Abstract
Yersinia pestis, the causative agent of deadly plague, is considered a reemerging infectious disease and a significant biological terrorism threat. The present project focused on epidemiological investigation of the genetic variability of well-documented strains of Y. pestis from the United States by pulsed-field gel electrophoresis (PFGE) and restriction fragment length polymorphism (RFLP) analysis with insertion sequences IS100 and IS285 as probes. We examined 37 U.S. Y. pestis strains and isolates of a single ribotype, ribotype B, recovered between 1939 and 1998 from patients, animals, and fleas. Our results showed that all isolates had similar PFGE patterns, but minor differences such as missing, additional, and shifted bands were found among almost all strains if they came from different parent strains. The 37 strains and isolates were divided into 26 PFGE types. RFLP analysis with IS100 as a probe divided these strains and isolates into 16 types, with 43% belonging to IS100 type 1. Typing with IS285 as a probe was less specific and led to only four RFLP types, with 81% belonging to type 1. Similarity analysis with BioNumerics software showed that all strains shared ≥80, 86, and 91% similarities on dendrograms prepared from digitized PFGE, IS100 RFLP analysis, and IS285 RFLP analysis images, respectively. Our results demonstrate that PFGE offers an increased ability to discriminate between strains (Simpson's index of diversity, 0.98) and therefore can significantly improve epidemiological studies related to the origin of new plague isolates.
Yersinia pestis is the causative agent of deadly plague. Plague circulates naturally among susceptible rodents and fleas in enzootic foci throughout the world. Y. pestis infections occasionally spill over into humans who come into contact with infected zoonotic agents. Plague can thus be considered a reemerging infectious disease in humans. This fact has been exemplified by increasing numbers of human plague cases since the early 1990s (4) and outbreaks of plague in Africa (29) and India (10). In the United States the number of human plague cases has increased from 3 yearly in the 1950s to 13 yearly in the 1990s (9). The appearance of multidrug-resistant strains of Y. pestis has raised concerns about control of the disease (12).
Recent emphasis on preparedness for biological terrorism threats (18) also led to renewed interest in examining Y. pestis, especially by methods that can determine the origin of the isolate. Classic methods have identified Y. pestis as comprising only one serotype, one phage type, and three biovars (27). These phenotypes provide limited information for tracing of the origin of the organism. Attempts at the establishment of a systematic method, including molecular biology-based techniques, of plague isolate classification have been under way, but the evaluation is incomplete. Recently, a variable-number tandem repeat (VNTR) technique (2) and ribotyping (14, 15) were used to type Y. pestis. The VNTR technique has a greater discrimination capacity than the ribotyping method, but strains from different areas were found to be identical types by the VNTR technique. Therefore, clinical laboratories have not routinely used any of the newly developed molecular biology-based techniques for plague surveillance (27).
Pulsed-field gel electrophoresis (PFGE), which separates DNA fragments upon digestion of the chromosome with restriction endonucleases that cleave infrequently (31), can facilitate a broad look at the whole genome of the organism. Although the method has been used to estimate the genome size and for detection of gross chromosome alterations (20, 30), it has not been used to systematically determine genetic relatedness between Y. pestis strains. It has also proved to be an effective method for qualitative evaluation of intraspecific genetic variation, permitting identification of individual isolates of a given species by comparison of their macrorestriction patterns (5).
Restriction fragment length polymorphism (RFLP) analysis provides information about the local genome environments of specific gene sequences on the basis of the probe used. Insertion sequence (IS) elements have been loosely defined as small (<2.5 kb), phenotypically cryptic segments of DNA with a simple genetic organization that are capable of inserting at multiple sites in a target molecule (21). ISs are involved in phenomena other than the acquisition of accessory functions. Many form an integral part of the chromosomes of most bacterial species to participate in chromosome rearrangements and promote plasmid integration. In contrast, some specific IS elements at defined places in the chromosome are sufficiently stable to allow them to be used as markers in RFLP analyses for species typing and epidemiological studies (21). Portnoy and Falkow (28) discovered an active IS element termed IS100, and Filippov et al. (11) discovered IS285. Both of these IS elements have been found in Yersinia spp. and thus were useful genetic markers for the typing of Y. pestis (1, 8, 11, 22). The utility of IS elements for the typing of Y. pestis is further suggested by the fact that this species contains more copies of IS elements than enteropathogenic Yersinia strains do (22).
The present project focused on the epidemiological investigation of the genetic variabilities of well-documented U.S. isolates of Y. pestis by PFGE and by RFLP analysis with IS100 and IS285 as probes. The recent infections with the biological warfare agent Bacillus anthracis have underscored the need to perform epidemiological studies with potential agents of biological terrorism. We compared our results with those obtained by the previously established method of ribosomal DNA (rDNA) restriction pattern typing (ribotyping) (14, 15). The group of isolates that we chose is particularly useful because the isolates were predominantly obtained from New Mexico and represent isolates obtained from similar regions during different years as well as from different regions of the state. Accordingly, the results of this project will give a clearer picture of the natural genomic variations among closely related strains of Y. pestis isolated in a local area of the United States and as such satisfies the need to test our typing method with a homogeneous group of strains (34). The determination of natural genetic variation and the development of genotyping methods for the comparison of isolates of Y. pestis may be useful for surveillance for potential newly emerging strains and determination of the origins of particular isolates.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
A total of 37 isolates made up of 30 independent strains and 7 subcultures designated as isolates of Y. pestis obtained between 1939 and 1999 from humans, animals, and fleas in the western United States were used in this study and are listed in Table 1. Our strains were predominantly isolated from New Mexico (81%; 30 strains and isolates); other strains were from Colorado (10.8%; 4 strains and isolates), Arizona (2.7%; 1 strain), and Texas (2.7%; 1 strain) (Table 1). All the strains are part of the collection of the Centers for Disease Control and Prevention, Ft. Collins, Colo. Five of our cultures were isolates (subcultures) from the same parent strains (Table 1). Y. pestis KIM5 (13) was obtained from the laboratory of Susan Straley, University of Kentucky, Lexington. The biotype information was provided by Patricia Worsham, U.S. Army Research Institute of Infectious Diseases, Frederick, Md. All U.S. isolates belong to biotype O (Orientalis). The KIM5 strains belongs to biotype M (Medievalis). For the preparation of DNA, the bacteria, which had been frozen at −80°C, were cultured on brain heart infusion (BHI) agar plates at 30°C for 48 h. DNA was prepared by inoculation of several colonies into 5 ml of BHI broth after incubation at 30°C overnight with shaking at 200 rpm.
TABLE 1.
Y. pestis isolates and characteristics
| Strainsa | Geographic origin | IS100 type | IS285 type | PFGE type | Source of isolation |
|---|---|---|---|---|---|
| A1122 | California | 8 | 1 | 23 | Spermophilus beecheyi |
| NM59-BENZ | Bernalilo, N.M. | 13 | 1 | 21 | Homo sapiens |
| NM61-DURAN | Santa Fe, N.M. | 1 | 1 | 4 | Homo sapiens |
| NM66Jaramillo | Bernalillo, N.M. | 1 | 1 | 13 | Homo sapiens |
| AZ70-130-1 | Apache, Ariz. | 1 | 1 | 14 | Flea pool |
| TX79-0209 | Canyon, Tex. | 1 | 1 | 11 | Cynomys ludovicianus |
| NM81-3387-684 | New Mexico (city unknown) N.M. | 7 | 1 | 6 | Flea pool |
| NM82-0395 | Rio Arriba, N.M. | 2 | 1 | 9 | Homo sapiens |
| NM83-0854 | San Miguel, N.M. | 12 | 1 | 26 | Homo sapiens |
| NM85-4298-585 | Bernalillo, N.M. | 1 | 1 | 13 | Flea pool |
| NM87-2981-614 | Cibola, N.M. | 1 | 1 | 7 | Flea pool |
| NM87-1298 | Rio Arriba, N.M. | 1 | 1 | 3 | Homo sapiens |
| NM87-2007 | McKinley, N.M. | 1 | 2 | 15 | Homo sapiens |
| NM95-1065 | Santa Fe, N.M. | 3 | 1 | 2 | Homo sapiens |
| NM95-1100-276 | Santa Fe, N.M. | 1 | 1 | 18 | Flea pool |
| CO96-3188A1 | Larimer, Colo. | 15 | 3 | 24 | Felis catus |
| CO96-3188B1 | Larimer, Colo. | 15 | 3 | 24 | Felis catus |
| NM96-3002-658 | Bernalillo, N.M. | 1 | 1 | 5 | Flea pool |
| NM96-3404 | Bernalillo, N.M. | 1 | 1 | 5 | Spermophilus variegatus |
| NM96-2970 | Bernalillo, N.M. | 1 | 1 | 5 | Homo sapiens |
| NM96-2968 | Bernalillo, N.M. | 5 | 1 | 17 | Homo sapiens |
| NM97-2129-3732 | Santa Fe, N.M. | 1 | 4 | 10 | Flea pool |
| NM97-2129-3742 | Santa Fe, N.M. | 1 | 4 | 10 | Flea pool |
| NM97-2070-3443 | Santa Fe, N.M. | 10 | 1 | 16 | Flea pool |
| NM97-2070-3453 | Santa Fe, N.M. | 10 | 1 | 16 | Flea pool |
| NM97-2064-338 | Santa Fe, N.M. | 1 | 1 | 28 | Flea pool |
| NM98-0152 | Albuquerque, N.M. | 14 | 1 | 22 | Canis familiaris |
| NM98-2993mp4 | Santa Fe, N.M. | 6 | 1 | 12 | Homo sapiens |
| NM98-2993org4 | Santa Fe, N.M. | 6 | 1 | 12 | Homo sapiens |
| NM98-2993sm4 | Santa Fe, N.M. | 6 | 1 | 12 | Homo sapiens |
| NM98-2993 1g4 | Santa Fe, N.M. | 6 | 1 | 12 | Homo sapiens |
| NM98-2252 | Santa Fe, N.M. | 16 | 1 | 25 | Homo sapiens |
| NM98-0510-86 | Santa Fe, N.M. | 1 | 1 | 1 | Flea pool |
| NM98-0511-87 | Santa Fe, N.M. | 1 | 1 | 1 | Flea pool |
| NM98-1714 | Santa Fe, N.M. | 4 | 1 | 4 | Homo sapiens |
| CO99-1214 | El Paso, Colo. | 8 | 3 | 19 | Homo sapiens |
| CO99-1133 | Larimer, Colo. | 9 | 3 | 20 | Homo sapiens |
| KIM5 | Iran | NAb | NA | NA | NA |
For all U.S. isolates (except isolate A1122, which was isolated in 1939), the first two capital letters represent states. The numbers following the state designation represent the last two digits of the year in which the organism was isolated. The number after the first dash represents the identification number for the last two digits of the patient or animal. Designations with two dashes indicate that the isolates were from fleas, with the number after the second dash indicating the flea identification number. Four groups of isolates are derivatives or subcultures from the same parent. The same superscript number on the right of the strain designation indicates isolates from the same parent: superscript 1, isolates CO96-3188A and CO96-3188B, which were prepared about 6 months apart; superscript 2, isolates NM97-2129-373 and NM97-2129-374, which were from different fleas but the same animal source; superscript 3, isolates NM97-2070-344 and NM97-2070-345, which were from different fleas but the same human source; superscript 4, isolates NM98-2993org, NM98-2993mp, NM98-2993sm, and NM2993Ig, which consisted of the original isolate (org), an isolate from mouse passage (mp) of the parent strain, a smaller colony (sm) of the parent strain, or a larger colony (lg) of the parent strain, respectively.
NA, not applicable.
PFGE.
The genomic DNAs of various strains of Y. pestis were prepared in agarose plugs as described previously (30), with a few modifications. Briefly, the pellets of overnight cultures were suspended in lysis buffer containing lysozyme, the suspension was mixed with an equal volume of 1.6% PFGE-grade melted agarose (Bio-Rad Laboratories, Richmond, Calif.), and that mixture was poured into plug molds (catalogue no., 1703706; Bio-Rad Laboratories). After 30 min at 4°C, the plugs were placed into sterile 50-ml blue-cap tubes and digested with proteinase K (1 mg/ml). Protein digestion was stopped by the addition of phenylmethylsulfonyl fluoride (0.5 mg/ml). Finally, the plugs were washed with 1× Tris-borate-EDTA buffer (TBE; 90 mM Tris-borate, 2 mM EDTA) before use. Following digestion overnight at 37°C with restriction endonuclease SpeI, NotI, or SfiI, the restriction fragments were resolved by PFGE with a CHEF-DRII apparatus (Bio-Rad Laboratories). Migration of DNA fragments was carried out in 0.5× TBE in 1% agarose gels at 200 V. The gels were maintained at 7°C during electrophoresis. Pulse times were ramped from 1 to 20 s over 48 h at 200 V. Isolates with identical restriction profiles were assigned to the same type, as reported previously (17, 19).
DNA digestion and transfer.
Y. pestis DNA for restriction digestion was also prepared from agarose plugs, as described above. The DNAs were digested with EcoRI or EcoRV overnight at 37°C and probed with 16S and 23S rDNAs (ribotyping) by the method described by Guiyoule et al. (14). RFLP analyses were carried out with HindIII-digested DNA in analyses with IS100 as the probe and EcoRI-digested DNA in analyses with IS285 as the probe. These restriction enzymes were selected because they do not cleave within IS DNA. The sample DNA was digested overnight at 37°C before it was loaded in a 0.7% agarose horizontal gel and subjected to electrophoresis for 15 h at 40 V in 1× TBE. Transfer of fractionated DNA onto nylon membranes was done by the method of Southern (32).
Preparation of probes and hybridization.
The 16S rDNA- and 23S rDNA-specific probes and the IS100 DNA- and IS285 DNA-specific probes were obtained by PCR amplification of Y. pestis KIM5 genomic DNA. The sequences of the primers used to amplify the 16S rDNA were AGTTTGATCATCGCTCAG (primer 8F) and CCATGGCGTGACGGGCAGTGTG (primer 1448R), as described previously (36). Other primers were chosen by use of the DNAStar software package (Lasergene, Madison, Wis.). The 23S rDNA primers sequences were CCGGCGAGGGGAGTGAAATAAATAGAA (primer 1623s 4) and TTTAAGCCCCAGGGAGACTCAT (primer 1623s5). The IS100-specific primer sequences were GCGCTGGCTGCACGATGTC (primer IS100-1) and CCCGAACGGCAGATTGGATGTC (primer IS100-2). The IS285-specific primers sequences were TTGCGGCTGAACTGGCTAAAG (primer IS285-1) and TAATAAAACGGCTCATCGCTAACC (primer IS285-2). Probes were labeled with alkaline phosphatase by use of a Gene Images AlkPhos Direct labeling and detection system (Amersham Pharmacia Biotech, Arlington Heights, Ill.). All hybridization processes were as described by the manufacturer. Briefly, the AlkPhos Direct hybridization buffer was prepared by adding NaCl to 0.5 M and blocking reagent to a final concentration of 4% (wt/vol). Prehybridization of blots was for at least 15 min at 55°C (0.125 to 0.25 ml of buffer/cm2 of membrane). Probe (5 to 10 ng/ml) was added, and hybridization was allowed to occur overnight at 55°C with gentle agitation. After hybridization, the membranes were washed with 55°C prewarmed primary wash buffer (2 M urea, 0.1% sodium dodecyl sulfate, 50 mM sodium phosphate, 150 mM NaCl, 1 mM MgCl2, 0.2% blocking reagent) twice for 10 min each time at 55°C. Final washes of the membranes were with secondary wash buffer (50 mM Tris base, 100 mM NaCl, 2 mM MgCl2 [pH 10.0]) twice for 5 min each time at room temperature. After the blots were washed, they were drained and developed by addition of CDP-Star detection reagent (30 to 40 μl/ml; Amersham Pharmacia Biotech) for 2 to 5 min at room temperature. The blots were wrapped in polyvinyl wrap and were exposed to Hyperfilm (Amersham Pharmacia Biotech). In this RFLP experiment the genotyping criteria depended on the band positions. In this study a strain with a shift, deletion, or addition of one band compared to the pattern for all other strains was considered to be a different RFLP type, as reported previously (33).
Computer-monitored fingerprinting analysis.
Computer analysis of the banding patterns obtained by PFGE and RFLP analyses with both IS100 and IS285 as probes was done with the BioNumerics software package (Applied Maths, Kortrijk, Belgium). The PFGE and RFLP banding patterns of all Y. pestis organisms were normalized by using a laboratory strain, Y. pestis KIM5, as the external standard. The images analyzed included three reference lanes with the external standard strain, Y. pestis KIM5. All images were compatible with one another after normalization, and complete PFGE or RFLP patterns were used for analysis. In general, bands were automatically assigned by the computer and were corrected manually after the original images were checked by eye. Only clearly resolved bands were counted. The Dice coefficient was used to analyze the similarities of the banding patterns. The unweighted pair group method with average linkages (UPGAMA) was used for cluster analysis. A slightly different position tolerance to allow 100% matching of the banding patterns obtained with duplicate samples on different images was selected for each method. The cophenetic correlation coefficient for the whole dendrogram was calculated for estimation of the faithfulness of the cluster analysis, using the BioNumerics software.
Discriminatory power analysis.
The discriminatory powers of the PFGE and RFLP techniques were evaluated by use of Simpson's index of diversity (discrimination index [D]), as defined by Hunter and Gaston (16), which was used as described previously (7). This index expresses the probability that two unrelated strains will be placed into different typing groups. D depends on the number of types defined by the test method and the relative frequencies of these types: D = 1 − {[(1/N) (N − 1)] × [Σ nj (nj − 1)]}, where N is the total number of unrelated strains and isolates, and nj is the number of strains and isolates that belong to the jth type.
RESULTS
RFLP profiles of Y. pestis isolates from the United States.
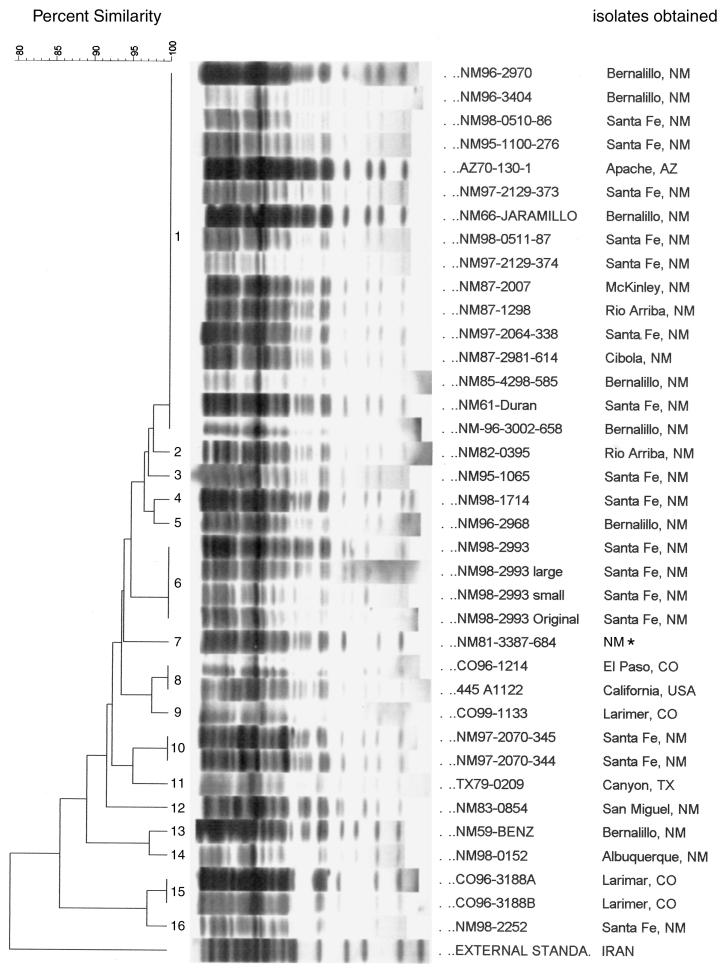
The 0.84-kb IS100 DNA fragment and the 1.15-kb IS285 DNA fragment were amplified by PCR, and the amplicons were used as probes to hybridize with the 37 U.S. Y. pestis strains and isolates. The most specific profile we observed by IS-based RFLP typing was with IS100 as the probe since about 20 distinguishable DNA fragments were hybridized. This allowed us to divide the 37 U.S. Y. pestis isolates into 16 types (Fig. 1). Sixteen of 37 isolates (43%) belonged to IS100 type 1 and had 100% similarity, as indicated by analysis with the BioNumerics software package (Fig. 1) (see Materials and Methods). Fifteen of 16 IS100 type 1 isolates were from New Mexico but were from different areas such as Bernalillo, Santa Fe, McKinley, Rio Arriba, and Cibola. One of the isolates from Arizona was also IS100 type 1. The other 21 isolates were divided into IS100 types 2 to type 16. Figure 1 shows that three groups of isolates subcultured from their parent strains shared the same IS100 type, designated IS100 type 6, type 10, and type 15 (Table 1). Although there are 16 types, the major patterns were similar. There was over 86% similarity among the 37 U.S. isolates by Dice coefficient analysis, but the external standard Y. pestis strain (strain KIM5) had its own IS100 RFLP pattern and shared less than 80% similarity with the U.S. strains and isolates. The cophenetic correlation value is 93 for the dendrogram derived from IS100 RFLP analysis.
FIG. 1.
Dendrogram derived from RFLP analysis with IS100 as a probe. HindIII-digested chromosomal DNAs of the 37 U.S. Y. pestis strains and isolates were hybridized with IS100 as the probe. The percent similarity scale is shown above the dendrogram. The Dice coefficient was used to calculate similarities, and UPGAMA (see Materials and Methods) was used for cluster analysis with BioNumerics software. The patterns are ordered from least similar to most similar (bottom to top). The IS100 RFLP types are marked with numbers. The position tolerance was 1.8%. The identification numbers and origins of the U.S. strains and isolates are listed on the right. The asterisk beside the designation for one strain indicates that no information on the specific location in New Mexico was available. The external standard was strain Y. pestis KIM5, which was used as an outgroup.
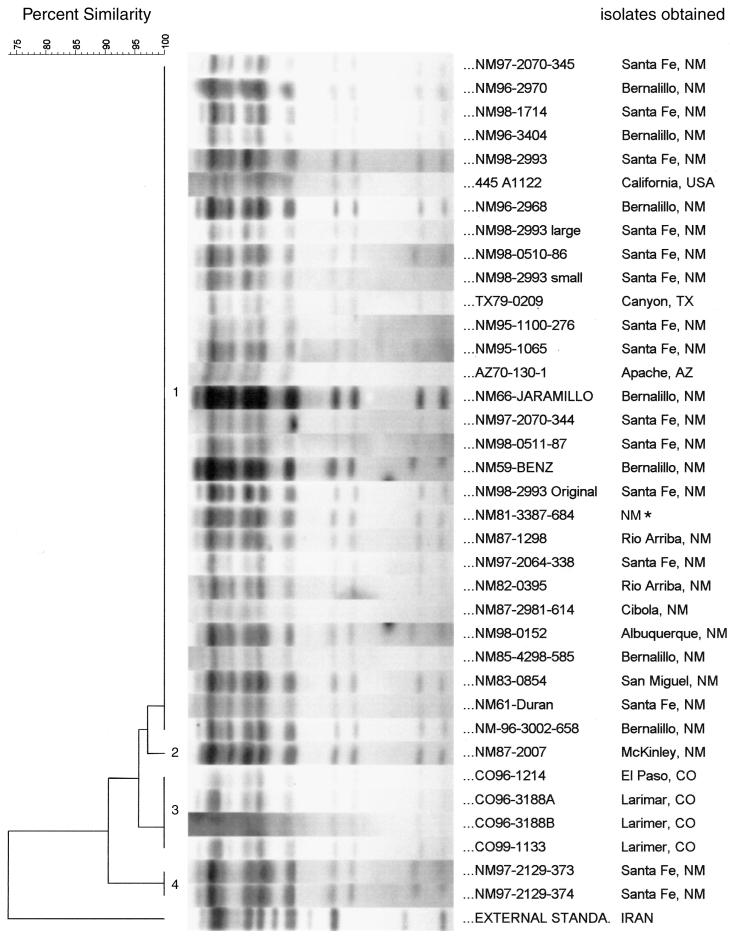
Since we wanted to investigate the possible use of IS-based RFLP analysis, we also used the other well-characterized mobile genetic element found in the Y. pestis genome, IS285, as a probe (11). Typing with IS285 as a probe was less specific than typing with IS100 as a probe, with only approximately about 12 visible bands observed from the genomes of the U.S. Y. pestis isolates (Fig. 2). Only four RFLP types, designated types 1, 2, 3, and 4, were found by IS285 RFLP analysis (Fig. 2 and Table 1). A total of 81% of the isolates (30 isolates) belonged to type 1. One type 2 isolate by IS285 RFLP analysis was obtained from McKinley, N.M. Four isolates from three different Y. pestis strains were type 3 by IS285 RFLP analysis. All of these isolates were obtained from Colorado, and three of them originated near Larimer, Colo. Finally, two isolates from the same parent strain were type 4 by IS285 RFLP analysis. The last two isolates came from Santa Fe, N.M. Similarity analysis by use of the Dice coefficient showed that all 37 strains and isolates within this group were at least 91% similar (Fig. 2), but the external standard Y. pestis strain (strain KIM5) had its own IS285 RFLP pattern and shared about 74% similarity with the U.S. strains and isolates. The cophenetic correlation value was 95 for the dendrogram derived from IS285 RFLP analysis.
FIG. 2.
Dendrogram derived from RFLP analysis with IS285 as a probe. EcoRI-digested chromosomal DNAs of the 37 U.S. Y. pestis strains and isolates were hybridized with IS285 as the probe. The percent similarity scale is shown above the dendrogram. The Dice coefficient was used to calculate similarities, and UPGAMA (see Materials and Methods) was used for cluster analysis with BioNumerics software. The IS285 RFLP types are marked with numbers. The patterns are ordered from least similar to most similar (bottom to top). The position tolerance was 1.8%. The identification numbers and origins of the strains and isolates are listed on the right. The asterisk beside the designation for one strain indicates that no information on the specific location in New Mexico was available. The external standard listed was strain Y. pestis KIM5, which was included as an outgroup.
Macrorestriction profiles of Y. pestis isolates from the United States.
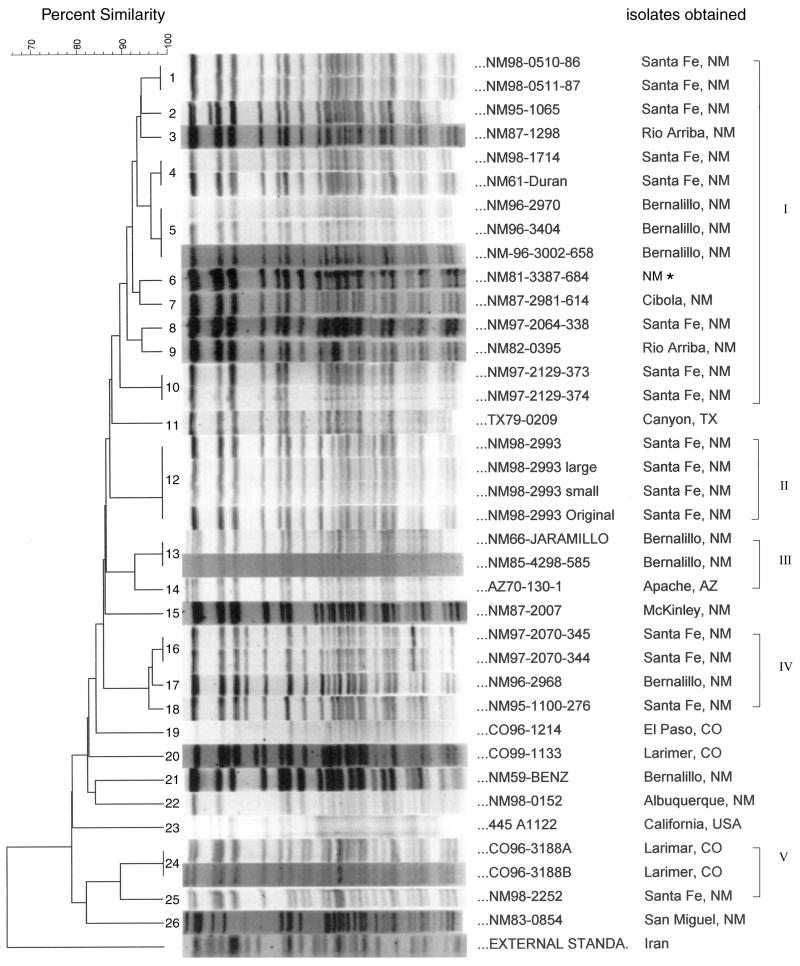
We tested the utility of PFGE for differentiation of the origins of individual Y. pestis strains within our homogeneous group of plague isolates from the United States. We reasoned that this technique might facilitate typing and epidemiological studies of the organism since it allows a broad look at the whole genome of the organism and does not rely on fragment visualization by IS probe hybridization. Initially, restriction enzymes SfiI, NotI, and SpeI were used to generate genomic restriction patterns (20). NotI digestion generated closely grouped bands of high molecular weight, as reported previously (20), and contained many comigrating fragments. In contrast, SfiI digestion produced many closely grouped smaller-molecular-weight bands. However, SpeI produced a relatively wider range of DNA fragments that could easily be resolved by our PFGE separation conditions (see Materials and Methods). Therefore, SpeI digestion was chosen for further study. Our electrophoresis conditions can resolve DNA fragments ranging between ≤48.5 and 339.5 kb. The profiles of the 37 U.S. isolates showed that they had similar major patterns (Fig. 3), but minor changes differentiated almost each individual strain even when the strains were recovered from the same local area and during the same year. Among the 37 U.S. isolates there were a total of 26 genotypes, in which each genotype had at least one band that was different from the band patterns of the other genotypes. The gels usually contained some weak or overlapping bands that we ignored for our analysis. We used only sharp and clearly visible bands for this analysis. Our electrophoresis conditions allowed identification of approximately 25 bands (≤48.5 to about 339.5 kb) that were useful for scoring and computer analysis. The dendrogram generated by computer-aided genotype analysis based on the PFGE patterns of all of the U.S. isolates showed ≥80% similarity by use of the Dice coefficient (Fig. 3). Among all 37 U.S. isolates, five clusters (designated clusters I, II, III, IV, and V) of isolates that shared over 90% similarity were found (Fig. 3). The first cluster (cluster I) contained 15 isolates, all of which were isolated from New Mexico between 1961 and 1998. However, those isolates were obtained from different areas of New Mexico such as Santa Fe, Bernalillo, Rio Arriba, and Cibola. Four New Mexico isolates that came from the same parent strain were found to be in cluster II (Fig. 3). These four isolates are subcultures of strain NM98-2993org. Cluster III included three strains, with one strain isolated from Apache, Ariz., in 1970; one strain isolated from Bernalillo N.M., in 1966; and one strain isolated from Bernalillo, N.M., in 1985. The two New Mexico strains in cluster III shared 100% similarity, even though they were isolated in different years. Cluster IV contains four isolates; three of them were obtained from Santa Fe, N.M., and one was obtained from Bernalillo, N.M., between 1995 and 1997. Two of the isolates in cluster IV have the same parent (Fig. 3). Cluster V contains two isolates obtained in Colorado in 1996 and one strain isolated in New Mexico in 1998. The other eight strains did not fit in any of the five clusters at similarity values ≥90%. Four nonclustered strains were from different areas of New Mexico. Among these four strains, one strain was recovered from San Miguel in 1983 (NM83-0854), one was recovered from Bernalillo in 1959 (NM59-Benz), one was recovered from Albuquerque in 1998 (NM98-0152), and one was recovered from McKinley in 1987 (NM87-2007). Two strains isolated in Colorado (strains CO99-1214 and CO99-1133), one strain isolated in Texas, and one strain isolated in California (strain A1122) could not be clustered by use of the criteria established in the present study. The external standard strain, strain KIM5 (from Iran), had its own genotype and shared only 65% similarity with the U.S. strains. The cophenetic correlation value is 88 for the dendrogram derived from PFGE.
FIG. 3.
Dendrogram derived from digitized PFGE patterns for the 37 U.S. Y. pestis strains and isolates whose chromosomal DNAs were digested with SpeI and constructed by similarity and clustering analysis by use of the Dice coefficient and UPGAMA (see Materials and Methods) with BioNumerics software. The percent similarity scale is shown above the dendrogram. The patterns are ordered from least similar to most similar (bottom to top). The PFGE types are marked on the left with numbers, and the clusters with a cutoff value at 90% similarity are marked on the right with roman numbers. The position tolerance was 1.1%. The identification numbers and origins of the U.S. strains and isolates are listed on the right. The asterisk beside the designation for one strain indicates that no information on the specific location in New Mexico was available. The external standard was strain Y. pestis KIM5, which was included as an outgroup.
Most patterns that we found to be 100% identical were for isolates of a single parent strain. Specifically, four groups of isolates that came from the same parent strain but that were collected after subculture or that had different colony types showed 100% identity. The isolates of specific strains with identical genotypes were as follows: strains NM97-2129-373 and NM97-2129-374; strains NM97-2070-344 and NM97-2070-345; strains NM98-2993ori (original parent strain), NM98-2993mp (mouse passage), NM98-2993lg (large colony), and NM98-2993sm (small colony); and strains CO96-3188A and CO96-3188B. Although there is a high degree of similarity between all U.S. isolates, most PFGE patterns revealed at least one band difference between isolates if they were from different parent strains (Fig. 3). However, four groups of independent isolates were found to have identical PFGE patterns. The first group consisted of strains NM96-2970, NM96-3002-658, and NM96-3404; the second group consisted of strains NM98-1714 and NM61-Duran; the third group consisted of strains NM66-Jaramillo and NM85-4298-585; and the last group consisted of strains NM98-0510-86 and NM98-0511-87 (Fig. 3). However, these independent but identical strains were all obtained from the same local area, and the first and fourth groups of strains were isolated in the same year (Table 1).
Ribotyping profiles of Y. pestis isolates from the United States.
In order to compare the RFLP and PFGE typing methods with the established ribotyping method (14), both 16S rDNA and 23S rDNA sequences were amplified from the Y. pestis KIM5 chromosome as described in Materials and Methods. The two probes were mixed and hybridized with EcoRI- and EcoRV-digested Y. pestis chromosomal DNAs from our group of 37 U.S. strains and isolates. The ribotype patterns of all U.S. strains belonged to ribotype B, as defined previously (14). Accordingly, we found ribotyping to be far less specific than PFGE or even IS-based RFLP analysis.
Relationship between PFGE profiles and RFLP profiles with IS100 and IS285 probes.
In order to know the ability of each of the three methods to differentiate between the strains and isolates, we calculated the D values. D has been used for evaluation of the discriminatory abilities (7, 15) of typing systems. The information used to calculate the D value and the results are shown in Table 2. PFGE typing proved to be the most discriminatory method (D = 0.98), followed by IS100 RFLP typing (D = 0.78). IS285 RFLP typing showed lowest the discrimination power (D = 0.38).
TABLE 2.
Comparison of discrimination powers of PFGE, IS-based RFLP, and ribotyping of U.S. Y. pestis strains and isolates
| DNA target | Method used | No. of patterns or genotypes | No. of bands detected | D valuea |
|---|---|---|---|---|
| Whole genome | PFGE | 26 | 25-28 | 0.98 |
| IS100 | RFLP analysis | 16 | 20-22 | 0.78 |
| IS285 | RFLP | 4 | 11-13 | 0.38 |
| 16S and 23S rRNA genes | RFLP | 1 | 5-6 | 0 |
See Materials and Methods for definition of D.
DISCUSSION
The purpose of the present study was to find an efficient, reliable method for discrimination between Y. pestis strains in order to enhance epidemiological studies. Several molecular biology-based methods for the typing of Y. pestis were reported previously. The most widely accepted and used method is ribotyping, in which 20 ribotypes, designated types A to T, were found for Y. pestis strains isolated on four continents (14, 15). However, our results indicated that all U.S. isolates had identical ribotypes, ribotype B. This result demonstrates that ribotypying is less applicable for epidemiological studies designed for a specific local region. Another modern technique which has recently been used for the typing of Y. pestis is VNTR analysis (2). VNTR analysis has a greater discrimination capacity than the ribotyping method. However, VNTR analysis examines a more limited region of the genome than PFGE does (2). PFGE can facilitate a broad look at the whole genome of the organism and has excellent discrimination power (35). Although PFGE has been used to characterize the Y. pestis genome (20), it has not been analyzed for its ability to support epidemiological studies. Accordingly, this is the first description of the use of PFGE for such studies to determine the relationships among Y. pestis strains.
In the present study, we used RFLP analysis and PFGE to type 37 U.S. Y. pestis strains and isolates. Since the genetic variability detected by IS285 RFLP analysis is very low, we do not consider it to be a good method for tracing of the origins of Y. pestis strains. IS100 RFLP analysis divided 37 U.S. strains and isolates into 16 types, with >43% of the isolates belonging to type 1. We used the restriction enzyme HindIII, which does not cleave within IS100. However, only four types were found in our primary study when we used EcoRV (data not shown), which has previously been used for IS100 RFLP analysis of Y. pestis (22). Our division of Y. pestis into more IS100 RFLP types may reflect the fact that HindIII does not cleave within the IS and is more appropriate for epidemiological studies. The IS100 and IS285 copy numbers in Y. pestis CO92 (a strain whose origin and time frame of collection are similar to those of our strain collection) are 44 and 21, respectively (26). However, the Southern blotting results showed only ∼20 visible bands for IS100 and ∼12 visible bands for IS285. These results may be caused by the comigration of bands or the presence of smaller bands (<2 kb), which were not resolved by the electrophoresis conditions used in our study.
Of all the methods that we tested, PFGE profiling showed the highest degree of variability in patterns since we observed 26 genotypes among the 37 U.S. strains and isolates. This resulted in the highest D value (D = 0.98) among the methods that we tested (Table 2). The majority of the isolates tested by PFGE had unique patterns even if they came from the same area and were recovered in the same year (Table 1 and Fig. 3). In most cases, identical PFGE patterns were observed only for isolates from the same strain; i.e., the colonies were isolates of one parent strain. However, a few strains from different parents still shared 100% identity if they were isolated in the same local area during the same year or during different times but from the same area (Fig. 3). The observation of identical genotypic patterns for different strains suggests that those strains came from the same source.
Strains and isolates that came from different states sometimes clustered together. For example, the clustering of strains in cluster III and cluster V (Fig. 3) suggested that geographic relationships exist between strains from Arizona and New Mexico and strains from New Mexico and Colorado, respectively. This result may suggest that a limited number of rearrangements occur within the Y. pestis genome and that these generate particular restriction patterns. Alternatively, these results may be due to the close geographic proximities of these states. However, the fact that the Y. pestis KIM5 strain from Iran, the external standard, showed much less similarity with U.S. strains supports the idea that the geographic relationship is correlated with the PFGE genotype. Interestingly, strains NM96-3002, NM96-3404, and NM96-2970 shared the exact same PFGE and RFLP patterns and were isolated in the same year and from the same local area, Bernalillo, N.M. These isolates were obtained from a flea pool and rodent and human sources, respectively, and may represent a cycle of plague in an area of endemicity. Accordingly, it is likely that these three strains were epidemiologically related. Taken together, our results generally indicate that the genetic variability of Y. pestis isolates observed by PFGE is related to the geographic distance of the strains. The mechanism of genomic plasticity that we observed remains unclear. However, the recent completion of the Y. pestis CO92 genome sequence suggests that this variability in restriction patterns may at least partially be due to inversion between ISs (26).
Our results also suggest that Y. pestis genotypic patterns may develop independently from each other. This was indicated by the fact that several isolates from the same region displayed relatively low levels of homology with each other. For example, we found that strains isolated from Santa Fe, N.M., were included in four of the five major PFGE clusters identified here (Fig. 3). Furthermore, one strain isolated from Santa Fe, N.M., in 1998, strain NM98-2252, was one of the strains that could not be grouped by PFGE with other strains from New Mexico. One possible explanation for these findings can be found in the life cycle of endemic plague in the United States. Plague is normally found in prairie dog colonies in the western United States. Generally, humans become infected following exposure to environments contaminated with infected prairie dog fleas. The transmission and spread of plague within prairie dog colonies are not well understood (3), but our results indicate that there has been little homogenization of the Y. pestis genome within the natural environment or life circle. Further studies will be required to determine the environmental factors that control the genotypic changes observed in different Y. pestis strains.
In contrast to the PFGE results reported previously (14), our PFGE results indicate that the chromosomal rearrangement rate is not obviously elevated during short-term laboratory maintenance since the subculture of the same parent strain always revealed exactly the same RFLP and PFGE patterns. Our findings agreed with the results obtained by VNTR analysis reported previously (2), in which low rates of mutation were shown by VNTR analysis during laboratory maintenance. However, recent PCR experiments (26) for examination of inversion of chromosomal regions between IS elements indicate that these regions do invert during short-term growth of Y. pestis. This indicates that the Y. pestis genome is fluid on a short-term scale but that these inversions may not be detected at the level of analysis achievable by PFGE, RFLP analysis, or VNTR analysis. Our observation that variations of smaller restriction fragments (e.g., HindIII and EcoRI digestion for RFLP analysis or ribotyping) are less than those of larger restriction fragments (e.g., SpeI digestion for PFGE) supports the idea that rearrangement of large genomic regions occurs in Y. pestis.
Y. pestis is closely related to Y. pseudotuberculosis at the DNA level and has even been thought to be a clone that evolved from Y. pseudotuberculosis (1). However, the numbers of IS elements harbored in the two species are very different. Specifically, Y. pestis carries significantly more copies of IS elements than Y. pseudotuberculosis (22, 24). IS elements of pathogenic bacteria have been reported to involve control of functions associated with virulence and immunogenicity (6) and may affect the stability of Y. pestis DNA (11, 25). Although the mechanism of genomic diversity remains unclear, our results as well as the results of others (23) indicate that the Y. pestis chromosome is generally unstable at the macrorestriction level. This genetic diversity may be useful for precise determination of the origin of a particular strain. For instance, our PFGE and IS100 RFLP patterns showed that strains isolated from the same local area and during the same year shared 100% similarity but that most other individual strains displayed unique patterns by PFGE (Fig. 1 and 3).
In conclusion, our results demonstrate that PFGE is able to resolve differences between most Y. pestis strains. PFGE offers significantly greater specificity than IS-based RPLP analysis techniques. Given the high degree of restriction fragment instability in the genome demonstrated here and the low level of nucleotide variability in individual genes of Y. pestis (1, 25), a technique such as PFGE will be best suited for the differentiation and analysis of new isolates. The combination of a technique such as PFGE, which analyzes the genotype of Y. pestis at the whole genome level, with a technique that analyzes regions of more limited variability may enhance our ability to trace the origins of strains. Further studies will be required to test this possibility.
Acknowledgments
We thank P. Worsham, U.S. Army Research Institute of Infectious Diseases, for biotyping information. Shaohua Zhao from the U.S. Food and Drug Administration and Sydney Lee gave advice and technical support in using BioNumeric software. Sarah Cohen critically read the manuscript and provided helpful discussions. Doug Tang of the Walter Reed Army Institute of Research gave advice on statistical analysis. L. Collins is gratefully acknowledged for artwork.
This work was supported by the U.S. Army Medical Research and Material Command.
REFERENCES
- 1.Achtman, M., K. Zurth, G. Morelli, G. Torrea, A. Guiyoule, and E. Carniel. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 96:14043-14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adair, D. M., P. L. Worsham, K. K. Hill, A. M. Klevytska, P. J. Jackson, A. M. Friedlander, and P. Keim. 2000. Diversity in a variable-number tandem repeat from Yersinia pestis. J. Clin. Microbiol. 38:1516-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, S. H., and E. S. Williams. 1997. Plague in a complex of white-tailed prairie dogs and associated small mammals in Wyoming. J. Wildl. Dis. 33:720-732. [DOI] [PubMed] [Google Scholar]
- 4.Anonymous. 1994. Human plague in 1992. Wkly. Epidemiol. Rec. 69:8-10. [PubMed] [Google Scholar]
- 5.Arbeit, R. D., M. Arthur, R. Dunn, C. Kim, R. K. Selander, and R. Goldstein. 1990. Resolution of recent evolutionary divergence among Escherichia coli from related lineages: the application of pulsed field electrophoresis to molecular epidemiology. J. Infect. Dis. 161:230-235. [DOI] [PubMed] [Google Scholar]
- 6.Barbour, A. G., C. J. Carter, N. Burman, C. S. Freitag, C. F. Garon, and S. Bergstrom. 1991. Tandem insertion sequence-like elements define the expression site for variable antigen genes of Borrelia hermsii. Infect. Immun. 59:390-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartie, K. L., M. J. Wilson, D. W. Williams, and M. A. Lewis. 2000. Macrorestriction fingerprinting of “Streptococcus milleri “ group bacteria by pulsed-field gel electrophoresis. J. Clin. Microbiol. 38:2141-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bobrov, A. G., and A. A. Filippov. 1997. Prevalence of IS285 and IS100 in Yersinia pestis and Yersinia pseudotuberculosis genomes. Mol. Gen. Mikrobiol. Virusol. 2:36-40. (In Russian.) [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention.1994. Human plague—United States, 1993-1994. Morb. Mortal. Wkly. Rep. 43:242-246. [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention. 1994. Update: human plague—India, 1994. Morb. Mortal. Wkly. Rep. 43:722-723. (Erratum, 43:763.) [PubMed] [Google Scholar]
- 11.Filippov, A. A., P. V. Oleinikov, V. L. Motin, O. A. Protsenko, and G. B. Smirnov. 1995. Sequencing of two Yersinia pestis IS elements, IS285 and IS100. Contrib. Microbiol. Immunol. 13:306-309. [PubMed] [Google Scholar]
- 12.Galimand, M., A. Guiyoule, G. Gerbaud, B. Rasoamanana, S. Chanteau, E. Carniel, and P. Courvalin. 1997. Multidrug resistance in Yersinia pestis mediated by a transferable plasmid. N Engl J. Med. 337:677-680. [DOI] [PubMed] [Google Scholar]
- 13.Goguen, J. D., J. Yother, and S. C. Straley. 1984. Genetic analysis of the low calcium response in Yersinia pestis Mu d1(Ap lac) insertion mutants. J. Bacteriol. 160:842-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guiyoule, A., F. Grimont, I. Iteman, P. A. Grimont, M. Lefevre, and E. Carniel. 1994. Plague pandemics investigated by ribotyping of Yersinia pestis strains. J. Clin. Microbiol. 32:634-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guiyoule, A., B. Rasoamanana, C. Buchrieser, P. Michel, S. Chanteau, and E. Carniel. 1997. Recent emergence of new variants of Yersinia pestis in Madagascar. J. Clin. Microbiol. 35:2826-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joo, Y. S., L. K. Fox, W. C. Davis, G. A. Bohach, and Y. H. Park. 2001. Staphylococcus aureus associated with mammary glands of cows: genotyping to distinguish different strains among herds. Vet. Microbiol. 80:131-138. [DOI] [PubMed] [Google Scholar]
- 18.Kortepeter, M., and G. Parker. 1999. Potential biological weapons threats. Emerg. Infect. Dis. 5:523-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lange, C., M. Cardoso, D. Senczek, and S. Schwarz. 1999. Molecular subtyping of Staphylococcus aureus isolates from cases of bovine mastitis in Brazil. Vet. Microbiol. 67:127-141. [DOI] [PubMed] [Google Scholar]
- 20.Lucier, T. S., and R. R. Brubaker. 1992. Determination of genome size, macrorestriction pattern polymorphism, and nonpigmentation-specific deletion in Yersinia pestis by pulsed-field gel electrophoresis. J. Bacteriol. 174:2078-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDonough, K. A., and J. M. Hare. 1997. Homology with a repeated Yersinia pestis DNA sequence IS100 correlates with pesticin sensitivity in Yersinia pseudotuberculosis. J. Bacteriol. 179:2081-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Najdenski, H., I. Iteman, and E. Carniel. 1995. The genome of Yersinia enterocolitica is the most stable of the three pathogenic species. Contrib. Microbiol. Immunol. 13:281-284. [PubMed] [Google Scholar]
- 24.Odaert, M., P. Berche, and M. Simonet. 1996. Molecular typing of Yersinia pseudotuberculosis by using an IS200-like element. J. Clin. Microbiol. 34:2231-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Odaert, M., A. Devalckenaere, P. Trieu-Cuot, and M. Simonet. 1998. Molecular characterization of IS1541 insertions in the genome of Yersinia pestis. J. Bacteriol. 180:178-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 27.Perry, R. D., and J. D. Fetherston. 1997. Yersinia pestis—etiologic agent of plague. Clin. Microbiol. Rev. 10:35-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Portnoy, D. A., and S. Falkow. 1981. Virulence-associated plasmids from Yersinia enterocolitica and Yersinia pestis. J. Bacteriol. 148:877-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ratsitorahina, M., S. Chanteau, L. Rahalison, L. Ratsifasoamanana, and P. Boisier. 2000. Epidemiological and diagnostic aspects of the outbreak of pneumonic plague in Madagascar. Lancet 355:111-113. [DOI] [PubMed] [Google Scholar]
- 30.Romalde, J. L., I. Iteman, and E. Carniel. 1991. Use of pulsed field gel electrophoresis to size the chromosome of the bacterial fish pathogen Yersinia ruckeri. FEMS Microbiol. Lett. 68:217-225. [DOI] [PubMed] [Google Scholar]
- 31.Smith, C. L., and G. Condemine. 1990. New approaches for physical mapping of small genomes. J. Bacteriol. 172:1167-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 33.Speijer, H., P. H. Savelkoul, M. J. Bonten, E. E. Stobberingh, and J. H. Tjhie. 1999. Application of different genotyping methods for Pseudomonas aeruginosa in a setting of endemicity in an intensive care unit. J. Clin. Microbiol. 37:3654-3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swaminathan, B., and G. M. Matar. 1993. Molecular typing methods: definition, applications, and advantages, p. 26-50. In D. H. Persing, T. F. Smith, F. C. Tenover, and T. J. White (ed.), Diagnostic molecular microbiology. ASM Press, Washington, D.C.
- 35.Tenover, F. C., R. D. Arbeit, R. V. Goering, et al. 1997. How to select and interpret molecular strain typing methods for epidemiological studies of bacterial infections: a review for healthcare epidemiologists. Infect. Control Hosp. Epidemiol. 18:426-439. [DOI] [PubMed] [Google Scholar]
- 36.Warner, C. K., and J. E. Dawson. 1996. Genus- and species-level identification of Ehrlichia species by PCR and sequencing, p. 100-105. In D. H. Persing (ed.), PCR protocols and emerging infectious diseases. ASM Press, Washington, D.C.