Abstract
Polyadenylation stimulates translation of capped eukaryotic mRNAs and those carrying picornaviral internal ribosome entry segments (IRESes) in vivo. Rabbit reticulocyte lysates (RRL) reproduce poly(A)-mediated translation stimulation in vitro after partial depletion of ribosomes and ribosome-associated factors. Here, we have evaluated the effects of varying different parameters (extent of extract depletion, cleavage of eIF4G, concentrations of KCl, MgCl2 and programming mRNA) on IRES-driven translation efficiency and poly(A)-dependency in ribosome-depleted RRL. For comparison, the study included a standard capped, polyadenylated mRNA. Dramatic differences were observed in the abilities of the different IRESes to direct translation in ribosome-depleted extracts. While the hepatitis A virus IRES was incapable of driving translation in physiological conditions in depleted RRL, mRNAs carrying the foot-and-mouth disease virus and hepatitis C virus IRESes were translated significantly better than a standard cellular mRNA in the same conditions. Indeed, the capacities of these IRESes to direct translation in ribosome-depleted RRL were similar to those reported previously in certain cell lines. Both the abilities of the IRESes to drive translation and their individual salt optima in ribosome-depleted extracts suggest that these elements have dramatically different affinities for some component(s) of the canonical translation machinery. Finally, using poliovirus as an example, we show that the ribosome-depleted system is well suited to the study of the translational capacity of naturally occurring IRES variants.
INTRODUCTION
Most eukaryotic mRNAs carry an m7GpppN cap at their 5′ end (1) and a poly(A) tail at their 3′ end (reviewed in 2,3). The initiation of protein synthesis on the majority of such mRNAs follows binding of the 40S ribosomal subunit near the capped 5′ end of the mRNA and subsequent migration of this subunit along the mRNA in a 5′ to 3′ direction until a suitable initiation codon is selected (reviewed in 4). Recognition of the mRNA 5′ end and 40S subunit recruitment requires the eukaryotic initiation factor (eIF) 4F complex (reviewed in 5,6). The eIF4F complex comprises the cap binding protein (eIF4E) and an ATP-dependent RNA helicase (eIF4A) bound, respectively, towards the N- and C-termini of a scaffold protein, eIF4G (reviewed in 5,7). The C-terminal half of eIF4G is also thought to associate with the eIF3 complex, which binds the 40S ribosomal subunit directly, thus bridging the gap between the mRNA 5′ end and the 40S subunit (reviewed in 8). While the cap or poly(A) tail alone enhance translation initiation, the two elements together co-operate to synergistically stimulate translation initiation in vivo (9–13). Cap–poly(A) synergy can be reproduced in a variety of in vitro cell-free extracts derived from eukaryotic cells (12,14–16). The majority of such systems exhibit synergy only in the presence of endogenous competitor mRNAs. In the absence of competition, the positive effects of capping and polyadenylation on translation are at best only additive (12,15–17). However, mammalian cell-free translation systems can be rendered poly(A)- dependent for translation in the absence of competitor mRNAs, by partial depletion of ribosomal subunits and those translation factors which associate tightly with ribosomes (16).
The uncapped, polyadenylated genomes of the animal picornaviruses are translated following an alternative mode of ribosome recruitment. Picornaviral RNAs contain an extensive (some 450 nt), heavily structured sequence within the 5′ non-coding region, known as the internal ribosome entry segment (IRES), which promotes direct internal entry of ribosomes some several hundred nucleotides from the RNA 5′ end (reviewed in 18). Thus, translation of the picornaviral RNAs is both cap- and 5′ end-independent. A similar mechanism of translation initiation has now been described for many other viruses, including the flavivirus, hepatitis C virus (HCV), whose uncapped and non-polyadenylated, positive strand RNA genome also carries an IRES (reviewed in 18) (19–21).
The picornaviral IRESes have been classified into three distinct groups on the basis of primary sequence and secondary structure conservation (reviewed in 18), and also their requirements for optimal internal initiation of translation in vitro (22). Type I IRESes [those of the enteroviruses and rhinoviruses; e.g. poliovirus (PV) and human rhinovirus (HRV)] are inefficient in driving translation initiation in the absence of specific cellular proteins which are absent or limiting in rabbit reticulocyte lysates (RRL), and are highly sensitive to modifications of KCl and MgCl2 concentrations. However, type I IRES efficiency is significantly enhanced in vitro in the presence of either the 2A or Lb protease, both of which cleave eIF4G (22,23). Conversely, type II IRESes [cardioviruses and aphthoviruses; e.g. encephalomyocarditis virus (EMCV) and foot-and-mouth disease virus (FMDV)] initiate translation efficiently in unsupplemented RRL, and are relatively insensitive to fluctuations in salt concentration. Furthermore, these elements are not dramatically affected by 2A or Lb protease-mediated cleavage of eIF4G (22). Hepatitis A virus (HAV; a type III IRES) IRES activity is relatively efficient in RRL, is not stimulated by supplementation with crude cell extracts, tolerates a wide range of salt concentrations, and is inhibited when eIF4G is cleaved by the 2A or Lb proteases (24–26). The HCV IRES constitutes a fourth type of viral IRES. While HCV IRES function shares many features with the picornaviral type II elements, including insensitivity to eIF4G cleavage and large variations in salt concentrations, and reasonable efficiency in unsupplemented RRL, 40S ribosomal subunit recruitment to the HCV IRES is independent of eIF4G (21).
It was recently demonstrated that polyadenylation increases the efficiency of picornaviral IRES-driven translation (16,27–29). Like cap–poly(A) synergy, poly(A)-mediated stimulation of IRES-driven translation is only evidenced when components of the translational machinery are functionally limiting (28). In addition, the positive effects of polyadenylation on both capped cellular and IRES-carrying mRNAs are most detectable when translation is carried out in physiological concentrations of added salt (28,30).
In the light of these recent data highlighting the importance of polyadenylation for picornaviral IRES activity, we report here a comprehensive study of the requirements for optimal picornaviral and HCV IRES activity and the detailed characterisation of the requirements for poly(A)-dependency of IRES-driven translation in ribosome-depleted RRL cell-free translation extracts.
MATERIALS AND METHODS
Plasmid constructions and in vitro transcriptions
The construction of the monocistronic pOp24, pPVp24, pEMCVp24, pHAVp24 and pHCVp24 plasmids has been described elsewhere (16,28). The pOp24 plasmids contain (under the control of the bacteriophage T7 φ10 promoter) a short oligonucleotide-derived 5′ untranslated region (UTR), followed by the region coding for the human immunodeficiency virus (HIV-1Lai) p24 protein and the influenza virus NS 3′ UTR (see Fig. 1). The pHRVp24 plasmid was constructed by introduction of the BamHI–SalI fragment of pXLJ 10–585 (31) (nucleotides 10–585 of HRV type 2) into pOp24 digested with the same enzymes. Similarly, the pFMDVp24 construct was generated by insertion of the end-filled EcoRI–SmaI fragment of pSP449 (32) (nucleotides 362–831 of FMDV O1K strain) into pOp24 which had been digested with BamHI and end-filled. Two versions of all of the plasmids differ only in the presence or absence of an A50 tract (or the 98 nt HCV 3′ X region for pHCVp24), inserted at the unique EcoRI site, 24 nt downstream of the authentic polyadenylation signal. The construction of truncated cDNAs corresponding to the PV type I Mahoney strain carrying the different Sabin mutations in domain V of the IRES have also been described previously (C.E.Malnou, A.M.Borman and K.M.Kean, manuscript submitted). The resulting plasmids contain (under the control of the bacteriophage T7 φ10 promoter) the full PV type 1 5′-UTR and authentic viral coding region, except for an internal deletion of nucleotides 2546–6304, with C469U (Sab3), A480G (Sab1) or G481A (Sab2) point mutations. All plasmids were verified by automatic sequencing. In vitro transcriptions, and quantification and purification of capped and uncapped in vitro transcripts were performed as described (16) using plasmids linearised by EcoRI.
Figure 1.
Schematic representation of plasmids used in this work. The HIV-1p24 coding region and the regions corresponding to the different IRESes are shown as open boxes. Numbers below the coding region refer to the first and last amino acids of HIV-1p24 and numbering below the IRESes denotes the first and last nucleotides of the corresponding viral genome sequences. The ATG codon initiating HIV-1p24 synthesis is shown in bold and underlined, the TGA stop codon is shown in bold. The NS 3′ UTR is depicted as a thick speckled line. Clones were constructed either in duplicate, differing only by the presence or absence of an A50 insertion (bracketed) at the EcoRI site used for linearisation prior to transcription, or in triplicate (pHCVp24) including, instead of the A50 oligonucleotide, 98 nt corresponding to the 3′ X region from the HCV genome.
Preparation of translation extracts and in vitro translations
Nuclease-treated RRL (Flexi-RRL; Promega) was partially depleted of ribosomes by ultracentrifugation in a Beckman Optimax benchtop ultracentrifuge essentially as described previously (16). In experiments aimed to evaluate the effects of different levels of ribosome-depletion on poly(A)-dependency, the speed and duration of ultracentrifugation were varied (90 000 r.p.m. for 15–25 min in a fixed-angle TL100 rotor; 1.1 ml volumes of RRL). The post-centrifugation supernatant was removed without disturbing the pellet of ribosomes/associated initiation factors, and was aliquoted and stored at –80°C. HeLa and IMR 32 (neuroblastoma) cell S10 extracts were prepared essentially as described previously (31), except that cells were not starved of methionine before extract preparation. S10 extracts were dialysed overnight against H100 buffer (10 mM HEPES–KOH pH 7.5, 100 mM KCl, 1 mM MgCl2, 0.1 mM EDTA and 7 mM β-mercaptoethanol), prior to treatment with micrococcal nuclease as described (33). FMDV Lb protease was expressed and purified exactly as described previously (34), and was dialysed against H100 buffer prior to use. Lb protease treatment of translation extracts was performed at 30°C for 15 min. Protease was inactivated by incubation with E-64 (500 µM final concentration; 10 min at 4°C) prior to the addition of mRNAs to translation extracts.
In vitro translation reactions were performed in the presence of 35S-methionine. Reactions contained 50% by volume of flexi-RRL or ribosome-depleted RRL, and 33% by volume of H100 buffer or HeLa cell extract in H100 buffer (2.5% S10 extract and 31.5% buffer). Reactions were programmed with the indicated concentrations of in vitro transcribed mRNAs. Final concentrations of added KCl and MgCl2 were 130 and 0.93 mM, respectively, unless otherwise stated. Translations were performed for 90 min at 30°C and the translation products were analysed by SDS–PAGE as described previously (35), using gels containing 23% w/v acrylamide. Dried gels were exposed to Biomax MR film (Kodak) for 1–10 days depending on the particular experiment. Densitometric quantification of translation products was as described previously (23) using multiple exposures of each gel to ensure that the linear response range of the film was respected and that low levels of translation could be accurately quantified.
RESULTS
Many new generation eukaryotic expression vectors utilise the capacity of IRESes to initiate translation internally to allow the expression of several genes from a single vector. Effectively, the ability of an RNA segment to direct the synthesis of the downstream open reading frame of a dicistronic mRNA when inserted between the two cistrons has long been the accepted test for IRES activity (reviewed in 18). We previously used such a dicistronic vector approach, with non-polyadenylated mRNAs, to compare the efficiencies of six different picornaviral IRESes and the flaviviral HCV IRES in directing internal ribosome entry in RRL (22). This comparison allowed the picornaviral IRESes to be divided into three groups on the basis of their functional requirements for directing optimal translation in vitro. Classification into the same three groups was also possible ex vivo, after introduction of the same dicistronic mRNAs into a variety of cultured cells of different origins (36). However, certain IRESes exhibited discrepancies between their potential to direct translation in vitro versus ex vivo. The HAV IRES was the most flagrant case: while it was relatively efficient in vitro, it showed no detectable IRES activity in any of the cultured cells tested. The recent demonstrations that picornaviral IRES-driven translation is extremely sensitive to the polyadenylation status of the IRES-carrying mRNA when the translation machinery is limiting (27–29) provides a potential explanation for such discrepancies.
In the light of these observations, the present study aimed to examine the properties of internal initiation of translation driven by different viral IRESes, as a function of the poly(A) status of the corresponding mRNAs. Since poly(A) tails can stimulate both capped cellular mRNA translation, and uncapped picornaviral IRES-driven translation, monocistronic mRNAs carrying the different IRESes were used for the present study. This avoided the possible complication of competition between a capped upstream cistron, and the downstream IRES-driven cistron for the poly(A) tail, which could not be excluded with dicistronic mRNAs. The HCV IRES was included in the study as an example of an IRES which is not stimulated by polyadenylation, but rather by an unusual conserved viral 3′ end sequence, called the X region (20,28). Previous results have suggested that the HCV IRES is very efficient in driving translation both in different cultured cells, and in a poly(A)-dependent in vitro translation system (28,36).
To allow a rigorous comparison, a series of transcription templates was constructed which differ only by the minimal region required for full activity of the different IRESes, fused in frame with a standard reporter gene (the p24 capsid coding region of HIV-1). Thus, for the EMCV, FMDV and HAV IRESes, in which essential sequence requirements extend up to the authentic initiation codon (37–40), the p24 coding region was fused after the authentic viral start site for translation (Fig. 1). For the HCV IRES, essential sequence extends past the authentic HCV initiation codon (41,42). Therefore, the sequence coding for the first nine amino acids of the HCV polyprotein were also included upstream of the p24 protein coding region. All plasmids contain an identical 3′-UTR (derived from the influenza virus NS gene), with or without an A50 tract inserted at the site used for linearisation prior to in vitro transcription. For pHCVp24 constructs, a third plasmid was generated in which the A50 tract was replaced by the authentic 98 nt viral X region which is conserved amongst all genomic HCV RNAs (Fig. 1).
The effects of KCl and MgCl2 concentration on the efficiency of viral IRES-driven translation in poly(A)-dependent RRL extracts
Protein synthesis in vitro has previously been shown to have a strong requirement for K+ ions (43). However, the absolute potassium concentration which is optimal for translation varies according to the mRNA species tested, with uncapped mRNAs in general exhibiting lower KCl optima than their capped counterparts (44). Furthermore, the individual salt optima were found to vary dramatically even amongst naturally uncapped mRNAs carrying the different picornaviral IRESes (22). Additionally, cap–poly(A) co-operativity with classical cellular mRNAs translated in ribosome-depleted RRL was reported to be greatest with near-physiological KCl and MgCl2 concentrations (30). Based on these data, we evaluated the effects of altering salt concentration on the efficiency of viral IRES-driven translation in poly(A)- dependent RRL by translating pIRESp24-derived mRNAs over a wide range of KCl and MgCl2 concentrations. The results of these experiments are summarised in Table 1.
Table 1. Requirements for optimal viral IRES-driven translation in ribosome depleted RRL.
| Virus | Optimal salt conditions | Maximal stimulation by poly(A) in depleted RRL | |||||
|---|---|---|---|---|---|---|---|
| KCl (mM) | MgCl2 (mM) | ||||||
| –Ac | +Ad | –Ac | +Ad | Factor | KCl (mM)e | MgCl2 (mM)e | |
| PVa | 97 (80) | 110 | 0.5 (0.2) | 0.5 | 5–10 × | 120 | 1.0 |
| HRV | <50 (65) | 50 | <0.3 (0.7) | <0.3 | 5–10 × | 80 | 0.5 |
| HAV | <75 (65) | <75 | <0.3 (0.7) | <0.3 | 20–40 × | 105 | 0.9 |
| EMCV | >130 (120) | >130 | <0.3 (0.7) | 0.6 | 2–4 × | >130 | >0.3 |
| FMDV | 85 (125) | 85 | 0.5 (0.7) | 0.7 | 2 × | 106 | 1.0 |
| HCV | >130 (110) | >130 | 0.5 (0.6) | 0.5 | 2–4 ×f | –g | –g |
| Op24b | 110 | 125 | <0.3 | 0.75 | 10–20 × | >130 | 0.9 |
aTranslation reactions were programmed with 5 µg/ml of IRESp24-derived mRNAs which had been synthesised in uncapped, non-polyadenylated (–A) or uncapped, polyadenylated form (+A).
bCapped pOp24-derived mRNAs with or without a poly(A) tail were translated at 3 µg/ml.
c,dThe values for optimal concentrations of added KCl and MgCl2 were adjusted to take into account differences in endogenous salt inherent to the batch of RRL used as compared to that used in Borman et al. (22). Values in parenthesesc are those reported in Borman et al. (22).
eThe values cited are the concentrations of KCl and MgCl2 allowing maximal stimulation of each type of mRNA by polyadenylation.
fHCV IRES activity is not stimulated by polyadenylation but is affected by the presence of the viral X region. Stimulation factor is given for translation with the viral X region.
gStimulation of HCV IRES activity by poly(A) is not evidenced under any salt concentrations tested. Conversely, stimulation mediated by the viral X region is insensitive to changes in salt concentrations within the ranges tested (50–140 mM KCl and 0.2–1.2 mM MgCl2).
The six viral IRESes examined exhibited significantly different KCl and MgCl2 optima for translation, in agreement with previous studies performed with non-polyadenylated mRNAs in standard RRL (Table 1) (22). However, notable differences in optima for certain IRESes were apparent between previous studies in standard RRL and the current comparison performed in ribosome-depleted RRL. While depletion of ribosomes did not significantly affect either the KCl or MgCl2 optima for the HCV element, the HRV, HAV, EMCV and FMDV IRESes exhibited significantly lower optima with respect to at least one of the salts in depleted as compared to standard RRL (Optimal salt conditions; Table 1). Conversely, the KCl optimum for PV IRES-carrying mRNAs in ribosome-depleted RRL was slightly higher than that measured with non-polyadenylated mRNAs in standard RRL (Table 1). In addition, for the five picornaviral IRESes examined, polyadenylation increased the optimal concentration of at least one of the salts tested, resulting in the fact that poly(A)-mediated stimulation of IRES activity was greatest at concentrations of KCl and MgCl2 in excess of those which are optimal for intrinsic IRES-driven translation (28) (Table 1). Since polyadenylation does not stimulate HCV IRES activity (28), it is not surprising that it had no effect on the salt optima for the HCV element. Finally, it is perhaps pertinent that the picornaviral IRESes whose activities are the most stimulated by polyadenylation (the HAV and HRV elements), are amongst those which exhibit the lowest salt optima in ribosome-depleted RRL (see Maximal stimulation by poly(A) in depleted RRL; Table 1).
The efficiency of IRES-driven translation in physiological conditions in ribosome-depleted RRL
Our previous comparison of viral IRES-driven translation in vitro employed dicistronic mRNAs which were transcribed in capped, non-polyadenylated form and translated in standard RRL (22). While this study allowed a detailed characterisation of the requirements for optimal IRES-driven translation, subsequent analyses performed with the same mRNAs in transfected mammalian cells highlighted the non-physiological nature of the standard RRL translation system (36). Our recent studies strongly suggest that RRL extracts which have been partially depleted of ribosomes and ribosome-associated translation factors represent a much more physiological alternative to standard RRL for the analysis of translation initiation. Significantly, translation of both capped and IRES-carrying mRNAs is poly(A)-dependent in depleted RRL extracts, whereas polyadenylation has little effect on the translation of such mRNAs in classical RRL systems (16,17,28).
Thus, in the present study, the efficiencies of the different IRESes to drive internal initiation of translation were compared as a feature of the poly(A) status of the monocistronic IRESp24 mRNAs in both standard and ribosome-depleted RRL. Limiting concentrations of HeLa cell S10 extract, which provide the non-canonical translation factors required by the PV and HRV IRESes, but also contain ribosomes and ribosome-associated translation factors and thus alter the absolute degree of depletion of the extracts, were included in all reactions. Since poly(A)-dependency of translation of a capped cellular mRNA is greatest in concentrations of KCl and MgCl2 approaching those encountered in a cellular environment (30), extracts received physiological concentrations of added salt (Fig. 2; results summarised in Table 2).
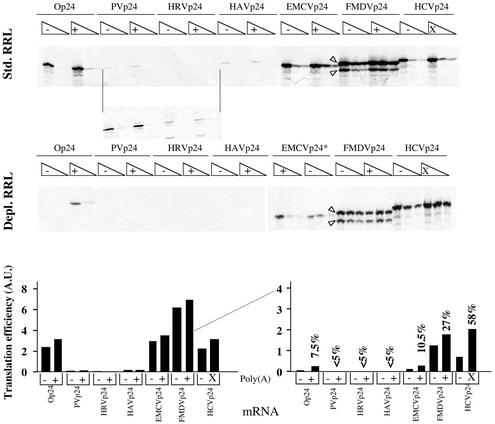
Figure 2.
Relative efficiencies of the different viral IRESes to drive translation in ribosome-depleted RRL in physiological concentrations of KCl and MgCl2. Translation reactions containing standard (upper 2 panels) or ribosome-depleted RRL (lower panel) were programmed with mRNAs derived from pOp24, or the different pIRESp24 constructs as indicated. Final RNA concentrations were 6, 3 and 1.5 µg/ml for Op24 mRNAs, and 10, 5 and 2.5 µg/ml for the IRESp24 mRNAs, as indicated by the open triangles above each series of three lanes. RNAs were transcribed in capped (for Op24) or uncapped (for IRESp24 mRNAs) form with (+ lanes) or without (– lanes) a poly(A) tail. All reactions contained 133 mM KCl and 0.93 mM MgCl2 and 2.5% vol/vol of nuclease- treated HeLa cell S10 extract. Translation reactions were analysed as described in Materials and Methods; the autoradiograph of the dried 23% polyacrylamide gel is shown. Dried gels were exposed to film for 48 h (top panel), 96 h (middle panel) or for 5 days (bottom panel). An asterisk in the EMCV lanes in ribosome-depleted RRL denotes the fact that the loading order for the polyadenylated and non-polyadenylated series of EMCVp24-based translation reactions is reversed. Translation efficiencies (in arbitrary units) of the various RNAs are shown below the three panels in either standard RRL (left-hand side) or ribosome-depleted RRL (right-hand side), and were determined densitometrically as described in Materials and Methods. Values in per cent indicate the efficiency of each IRES in driving translation of polyadenylated mRNA (or mRNA with a 3′ X region for the HCV IRES) in depleted as opposed to standard RRL. For FMDV IRES-driven translation, values for translation efficiency represent the sum of both major p24-related protein products (marked with arrow-heads), which are the equivalents of the authentic viral Lab and Lb products (see Discussion).
Table 2. Efficiency of viral IRES-driven translation in standard and ribosome-depleted RRL.
| Virus | Relative efficiency in physiological conditionsc | Relative efficiency in optimal conditionsd | ||||||
|---|---|---|---|---|---|---|---|---|
| std RRL | depl. RRL | std RRL | depl. RRL | |||||
| –Ae | +A | –A | +A | –Ae | +A | –A | +Af | |
| PVa | 1 (45) | 1.5 | 0.2 | 1.0 | 6.9 (53) | 8.8 | 0.1 | 0.9 (10.5) |
| HRV | 0.4 (12) | 0.7 | 0.1 | 0.5 | 37 (24) | 39 | 4.1 | 17 (22.5) |
| HAV | 1.9 (20) | 2.2 | <0.05 | 0.1 | 30 (26) | 29 | 1.0 | 5.5 (11) |
| EMCV | 46 (100) | 53 | 6.8 | 15 | 44 (100) | 52 | 2.4 | 4.9 (7) |
| FMDV | 89 (90) | 100 | 65 | 92 | 100 (98) | 98 | 71 | 100 (65) |
| HCV | 41 (14) | 48 | 36 | 100 | 45 (19) | 54 | 37 | 67 (66) |
| Op24b | 38 (nd) | 50 | 2.1 | 17.2 | 38 (nd) | 73 | 0.6 | 7.5 (8) |
aTranslation reactions were programmed with 5 µg/ml of IRES-p24 derived uncapped mRNAs in non-polyadenylated (–A) or polyadenylated form (+A).
bpOp24-derived mRNAs were translated at 3 µg/ml in capped form with or without a poly(A) tail.
cPhysiological concentrations of KCl amd MgCl2 for this particular batch of RRL were 130 and 0.93 mM, respectively. These values are adjusted to take into account the salt contained in the 2.5% by volume of HeLa cell extract included in each translation reaction.
dThe values for optimal KCl and MgCl2 concentrations were those cited in Table 1.
eThe values in parentheses are the calculated efficiencies of IRES-driven translation published in Borman et al. (22).
fThe values in parentheses represent the percent translation efficiency retained by the polyadenylated version of each mRNA (or that with the X region for HCVp24) in depleted as opposed to standard RRL.
nd = not done.
In agreement with previous comparisons, the EMCV and FMDV IRESes were the most efficient of the viral IRESes examined in driving translation initiation in standard RRL. However, the relative efficiency of certain of the IRESes was significantly different in the current study as compared to that reported previously. Notably, the efficiencies of the EMCV, HAV, HRV, PV and HCV elements were respectively 2-, 10-, 25- and 40-fold lower, and 3-fold higher in the current study as compared to that published previously. Since IRES-driven translation is not significantly stimulated by polyadenylation in standard RRL (28) (Table 2), the efficiencies observed in the present analysis were largely unaffected by the polyadenylation status of the test mRNAs. We presume that the inefficiency of the PV, HRV and HAV elements results at least in part from the elevated KCl concentration employed in the current study (110 mM compared to 80 mM used in 22). In addition, the inclusion of only minimal volumes of HeLa cell S10 extract in the translation reactions employed here (2.5% by volume as opposed to 20% used in 22) almost certainly contributes to the inefficiency of the HRV and PV elements (22).
In ribosome-depleted RRL, translation of the same mRNAs in physiological salt revealed even more dramatic differences in relative translation efficiencies (Fig. 2; Table 2). Effectively, the HCV and FMDV IRESes remained capable of driving translation initiation with elevated efficiencies in the depleted system. Translation of HCVp24 mRNA, which was stimulated ∼3-fold by the presence of the 3′ X region, approached 60% of the efficiency evidenced with the same mRNA in non-depleted RRL. While polyadenylation did not dramatically improve the translation of FMDVp24 mRNA, the FMDV IRES remained approximately 30% as efficient in depleted as compared to standard RRL. EMCV IRES-driven translation, while modestly stimulated by polyadenylation in physiological conditions in depleted extracts, was nonetheless reduced some 10-fold in depleted as opposed to standard RRL. While it was evident after prolonged exposure of the autoradiographs that translation driven by the PV, HRV and HAV IRESes was significantly enhanced by the presence of a 3′ poly(A) tail (Table 2), overall efficiency of these three elements to direct translation was extremely low (<5% of the corresponding translation capacities observed in standard RRL, and <1% of the activity of the HCV IRES in depleted RRL; Fig. 2 and Table 2). Importantly, translation of the capped and polyadenylated ‘cellular’ mRNA was also reduced 10-fold in depleted versus standard RRL (Op24 lanes, Fig. 2). Thus, FMDV and HCV IRES-carrying mRNAs were translated significantly more efficiently than the cellular mRNA in ribosome-depleted RRL. Finally, it is also noteworthy that the IRESes which were most efficient in directing translation in the depleted system were those which were least sensitive to the polyadenylation status of the mRNA in the particular translation conditions used for the assay (Table 2).
The elevated salt concentrations which are physiological for this batch of depleted RRL (130 mM KCl and 0.9 mM MgCl2) are far from optimal for translation driven by the HRV, HAV and PV IRESes in the same extract. Since these IRESes were the least efficient elements in physiological conditions (see above), the above comparison was repeated in both standard and depleted RRL in the salt conditions which are optimal for translation of the polyadenylated form of each IRESp24 mRNA (see Table 1). In standard RRL, the calculated relative efficiencies of IRES-driven translation were globally similar to those reported previously (Relative efficiency in optimal conditions; Table 2). However, the EMCV and PV IRESes were, respectively, 2- and 7-fold less efficient relative to FMDV in the present study as compared to that published previously. Conversely, the HCV element was 2-fold more efficient (relative to FMDV) in the present study as compared to the previous analysis. Thus, we cannot exclude that replacing the NS coding region with that of HIVp24 has altered the intrinsic efficiency of certain viral IRESes to direct translation in vitro. Next the same mRNAs were translated in ribosome-depleted RRL in the same salt conditions. While significant differences were still apparent between the various IRESes in their ability to drive translation initiation (Table 2), the HAV and especially HRV elements were now relatively efficient in driving translation initiation on polyadenylated mRNAs. Indeed, HRV IRES driven translation exceeded that observed with the standard capped, polyadenylated cellular mRNA in optimal salt concentrations. However, the PV and EMCV were once again less efficient in driving internal initiation of translation than could have been predicted from previous studies.
Evaluation of viral IRES-driven translation efficiency and poly(A) dependency in ribosome-depleted RRL as a feature of extract depletion and programming mRNA concentration
The experiments described above demonstrate that partial depletion of ribosomes/ribosome-associated translation factors from RRL perturbs translation efficiency to different degrees depending on the particular viral IRES studied. To further assess poly(A) dependency and IRES-driven translation efficiency as a function of the level of depletion of RRL, five aliquots of RRL were depleted of ribosomes by different conditions of ultracentrifugation (Fig. 3; extracts A–E in order of decreasing extract depletion). First, two different versions of the pOp24-derived mRNAs (capped or capped and polyadenylated) were translated in the various resulting post-centrifugation supernatants, to evaluate the degree of cap–poly(A) co-operativity on a standard cellular mRNA in these extracts. Significant cap–poly(A) synergy was observed in all five extracts tested (Fig. 3). As the degree of depletion diminished, the efficiency of both capped and capped, polyadenylated mRNA translation increased. However, this increase was greater for capped as compared to capped, polyadenylated mRNA translation (8-fold increase in capped mRNA translation efficiency in extract E compared to A, as opposed to a 4-fold increase for capped, polyadenylated mRNA translated in the same extracts; Fig. 3), with the result that calculated cap–poly(A) co-operativity decreased markedly as the level of extract depletion decreased (synergy of 3.3- and 8.9-fold in extracts E and B, respectively).
Figure 3.

Cap–poly(A) synergy on a cellular mRNA as a function of the degree of extract depletion. Aliquots (A–E) of ribosome-depleted RRL were prepared under different centrifugation conditions as follows using 1.1 ml volumes of RRL: centrifugation in a fixed-angle TL100 rotor at 90 000 r.p.m. for 25 min (extract A), or 90 000 r.p.m. for 15 min (extract F). Aliquots B through E were derived by mixing aliquots A and F, in the ratios 98.5:1.5, 97:3, 95.5:4.5 and 93:7, respectively. Translation reactions containing ribosome-depleted RRL extracts A–E were programmed with 6.3 µg/ml of pOp24-derived mRNAs transcribed in the form indicated above each lane, and contained 2.5% by volume of HeLa cell S10 extract. Final concentrations of added KCl and MgCl2 were 130 and 0.93 mM, respectively. Translation reactions were analysed as described in Materials and Methods; the autoradiograph of the dried 23% polyacrylamide gel is shown. Translation efficiency was determined densitometrically as described in Materials and Methods, and is plotted in arbitrary units. Closed and open squares represent capped, polyadenylated and capped, non-polyadenylated mRNAs, respectively. Stimulation by poly(A) is also plotted (open circles).
Next, mRNAs carrying the HRV, HAV, PV or EMCV IRESes were translated in uncapped and uncapped, polyadenylated form in each of the five extracts, at a range of final mRNA concentrations (Fig. 4). The concentrations of KCl and MgCl2 added to translations of each IRES-carrying mRNA were those which permit the maximal poly(A)-mediated stimulation of translation driven by each IRES. The HCV and FMDV elements were omitted from the analysis since polyadenylation has little (for FMDV) or no (for HCV) effect on translation driven by these elements (see Table 1). Similarly, although HCV IRES-driven translation is stimulated by the cognate viral 3′ X region, this stimulation is not particularly dependent on the degree of depletion of the translation extract used (data not shown).
Figure 4.
Poly(A)-dependency of viral IRES-driven translation is sensitive to the extent of extract depletion. (A) Ribosome-depleted RRL reactions derived from extracts A–E (as indicated, see legend to Fig. 3) containing 2.5 % vol/vol of nuclease-treated HeLa cell S10 extract were programmed with 10, 5 or 2.5 µg/ml of HRV-p24 mRNAs (as indicated by triangles above each series of three lanes) synthesised with or without an A50 poly(A) tail, (+ and – lanes, respectively). Final concentrations of added KCl and MgCl2 (in mM) were those which allow maximal poly(A) stimulation of IRES-driven translation (see Table 1). Analysis of the translation products was as described in the legend to Figure 2. The position of the authentic p24 product is indicated by an arrow. The doublet of products which migrate slower than p24 derive from initiation of translation at AUGs at positions 507 and 522 in the HRV IRES, as described previously (32). (B) Quantification of translation products was as described in Materials and Methods. Translation efficiency of non-polyadenylated (open squares) and polyadenylated HRVp24 mRNAs (black squares) are given in arbitrary units (left-hand side), for translation reactions programmed with 10 µg/ml of RNA. The calculated stimulation of HRV IRES-driven translation by polyadenylation in each extract is plotted for 10 µg/ml (white circles), 5 µg/ml (grey circles) and 2.5 µg/ml (black circles) of programming HRVp24 mRNAs (right-hand side). The experiment shown in (A) was repeated using PVp24 mRNAs (C), HAVp24 mRNAs (D) or EMCVp24 mRNAs (E). Only the calculated translation efficiencies (with 10 µg/ml of each programming mRNA; open and closed squares represent respectively non-polyadenylated and polyadenylated IRESp24 mRNAs) are plotted, and the poly(A)-mediated stimulation of translation driven by each IRES with the same concentration of mRNA is shown on the right-hand y axis (open circles). Note the variable scales of the right-hand y axes.
In agreement with previous studies, the degree of stimulation of picornaviral IRES activity by poly(A) varied widely, from only 2- to 3-fold for the EMCV element, to nearly 20-fold for the HAV IRES (Fig. 4; Table 1) (27,28). In addition, with the exception of EMCV IRES-driven translation, which was stimulated between 2- and 3-fold by polyadenylation in all of the extracts tested, the degree of poly(A)-mediated stimulation was dependent on the degree of extract depletion. Thus, poly(A) stimulated translation driven by the different IRESes between 2- and 7-fold (for HRV), between 3- and 9-fold (for PV) and between 5- and 15-fold (for HAV) in the various extracts (Fig. 4). Moreover, the degree of poly(A)-mediated stimulation was largely unaffected by the concentration of IRESp24 mRNA used to programme the different extracts irrespectively of the IRES studied (e.g. Fig. 4B; right-hand panel), in contrast to cap–poly(A) synergy on cellular mRNAs, which is greatest at low RNA concentrations (30).
Of more import, while translation driven by each IRES increased in efficiency as the degree of extract depletion decreased, the extract in which poly(A)-mediated translation stimulation was greatest varied according to the IRES tested (Fig. 4). This IRES-specific variation in calculated poly(A)-dependency is due to the variable response of non-polyadenylated as compared to polyadenylated mRNA translation to decreasing extract depletion. Effectively, while the efficiency of translation of the non-polyadenylated forms of the EMCV and HRVp24 mRNAs increased dramatically (for EMCV) or gradually (for HRV) in extracts A through E, non-polyadenylated HAV and especially PVp24 mRNA translation only increased significantly in efficiency in extracts D or E. We presume that these differential responses of the non-polyadenylated versus polyadenylated forms of the various IRESp24 mRNAs reflect inherent differences in affinities of the IRESes for some component of the translation machinery which is limiting in depleted RRL (see Discussion).
Effects of cleavage of eIF4G by FMDV Lb protease on the efficiency of IRES-driven translation in ribosome-depleted RRL in physiological salt concentrations
The vastly different efficiencies of the six viral IRESes to drive translation in physiological conditions in ribosome-depleted RRL were reminiscent of their capacities to direct translation in certain cell lines (36) (summarised in Table 3). While these viral IRESes (with the exception of HAV) drove translation efficiently in certain cell types (permissive cells; Table 3), in other cell lines (non-permissive cells; Table 3) only the FMDV, HCV and to a lesser extent EMCV elements exhibited detectable IRES activity. The PV and HRV IRESes could however be trans-activated in such cells by expression of PV 2A protease (Table 3). Thus, the experiment depicted in Figure 2 was repeated in extracts which had been pre-treated with the FMDV Lb protease, which cleaves eIF4G in a position similar to that recognised by the enteroviral 2A protease and which also stimulates PV and HRV IRES activity (23,24,34).
Table 3. The effects of picornaviral proteases on mRNA translation efficiency in poly(A)-dependent RRL and non-permissive mammalian cell lines.
| Translation efficiency (%) | IRES/mRNA | ||||||
|---|---|---|---|---|---|---|---|
| PV | HRV | HAV | EMCV | FMDV | HCV | Op24f | |
| In permissive cellsa | 43 (± 5) | 26 (± 14) | 5 (± 2) | 84 (± 12) | 90 (± 8) | 70 (± 13) | nd |
| In non-permissive cellsb | 4 (± 5) | 1 (± 1.4) | 0.1 (± 0.2) | 39 (± 19) | 92 (± 11) | 88 (± 9) | nd |
| In Neuro-2A cells – proteasec | 5 | 2.4 | 0.1 | 56 | 100 | 82 | nd |
| In Neuro-2A cells + proteasec | 45 (12) | 52 (16) | 0.2 (1.3) | 84 (1.6) | 100 (1.3) | 67 (0.9) | nd |
| In depleted RRL and physiological saltd | 0.6 | 0.2 | 0.03 | 28 | 91 | 91 | 33 |
| In depleted RRL and physiological salte + protease | 4.2 (7) | 0.9 (4) | 0.03 (1.0) | 71 (2.5) | 100 (1.1) | 85 (1.0) | 1.7 (0.05) |
a,bValues represent the average relative efficiencies of each IRES (± standard deviation) in HeLa, HepG2 and FRhK4 cells which are permissive for entero- and rhinoviral IRES activitya or in Neuro-2A, SKNBE, and BHK-21 cells which are not permissive for the type I IRESesb [adapted from Borman et al. (36)].
cThe values for translation efficiency in Neuro-2A cells were taken from Borman et al. (36). Values in parentheses represent the fold stimulation of IRES activity upon expression of 2A protease in these cells.
d,eValues are the averages of the results depicted in Figure 5 for the different polyadenylated IRES-p24 mRNAs translated at 10, 5 and 2.5 µg/ml final RNA concentration (expressed as percent of the most efficient mRNA).
eValues in parentheses represent the fold stimulation of translation efficiency mediated by inclusion of the Lb protease.
fValues are the averages of the results obtained with Op24-derived mRNAs translated in capped and polyadenylated form at 6, 3 and 1.5 µg/ml.
nd = not done.
In agreement with previous reports (16,22,25,26,28), cleavage of eIF4G dramatically reduced capped, polyadenylated mRNA translation, had little effect on translation driven by type II or type IV IRESes, but significantly stimulated PV IRES activity (Fig. 5; Table 3). Thus, the effects of the Lb protease on translation in ribosome-depleted RRL qualitatively resemble those of the 2A protease in certain cell lines, in which type I IRES activity is undetectable in the absence of protease. It should be noted that Lb protease-mediated cleavage of eIF4G was complete, as analysed by western blotting (data not shown). Thus, partial depletion of eIF4E from RRL does not appear to significantly affect the efficiency of eIF4G cleavage, presumably because the majority of the eIF4E which remains after ribosome depletion is in association with intact eIF4F (30). However, trans-rescue of PV IRES activity by protease in the current report was significantly less efficient than that observed in non-permissive cells. It remains to be determined whether this discrepancy reflects an undefined inherent difference between the in vitro and ex vivo translational environments, or rather results from the use of a different picornaviral protease in the two studies. In this respect, it should be noted that while the 2A protease is capable, albeit inefficiently, of cleaving PABP, the Lb protease used in the current study is bereft of such activity (26). However, given that 2A protease-mediated cleavage of PABP is much less efficient than that of eIF4G, and that the protease expression vectors used in the previous study mediated cap-dependent expression of 2A (which would be abolished as soon as protease accumulated to sufficient levels to cleave eIF4G), we feel that it is unlikely that PABP concentrations would have been significantly affected by the quantities of 2A protease expressed in this previous study. Furthermore, it is not obvious what additional advantage specifically for PV IRES activity might result from cleavage of PABP. These considerations aside, even though protease-mediated rescue of PV IRES-driven translation was incomplete (PV IRES activity was some 20-fold less than that of the most active IRES in the presence of protease as compared to 100-fold less efficient in the absence of Lb), PV IRES-driven translation was some 2–3-fold more efficient than classical mRNA translation upon eIF4G cleavage (Table 3).
Figure 5.
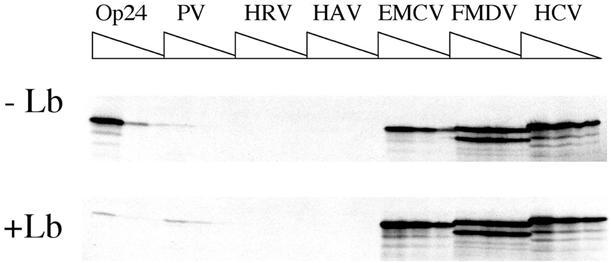
The effects of viral protease-mediated cleavage of eIF4G on the efficiency of viral IRESes to direct translation in ribosome-depleted RRL. Ribosome-depleted RRL translation reactions containing physiological concentrations of added KCl and MgCl2 (see legend to Fig. 2), were pre-treated with Lb protease in H100 buffer (10 µg/ml final concentration; +Lb panel) or H100 buffer for 15 min at 30°C (–Lb panel). Following inactivation of protease with E-64, reactions were programmed with the indicated forms of pOp24- or pIRESp24-derived mRNAs (final RNA concentrations 6.6, 3.3 and 1.65 µg/ml for Op24 mRNAs and 10, 5 and 2.5 µg/ml for IRESp24 mRNAs). Translation products were analysed as described in the legend to Figure 2.
The effects of mutations on IRES activity can be evaluated in ribosome-depleted RRL
To further validate the depleted RRL translation system, we examined whether it was suitable for evaluating the impact of IRES mutations on translation efficiency. Effectively, given the low translation efficiency of certain of the wild-type viral IRESes studied here, it was not obvious that even lower levels of translation could be adequately quantified. To this end, a series of polyadenylated mRNAs were synthesised to contain PV IRESes carrying single point mutations in domain V of the IRES (Fig. 6). These point mutations, derived from the Sabin vaccine PV strains, have previously been shown to provoke translational defects of varying severity in vitro (C.E.Malnou, A.M.Borman and K.M.Kean, manuscript submitted) (45). RNAs carrying the wild-type and three mutant PV IRESes were translated at 5 μg/ml (final RNA concentration) in ribosome-depleted RRL containing increasing volumes of S10 extract prepared from a neuroblastoma (IMR 32) cell line (Fig. 6). The RNAs employed correspond to genomic length PV RNA lacking a substantial portion of the region coding for the viral non-structural proteins. Translation of such RNAs thus generates a single protein product which is not proteolytically processed (C.E.Malnou, A.M.Borman and K.M.Kean, manuscript submitted).
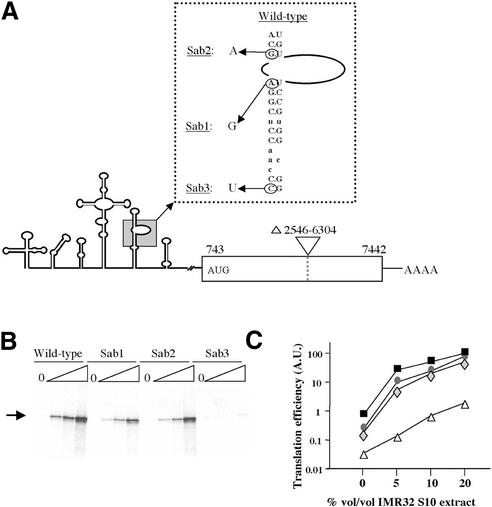
Figure 6.
Translation of mutated PV IRES-carrying mRNAs in ribosome-depleted RRL. (A) Schematic representation of the mutant PV IRES mRNAs. The predicted secondary structure of the PV 5′ UTR is shown. The PV-derived coding region is shown as an open box. Numbers below the coding region refer to the first and last nucleotide of PV sequence. The AUG codon initiating protein synthesis is shown. The natures of the domain V PV IRES mutations (Sab1 at nucleotide 480; Sab2 at nucleotide 481 and Sab3 at nucleotide 469) are indicated in the expanded, boxed inset. (B) Ribosome-depleted translation reactions containing 0, 5, 10 or 20% by volume of IMR 32 cell S10 extract (triangles) and 97 mM and 0.5 mM of KCl and MgCl2, respectively, were programmed with 5 µg/ml (final concentrations) of wild-type or Sab1-3 mutant PV IRES-carrying mRNAs. The position of the authentic PV coding region-derived translation product is indicated. Translation products were analysed as described in the legend to Figure 2. (C) The relative efficiencies of the wild-type and mutant PV IRES to drive translation initiation were calculated with respect to the wild-type mRNA translated in reactions containing 20% of S10 extract (which was arbitrarily taken as 100). The curves represent wild-type (black squares), Sab1 (grey diamonds), Sab2 (grey circles) and Sab3 (open triangles) mRNAs.
The Sab mutant PV IRESes were all less efficient than the parental PV element in driving translation initiation. The various mutations provoked a 2–3-fold (Sab2), a 4–5-fold (Sab1) and a 50–100-fold (Sab3) reduction in translation efficiency with respect to the wild-type element. However, translation from all four PV IRESes was still detectable, and notably, quantifiable after prolonged exposure of the autoradiographs (Fig. 6). Furthermore, the relative translation efficiencies of the mutants in ribosome-depleted RRL were quantitatively similar to those previously reported in standard RRL, with the exception that the translation defects were 2–3-fold greater for the Sab 1 and 2 mutants translated in depleted as opposed to standard RRL (compare Fig. 6 and data in C.E.Malnou, A.M.Borman and K.M.Kean, manuscript submitted).
DISCUSSION
The aims of this study were several-fold: (i) to re-examine the capacities of the different picornaviral IRESes to drive translation in a poly(A)-dependent in vitro system; (ii) to gain further insight into the mechanism of IRES-driven translation, notably with respect to those IRESes which drive translation efficiently in vitro, but poorly in certain cell types (the PV, HRV and especially HAV elements); (iii) to attempt to correlate global IRES-driven translation efficiency with poly(A)-dependency; and (iv) to evaluate the ribosome-depleted RRL system as a more physiological in vitro alternative to standard RRL for evaluating translation efficiency.
The current comparison revealed dramatic inherent differences in the capacities of the various viral IRESes to direct translation (see Table 2; Fig. 2). In ribosome-depleted RRL in physiological salt concentrations, translation was initiated some 2–4 logs more efficiently from the FMDV and HCV IRESes than from the PV, HRV and HAV elements. While EMCV IRES-driven translation was easily detectable, it was still some 6-fold less efficient than that driven by the HCV and FMDV elements. Thus, the viral IRESes tested can be classified in the order FMDV=HCV>>>EMCV>>>PV=HRV>>HAV in terms of decreasing translational capacity. Previous in vitro comparisons of the same IRESes failed to detect such major differences between the FMDV and HCV elements on the one hand and the EMCV and other IRESes on the other. Evidently, the dramatically reduced efficiencies of the latter elements in this study stems for a large part from the ribosome-depleted translation extracts used, in which only the most efficient mRNAs are capable of recruiting the machinery necessary for their translation. The stringent salt concentrations required to render RRL ‘physiological’ also contribute to the much diminished translation capacities at least of the PV, HRV and HAV IRESes (see Tables 1 and 2). It should also be noted that the contrast in efficiencies between the PV or HRV IRESes and the FMDV element is likely to be slightly exaggerated by the current study. First, so as to avoid altering the degree of extract depletion, the concentrations of HeLa cell S10 extract employed here were very low, and are sub-optimal for maximal entero- and rhinovirus IRES-driven translation. Second, while the FMDV construct used in the current study allowed the detection of the two major initiation products of FMDV IRES activity [the equivalents of the viral La and Lb products (46)], previous studies used a construct which did not possess a second AUG codon capable of recruiting those ribosomes which had failed to initiate translation at the upstream site (22). However, we do not believe that these relatively minor differences in experimental design alone could account for the 2–4 log greater activity of the FMDV IRES as compared to its PV, HRV and HAV counterparts, as observed here.
The above classification of IRES activity in ribosome-depleted RRL is very similar to that observed when the same IRESes were evaluated in certain cell lines (Table 3). Effectively, while all of the IRESes tested here were of similar efficiency in driving translation in some cell lines, in other cell lines the FMDV and HCV elements were reproducibly better than the other IRESes in driving translation. While the nature of the block to translation driven by certain IRESes ex vivo remains unknown, the PV, HRV and to a lesser extent EMCV IRES activity could be rescued in cells non-permissive for activity of these IRESes by expression of the PV 2A protease (36) (Table 3; see below). Here, we have shown that type I IRES activity could also be partially rescued by the Lb protease in depleted RRL. This is apparently at odds with our recent data concerning the effects of eIF4G cleavage on poly(A)-dependent translation driven by the PV IRES in the same system, in which it was demonstrated that protease- and poly(A)-mediated translation stimulation were mutually exclusive (28). In effect, the positive effects of eIF4G cleavage on IRES activity were negated by abolition of the positive effects of polyadenylation on IRES-driven translation (which require the interaction between intact eIF4G and PABP). However, this previous study was performed in salt conditions which are optimal for PV IRES-driven translation, rather than those which are physiological. Thus, it appears that the relative contributions of eIF4G cleavage and polyadenylation to PV IRES activity vary according to the translation conditions used.
Another important feature of the experiments described here concerns the relationship between overall translation efficiency, the different optima for efficient translation and the degree of poly(A) dependency. Effectively, those IRESes which drove translation most efficiently in depleted RRL in physiological conditions were those which had the highest KCl and/or MgCl2 optima, and were the least sensitive to polyadenylation (Tables 1 and 2). Thus, translation driven by the HAV IRES can routinely be stimulated some 20-fold by polyadenylation, but remains some 4-logs less efficient than FMDV IRES-driven translation, which is stimulated <2-fold by polyadenylation. Taken together, these data imply that the dramatic differences in basal translation efficiency of the different picornaviral IRESes are due to intrinsically different affinities of these elements for components of the canonical translation machinery, and that these affinities are very sensitive to the polyadenylation status of the mRNAs. This hypothesis is further supported by the results presented in Figure 4. Significant quantitative differences exist between the way that translation of the polyadenylated as compared to the non-polyadenylated versions of the HAV and PV IRES-driven mRNAs respond to increasing extract depletion. Conversely, for the more efficient EMCV IRES [which the hypothesis predicts would have a greater affinity for the translation machinery irrespective of the poly(A) status of the mRNA], the polyadenylated and non-polyadenylated EMCVp24 mRNAs are similarly affected by increased degrees of depletion. Further studies will be required to identify those specific IRES-protein interactions which are sensitive to poly(A). However, it is interesting to speculate that the PABP-eIF4G interaction, which is required for poly(A)-mediated stimulation of both cellular and IRES-carrying mRNA translation, is involved. Indeed, although we have no data pertaining to the differences in depletion levels of certain factors between the five depleted extracts used in this study, we have previously shown that eIF4G and eIF4E concentrations are significantly lower in depleted as compared to standard RRL extracts (30).
Finally, we have shown that the depleted RRL system is appropriate for studying the effects of mutations on IRES-driven translation efficiency. Mutations introduced into the PV IRES at the same position as the naturally occurring attenuating mutations identified in the Sabin vaccine strains had profound effects on IRES-driven translation efficiency in the depleted RRL system. Moreover, translation was still detectable with the most affected (Sab3) mutant, even though this mutant IRES directed translation some 50-fold less efficiently than the wild-type PV IRES (which itself is one of the least efficient of the picornaviral IRESes in the depleted RRL system; see Table 2). Thus, based on all of the data presented here, we propose that the ribosome-depleted RRL system employed in this study represents a more physiological alternative to standard RRL-based translation systems, and that the system is appropriate for analysing the impact of mutations on IRES-driven translation.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Regina Kirchweger and Tim Skern for the gift of purified FMDV Lb protease. We are also grateful to Mélanie Quesnoit and Richard Paul for their interest in this work. This work was funded in part by a grant from the MENRT (Réseau National hépatite C) to K.M.K. Y.M.M. and C.E.M. are supported by doctoral fellowships from the CANAM and the CNRS, respectively.
REFERENCES
- 1.Banerjee A.K. (1980) 5′ terminal cap structure in eukaryotic messenger ribonucleic acids. Microbiol. Rev., 44, 175–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson R.J. (1993) Cytoplasmic regulation of mRNA function: the importance of the 3′ untranslated region. Cell, 74, 9–14. [DOI] [PubMed] [Google Scholar]
- 3.Rhoads R.E. (1988) Cap recognition and the entry of mRNA into the protein synthesis initiation cycle. Trends Biochem. Sci., 13, 52–56. [DOI] [PubMed] [Google Scholar]
- 4.Kozak M. (1999) Initiation of translation in prokaryotes and eukaryotes. Gene, 234, 187–208. [DOI] [PubMed] [Google Scholar]
- 5.Morley S.J., Curtis,P.S. and Pain,V.M. (1997) Translation’s mystery factor begins to yield its secrets. RNA, 3, 1085–1104. [PMC free article] [PubMed] [Google Scholar]
- 6.Sachs A.B., Sarnow,P. and Hentze,M.W. (1997) Starting at the beginning, middle and end: translation initiation in eukaryotes. Cell, 89, 831–838. [DOI] [PubMed] [Google Scholar]
- 7.Gingras A.-C., Raught,B. and Sonenberg,N. (1999) eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem., 68, 913–963. [DOI] [PubMed] [Google Scholar]
- 8.Hershey J.W.B., and Merrick,W.C. (2000) Pathway and mechanism of initiation of protein synthesis. In Hershey,J.W.B., Mathews,M.B. and Sonenberg,N. (eds), Translational Control Of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 33–88.
- 9.Doel M.T. and Carey,N.H. (1976) The translational capacity of deadenylated ovalbumin messenger RNA. Cell, 8, 51–58. [DOI] [PubMed] [Google Scholar]
- 10.Gallie D.R. (1991) The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev., 5, 2108–2116. [DOI] [PubMed] [Google Scholar]
- 11.Gallie D.R. (1998) A tale of two termini: a functional interaction between the termini of an mRNA is a prerequisite for efficient translation initiation. Gene, 216, 1–11. [DOI] [PubMed] [Google Scholar]
- 12.Preiss T. and Hentze,M.W. (1998) Dual function of the messenger RNA cap structure in poly(A)-tail-promoted translation in yeast. Nature, 392, 516–520. [DOI] [PubMed] [Google Scholar]
- 13.Tarun S.Z. Jr and Sachs,A.B. (1995) A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev., 9, 2997–3007. [DOI] [PubMed] [Google Scholar]
- 14.Iizuka N., Najita,L., Franzusoff,A. and Sarnow,P. (1994) Cap-dependent and cap-independent translation by internal initiation of mRNAs in cell extracts prepared from Saccharomyces cerevisiae. Mol. Cell. Biol., 14, 7322–7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gebauer F., Corona,D.F.V., Preiss,T., Becker,P.B. and Hentze,M.W. (1999) Translational control of dosage compensation in Drosophila by Sex-lethal: cooperative silencing via the 5′ and 3′ UTRs of msl-2 mRNA is independent of the poly(A) tail. EMBO J., 18, 6146–6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michel Y.M., Poncet,D., Piron,M., Kean,K.M. and Borman,A.M. (2000) Cap-poly(A) synergy in mammalian cell-free extracts: investigation of the requirements for poly(A)-mediated stimulation of translation initiation. J. Biol. Chem., 275, 32268–32276. [DOI] [PubMed] [Google Scholar]
- 17.Munroe D. and Jacobson,A. (1990) mRNA poly(A) tail, a 3′ enhancer of translational initiation. Mol. Cell. Biol., 10, 3441–3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson R.J. and Kaminski,A. (1995) Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA, 1, 985–1000. [PMC free article] [PubMed] [Google Scholar]
- 19.Tsukiyama-Kohara K., Iizuka,N., Kohara,M. and Nomoto,A. (1992) Internal ribosome entry site within hepatitis C virus RNA. J. Virol., 66, 1476–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito T., Tahara,S.M. and. Lai,M.C (1998) The 3′ untranslated region of hepatitis C virus enhances translation from an internal ribosomal entry site. J. Virol., 72, 8789–8796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pestova T.V., Shatsky,I.N., Fletcher,S.P., Jackson,R.J. and Hellen,C.U.T. (1998) A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev., 12, 67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borman A.M., Bailly,J.-L., Girard,M. and Kean,K.M. (1995) Picornavirus internal ribosome entry segments: comparison of translation efficiency and the requirements for optimal internal initiation in vitro. Nucleic Acids Res., 23, 3656–3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borman A.M., Kirchweger,R., Ziegler,E., Rhoads,R.E., Skern,T. and Kean,K.M. (1997) eIF4G and its proteolytic cleavage products: effect on initiation of protein synthesis from capped, uncapped and IRES-containing mRNAs. RNA, 3, 186–196. [PMC free article] [PubMed] [Google Scholar]
- 24.Borman A.M., and Kean,K.M. (1997) Intact eukaryotic initiation factor 4G is required for hepatitis A virus internal initiation of translation. Virology, 237, 129–136. [DOI] [PubMed] [Google Scholar]
- 25.Ali I.K., McKendrick,L., Morley,S.J. and Jackson,R.J. (2001) Activity of the hepatitis A virus IRES requires association between the cap-binding translation initiation factor (eIF4E) and eIF4G. J. Virol., 75, 7854–7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borman A.M., Michel,Y.M. and Kean,K.M. (2001) Detailed analysis of the requirements of hepatitis A virus internal ribosome entry segment for the eukaryotic initiation factor complex eIF4F. J. Virol., 75, 7864–7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bergamini G., Preiss,T. and Hentze,M.W. (2000) Picornavirus IRESes and the poly(A) tail jointly promote cap-independent translation in a mammalian cell-free system. RNA, 6, 1773–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michel Y.M., Borman,A.M., Paulous,S. and Kean,K.M. (2001) Eukaryotic initiation factor 4G-poly(A) binding protein interaction is required for poly(A) tail-mediated stimulation of picornavirus internal ribosome entry segment-driven translation but not for X-mediated stimulation of hepatitis C virus translation. Mol. Cell. Biol., 21, 4097–4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Svitkin Y.V., Imataka,H., Khaleghpour,K., Kahvejian,A., Liebig,H.-D. and Sonenberg,N. (2001) Poly(A)-binding protein interaction with elF4G stimulates picornavirus IRES-dependent translation. RNA, 7, 1743–1752. [PMC free article] [PubMed] [Google Scholar]
- 30.Borman A.M., Michel,Y.M. and Kean,K.M. (2000) Biochemical characterisation of cap-poly(A) synergy in rabbit reticulocyte lysates: the eIF4G-PABP interaction increases the functional affinity of eIF4E for the capped mRNA 5′ end. Nucleic Acids Res., 28, 4068–4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borman A. and Jackson,R.J. (1992) Initiation of translation of human rhinovirus RNA: mapping the internal ribosome entry site. Virology, 188, 685–696. [DOI] [PubMed] [Google Scholar]
- 32.Luz N. and Beck,E. (1990) A cellular 57 kDa protein binds to two regions of the internal translation initiation site of foot-and-mouth disease virus. FEBS Lett., 269, 311–314. [DOI] [PubMed] [Google Scholar]
- 33.Jackson R.J. and Hunt,T. (1983) Preparation and use of nuclease-treated rabbit reticulocyte lysates for the translation of eukaryotic messenger RNA. Methods Enzymol., 96, 50–74. [DOI] [PubMed] [Google Scholar]
- 34.Kirchweger R., Ziegler,E, Lamphear,B.J., Waters,D., Liebig,H.D., Sommergruber,W., Sobrino,F., Hohenadl,C., Blaas,D., Rhoads,R.E. and Skern,T. (1994) Foot-and-mouth disease virus leader proteinase: purification of the Lb form and determination of its cleavage site on eIF4G. J. Virol., 68, 5677–5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dasso M.C. and Jackson,R.J. (1989) Efficient initiation of mammalian mRNA translation at a CUG codon. Nucleic Acids Res., 17, 6485–6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borman A.M., Le Mercier,P., Girard,M. and Kean,K.M. (1997) Comparison of picornaviral IRES-driven internal initiation of translation in cultured cells of different origins. Nucleic Acids Res., 25, 925–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaminski A., Howell,M.T. and Jackson,R.J. (1990) Initiation of encephalomyocarditis virus RNA translation: the authentic initiation site is not selected by a scanning mechanism. EMBO J., 9, 3753–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jang S.K. and Wimmer,E. (1990) Cap-independent translation of encephalomyocarditis virus RNA: structural elements of the internal ribosomal entry site and involvement of a cellular 57-kD RNA-binding protein. Genes Dev., 4, 1560–1572. [DOI] [PubMed] [Google Scholar]
- 39.Tesar M., Harmon,S.A., Summers,D.F. and Ehrenfeld,E. (1992) Hepatitis A virus polyprotein synthesis initiates from two alternative AUG codons. Virology, 186, 609–618. [DOI] [PubMed] [Google Scholar]
- 40.Glass M.J., Jia,X.-J. and Summers,D.F. (1993) Identification of the hepatitis A virus internal ribosome entry site: in vivo and in vitro analysis of bicistronic RNAs containing the HAV 5′ noncoding region. Virology, 193, 842–852. [DOI] [PubMed] [Google Scholar]
- 41.Reynolds J.E., Kaminski,A., Kettinen,H.J., Grace,K. Clarke,B.E., Carroll,A.R., Rowlands,D.J. and Jackson,R.J. (1995) Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J., 14, 6010–6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kettinen H., Grace,K., Grunert,S., Clarke,B., Rowlands,D. and Jackson,R.J. (1994) Unique features of hepatitis C virus translation. In Nishioka,K., Suzuki,H., Mishiro,S and Oda,T. (eds), Viral Hepatitis and Liver Disease. Springer Verlag, Tokyo, Japan, pp. 125–131.
- 43.Pelham H. and Jackson,R.J. (1976) An efficient mRNA-dependent translation system from reticulocyte lysates. Eur. J. Biochem., 67, 247–256. [DOI] [PubMed] [Google Scholar]
- 44.Jackson R.J. (1991) Potassium salts influence the fidelity of mRNA translation initiation in rabbit reticulocyte lysates: unique features of encephalomyocarditis virus RNA translation. Biochim. Biophys. Acta, 1088, 345–358. [DOI] [PubMed] [Google Scholar]
- 45.Svitkin Y.V., Pestova,T.V., Maslova,S.V. and Agol,V.I. (1988) Point mutations modify the response of poliovirus RNA to a translation initiation factor: a comparison of neurovirulent and attenuated strains. Virology, 166, 394–404. [DOI] [PubMed] [Google Scholar]
- 46.Medina M., Domingo,E., Brangwyn,J.K. and Belsham,G.J. (1993) The two species of the foot-and-mouth disease virus leader protein, expressed individually, exhibit the same activities. Virology, 194, 355–359. [DOI] [PubMed] [Google Scholar]