (7 , 8). Twenty-four of 27 specimens contained spirochetes of a single RFLP type, and the analysis was limited to these 24 specimens. The number of B. burgdorferi organisms in EM lesions showed a significant association (P = 0.008) with the RFLP type of the infecting spirochete cultured from the skin of patients (Table 5). The average number of spirochetes in 2-mm biopsy specimens of the EM lesions of patients infected with RFLP type 1 organisms was nearly three times higher than that in patients infected with either RFLP type 2 or 3 organisms. Multiple linear regression analyses of RFLP type, EM size, and duration of EM showed that RFLP subtype 1 (P = 0.002) and EM duration (P = 0.015) were independent predictors of the number of spirochetes detected in the 2-mm sample of the EM lesions.
TABLE 5.
Correlation of B. burgdorferi RFLP type recovered in skin biopsy specimen culture and spirochete number
| RFLP type | No. of specimens | Median spirochete no. |
|---|---|---|
| 1 | 9 | 4,000 |
| 2 | 12 | 1,450 |
| 3 | 3 | 1,200 |
| P value for median | 0.008a |
Kruskal-Wallis test.
DISCUSSION
In an effort to understand better the pathogenesis of B. burgdorferi infection and to evaluate the performances of diagnostic assays, we analyzed skin biopsy specimens from 50 untreated adult Lyme disease patients with EM lesions by culture, nested PCR, and qPCR. The first description of qPCR to determine the number of B. burgdorferi organisms in tissue specimens used radioactive isotopes, which would preclude its general application in diagnostic laboratories (27). In the present study we used a LightCycler real-time PCR instrument for quantitation of B. burgdorferi sensu stricto. The method used was an adaptation of one previously reported by Morrison et al. (12) for use with mouse tissues. While previous studies have used the LightCycler instrument to type B. burgdorferi sensu lato (11, 17, 18), to our knowledge the present study is the first one in which qPCR has been used to enumerate the B. burgdorferi organisms in specimens from Lyme disease patients and the first to attempt to correlate spirochete numbers with clinical findings and other laboratory results. In addition to qPCR, conventional nested PCR and culture were also performed with the same skin biopsy specimens.
Among the three detection methods, qPCR was the most sensitive (positivity rate, 80%), followed by nested PCR (positivity rate, 64%) and culture (positivity rate, 54%) (also see reference 16). The sensitivities of the last two diagnostic modalities are comparable to those that we previously reported in 1992 (19), in which the sensitivity of conventional PCR sensitivity with 2-mm EM biopsy specimens was 59%, whereas the sensitivity of culture was 57%. The higher sensitivity of qPCR over that of nested PCR may be attributed to the fact that sample amplification and quantitative analysis were monitored in real time by a fluorometric assay with the double-stranded DNA-specific dye SYBR Green I. Amplicon detection was performed at the end of each cycle, not at the end point of amplification, as was done for nested PCR. The apparent discrepancy between the two PCR amplification methods was most likely due to inherent differences in amplicon detection technology and not to target amplification differences, as noted previously (11, 24). Both PCR amplification procedures targeted single-copy chromosomal genes with comparable specificities and sensitivities. The estimated sensitivities ranged from 1 to 10 copies of the target gene (flaB) for nested PCR and a single copy for qPCR. Real-time detection of qPCR amplification products by continuous fluorescence, followed by Tm analysis, is a more streamlined approach than conventional nested PCR, which involves two consecutive amplification reactions and which requires an additional gel electrophoresis step for detection after the completion of amplification (24, 25).
The 2-mm skin biopsy samples which were positive by both PCR methods had an average of 3,381 ± 544 spirochetes per biopsy specimen. In contrast, specimens in which spirochetes were not detected by conventional PCR but which were qPCR positive had an average of 1,580 ± 784 spirochetes per biopsy specimen (P = 0.13). This suggests that the lower number of spirochetes in these samples was likely a contributing factor in the failure of conventional PCR. Similarly, the number of spirochetes in culture-positive specimens was more than double that detected in culture-negative biopsy specimens (3,940 versus 1,642 spirochetes [P = 0.019]). A total of 16 samples (40%) which were qPCR positive yielded no growth of B. burgdorferi in culture. The presence of nonviable spirochetes in the specimens could account for this discrepancy. In three of the culture-positive skin specimens, spirochetes were not detectable by qPCR. The presence of PCR inhibitors in these specimens was ruled out because addition of B. burgdorferi DNA to the samples yielded positive PCR amplifications. The explanation for the culture-positive, qPCR-negative results for some specimens is not known, but it may be attributable to low spirochete numbers or sampling error. It should be noted that the data presented here are based on the results of tests with single 2-mm biopsy specimens taken from the leading edges of EM lesions. It is possible that heterogeneity in spirochete density may exist throughout a lesion. This should not have an impact on comparisons of culture yield and PCR positivity since all assays were performed with material from the same 2-mm biopsy specimen.
EM lesions, which are characteristic of early Lyme disease, expand rapidly over time (4, 14). Several studies have shown that the sizes of EM lesions directly correlate with the duration of the lesion (4, 13). We have previously reported that an inverse relationship between the recovery of B. burgdorferi from a skin specimen and EM lesion duration exists (13). The present study suggests that the explanation for this observation is that the number of spirochetes is significantly greater in skin lesions of shorter duration (P = 0.015) and, consequently, of smaller diameter (P = 0.006). Apparently, the number of organisms in the skin is reduced over time even in untreated patients, making recovery of spirochetes in culture less likely. The exact mechanism for the reduction of spirochete numbers in patient skin is unclear but may be related to elimination of spirochetes by the host immune system since development of a serologic response to B. burgdorferi has also been shown to be related to the duration of infection (1).
Differential dissemination of B. burgdorferi genotypes via blood has been reported previously (26). Highly significant associations between the presence of a particular RFLP type in skin (RFLP type 1) and the presence of spirochetemia (P < 0.001) or multiple EM lesions (P = 0.045) were observed. In the present study, there was a significant correlation between the RFLP type of the infecting spirochete and the number of spirochetes in the skin, with the highest number of organisms being present in patients with type 1 infections (P = 0.008). Whether the hematogenous dissemination observed in association with type 1 B. burgdorferi skin infections is caused by high spirochete numbers in the EM lesions merits further investigation.
No association between patient symptoms and the number of spirochetes detected by qPCR in the EM lesions was found, suggesting that the number of spirochetes in skin is not, per se, the cause for patient complaints. In a prior study, aside from a history of fever or chills, symptoms were not significantly associated with the particular B. burgdorferi genotype (RFLP type) causing the cutaneous infection (26). Clinical disease manifestations may depend on as yet unknown pathogenic factors that certain strains of B. burgdorferi may express or on host genetic determinants associated with the inflammatory response to B. burgdorferi.
In summary, this study is the first to measure the number of spirochetes in EM lesions of untreated adult Lyme disease patients. Larger numbers of B. burgdorferi organisms were significantly associated with shorter-duration, smaller skin lesions and infection with RFLP type 1 B. burgdorferi but not with the number or severity of symptoms. Culture positivity of skin biopsy specimens was significantly associated with skin lesions containing higher numbers of spirochetes. The duration of EM and the presence of RFLP subtype 1 B. burgdorferi were found to be independently associated with the number of spirochetes that could be detected by qPCR.
Acknowledgments
This work was supported in part by a grant from the National Institutes of Health (grant AR41511).
We gratefully acknowledge the investigators in the Lyme Disease Study Group, including L. Frank Cavaliere, Maria Aguero-Rosenfeld, Diane Holmgren, Kathy O'Keefe, Susan Bittker, Denise Cooper, Charles Pavia, Mohammed Bagheri, Jennifer Geiger, Anne Hardick, Matthew Harris, Pamela Jakubowicz, Doug Melman, Jonathan Nelson, Alexander Nicolaides, Daniel Radin, Jeffrey Rebish, and Karen Stolman.
REFERENCES
- 1.Aguero-Rosenfeld, M. E., J. Nowakowski, S. Bittker, D. Cooper, R. B. Nadelman, and G. P. Wormser. 1996. Evolution of the serologic response to Borrelia burgdorferi in treated patients with culture-confirmed erythema migrans. J. Clin. Microbiol. 34:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbour, A. G., and D. Fish. 1993. The biological and social phenomenon of Lyme disease. Science 260:1610-1616. [DOI] [PubMed] [Google Scholar]
- 3.Barbour, A. G., G. O. Maupin, G. J. Teltow, C. J. Carter, and J. Piesman. 1996. Identification of an uncultivable Borrelia species in the hard tick Amblyomma americanum: possible agent of a Lyme disease-like illness. J. Infect. Dis. 173:403-409. [DOI] [PubMed] [Google Scholar]
- 4.Berger, B. W. 1989. Dermatologic manifestations of Lyme disease. Rev. Infect. Dis. 11:S1475-S1481. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 1997. Case definitions for infectious conditions under public health surveillance: Lyme disease (revised 9/96). Morb. Mortal. Wkly. Rep. 46:1-55. [PubMed] [Google Scholar]
- 6.Foretz, M., D. Postic, and G. Baranton. 1997. Phylogenetic analysis of Borrelia burgdorferi sensu stricto by arbitrarily primed PCR and pulsed-field gel electrophoresis. Int. J. Syst. Bacteriol. 47:11-18. [DOI] [PubMed] [Google Scholar]
- 7.Liveris, D., A. Gazumyan, and I. Schwartz. 1995. Molecular typing of Borrelia burgdorferi sensu lato by PCR-restriction fragment length polymorphism analysis. J. Clin. Microbiol. 33:589-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liveris, D., S. Varde, R. Iyer, S. Koenig, S. Bittker, D. Cooper, D. McKenna, J. Nowakowski, R. B. Nadelman, G. P. Wormser, and I. Schwartz. 1999. Genetic diversity of Borrelia burgdorferi in Lyme disease patients as determined by culture versus direct PCR with clinical specimens. J. Clin. Microbiol. 37:565-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luft, B. J., R. J. Dattwyler, R. C. Johnson, S. W. Luger, E. M. Bosler, D. W. Rahn, E. J. Masters, E. Grunwaldt, and S. D. Gadgil. 1996. Azithromycin compared with amoxicillin in the treatment of erythema migrans: a double-blind, randomized, controlled trial. Ann. Intern. Med. 124:785-791. [DOI] [PubMed] [Google Scholar]
- 10.Mathiesen, D. A., J. H. Oliver, Jr., C. P. Kolbert, E. D. Tullson, B. J. Johnson, G. L. Campbell, P. D. Mitchell, K. D. Reed, S. R. Telford III, J. F. Anderson, R. S. Lane, and D. H. Persing. 1997. Genetic heterogeneity of Borrelia burgdorferi in the United States. J. Infect. Dis. 175:98-107. [DOI] [PubMed] [Google Scholar]
- 11.Mommert, S., R. Gutzmer, A. Kapp, and T. Werfel. 2001. Sensitive detection of Borrelia burgdorferi sensu lato DNA and differentiation of Borrelia species by LightCycler PCR. J. Clin. Microbiol. 39:2663-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morrison, T. B., Y. Ma, J. H. Weis, and J. J. Weis. 1999. Rapid and sensitive quantification of Borrelia burgdorferi-infected mouse tissues by continuous fluorescent monitoring of PCR. J. Clin. Microbiol. 37:987-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nadelman, R. B., J. Nowakowski, G. Forseter, S. Bittker, D. Cooper, N. Goldberg, D. McKenna, and G. P. Wormser. 1993. Failure to isolate Borrelia burgdorferi after antimicrobial therapy in culture-documented Lyme borreliosis associated with erythema migrans: report of a prospective study. Am. J. Med. 94:583-588. [DOI] [PubMed] [Google Scholar]
- 14.Nadelman, R. B., J. Nowakowski, G. Forseter, N. S. Goldberg, S. Bittker, Cooper, M. Aguero-Rosenfeld, and G. P. Wormser. 1996. The clinical spectrum of early Lyme borreliosis in patients with culture-confirmed erythema migrans. Am. J. Med. 100:502-508. [DOI] [PubMed] [Google Scholar]
- 15.Nadelman, R. B., and G. P. Wormser. 1998. Lyme borreliosis. Lancet 352:557-565. [DOI] [PubMed] [Google Scholar]
- 16.Nowakowski, J., I. Schwartz, D. Liveris, G. Wang, M. E. Aguero-Rosenfeld, G. Girao, D. McKenna, R. B. Nadelman, L. F. Cavaliere, and G. P. Wormser. 2001. Laboratory diagnostic techniques for patients with early Lyme disease associated with erythema migrans: a comparison of different techniques. Clin. Infect. Dis. 33:2023-2027. [DOI] [PubMed] [Google Scholar]
- 17.Pahl, A., U. Kuhlbrandt, K. Brune, M. Rollinghoff, and A. Gessner. 1999. Quantitative detection of Borrelia burgdorferi by real-time PCR. J. Clin. Microbiol. 37:1958-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pietila, J., Q. He, J. Oksi, and M. K. Viljanen. 2000. Rapid differentiation of Borrelia garinii from Borrelia afzelii and Borrelia burgdorferi sensu stricto by LightCycler fluorescence melting curve analysis of a PCR product of the recA gene. J. Clin. Microbiol. 38:2756-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwartz, I., G. P. Wormser, J. J. Schwartz, D. Cooper, P. Weissensee, A. Gazumyan, E. Zimmermann, N. S. Goldberg, S. Bittker, G. L. Campbell, and C. S. Pavia. 1992. Diagnosis of early Lyme disease by polymerase chain reaction amplification and culture of skin biopsies from erythema migrans lesions. J. Clin. Microbiol. 30:3082-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spach, D. H., W. C. Liles, G. L. Campbell, R. E. Quick, D. E. Anderson, Jr., and T. R. Fritsche. 1993. Tick-borne diseases in the United States. N. Engl. J. Med. 329:936-947. [DOI] [PubMed] [Google Scholar]
- 21.Steere, A. C., R. L. Grodzicki, A. N. Kornblatt, J. E. Craft, A. G. Barbour, W. Burgdorfer, G. P. Schmid, E. Johnson, and S. E. Malawista. 1983. The spirochetal etiology of Lyme disease. N. Engl. J. Med. 308:733-740. [DOI] [PubMed] [Google Scholar]
- 22.Steere, A. C., R. T. Schoen, and E. Taylor. 1987. The clinical evolution of Lyme arthritis. Ann. Intern. Med. 107:725-731. [DOI] [PubMed] [Google Scholar]
- 23.Wang, G., C. Ojaimi, R. Iyer, V. Saksenberg, S. A. McClain, G. P. Wormser, and I. Schwartz. 2001. Impact of genotypic variation of Borrelia burgdorferi sensu stricto on kinetics of dissemination and severity of disease in C3H/HeJ mice. Infect. Immun. 69:4303-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wittwer, C. T., M. G. Herrmann, A. A. Moss, and R. P. Rasmussen. 1997. Continuous fluorescence monitoring of rapid cycle DNA amplification. BioTechniques 22:130-138. [DOI] [PubMed] [Google Scholar]
- 25.Wittwer, C. T., K. M. Ririe, R. V. Andrew, D. A. David, R. A. Gundry, and U. J. Balis. 1997. The LightCycler: a microvolume multisample fluorimeter with rapid temperature control. BioTechniques 22:176-181. [DOI] [PubMed] [Google Scholar]
- 26.Wormser, G. P., D. Liveris, J. Nowakowski, R. B. Nadelman, L. F. Cavaliere, D. McKenna, D. Holmgren, and I. Schwartz. 1999. Association of specific subtypes of Borrelia burgdorferi with hematogenous dissemination in early Lyme disease. J. Infect. Dis 180:720-725. [DOI] [PubMed] [Google Scholar]
- 27.Yang, L., J. H. Weis, E. Eichwald, C. P. Kolbert, D. H. Persing, and J. J. Weis. 1994. Heritable susceptibility to severe Borrelia burgdorferi-induced arthritis is dominant and is associated with persistence of large numbers of spirochetes in tissues. Infect. Immun. 62:492-500. [DOI] [PMC free article] [PubMed] [Google Scholar]