Abstract
The U3271C mutation affecting the human mitochondrial transfer RNALeu(UUR) (hs mt tRNA) is correlated with diabetes and mitochondrial encephalopathies. We have explored the relationship between the structural effects of this mutation and its impact on function using chemical probing experiments and in vitro aminoacylation assays to investigate a series of tRNA constructs. Chemical probing experiments indicate that the U3271C substitution, which replaces an AU pair with a CA mispair, significantly destabilizes the anticodon stem. The introduction of a compensatory A3261G mutation reintroduces base pairing at this site and restores the structure of this domain. In fact, the anticodon stem of the A3261G/U3271C mutant appears more structured than wild-type (WT) hs mt tRNALeu(UUR), indicating that the entirely AU stem of the native tRNA is intrinsically weak. The results of the chemical probing experiments are mirrored in the aminoacylation activities of the mutants. The U3271C substitution decreases aminoacylation reactivity relative to the WT tRNA due to an increase in Km for the pathogenic mutant. The binding defect is a direct result of the structural disruption caused by the pathogenic mutation, as the introduction of the stabilizing compensatory mutation restores aminoacylation activity. Other examples of functional defects associated with the disruption of weak domains in hs mt tRNAs have been reported, indicating that the effects of pathogenic mutations may be amplified by the fragile structures that are characteristic of this class of tRNAs.
INTRODUCTION
The human mitochondrial (hs mt) genome encodes 13 proteins, two ribosomal RNAs and 22 transfer RNAs (tRNAs) (1,2). The hs mt tRNAs have sequences and structures that differ significantly from canonical bacterial and cytoplasmic tRNAs (3). Moreover, the tRNAs functioning in human mitochondria are less thermodynamically stable, as they generally contain higher numbers of mismatched and AU base pairs (4).
The structural instability of the hs mt tRNAs may contribute to their involvement in mitochondrial diseases. Almost 50% of the 145 pathology-associated mutations currently identified within the hs mt genome affect tRNAs (http://www.mitomap. org), despite the fact that the regions coding for these molecules comprise only ∼10% of the genome. Indeed, molecular-level studies of hs mt tRNALeu(UUR) and tRNAIle have revealed that pathogenic mutations produce severe structural perturbations causing functional defects (5–7). In particular, hs mt tRNAIle mutants containing mismatch-producing substitutions in regions of secondary structure displayed markedly reduced aminoacylation (6,7). The fact that these functional defects were structural in origin was confirmed with compensatory mutations, which completely restored activity by reintroducing Watson–Crick base pairs (6).
To date, 18 pathogenic mutations have been identified within the gene encoding hs mt tRNALeu(UUR) (http://www.mitomap.org). Based on the canonical tRNA folding pattern, three of these mutations are predicted to alter the tertiary structure of hs mt tRNALeu(UUR), eight mutations would affect its secondary structure, and seven mutations do not have an obvious structural impact on hs mt tRNALeu(UUR). One mutation predicted to disrupt tertiary structure, the A3243G substitution associated with maternally inherited diabetes and deafness (MIDD) and mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes (MELAS), was recently shown to attenuate aminoacylation (5). The loss of function was linked to the formation of an inactive dimeric complex in addition to the disruption of a tertiary contact (5).
Studies of pathogenic hs mt tRNA mutants correlated with disease have provided insight into potential functional defects that could impede cellular function (5–15). Many different aspects of tRNA function can be impaired by point mutations, including aminoacylation, processing, post-transcriptional modification and participation in ribosomal protein synthesis. Understanding how disruptions in these activities are related to the structural effects of pathogenic mutations presents a means to understand the molecular-level defects that may contribute to disease.
To evaluate the relationship of structural defects caused by disease-related tRNA mutations to function, we have investigated the MELAS- and MIDD-related U3271C (U40C) substitution in hs mt tRNALeu(UUR) that introduces a CA mispair into the anticodon stem (Fig. 1) (16,17). This mutation has been the subject of a variety of studies that have elucidated losses in processing (8), modification levels (9) and cellular levels of tRNALeu(UUR) (9). However, none of these studies investigated whether a conformational disruption induced by the U3271C mutation was responsible for the loss in function. Given the high AU content of the anticodon stem of hs mt tRNALeu(UUR), the U3271C mutation may induce a significant structural perturbation and interfere with the various aspects of tRNA function. Indeed, using aminoacylation assays and structural probing experiments, we have uncovered that the U3271C mutation causes a severe structural defect within the anticodon stem. The anticodon stem of this mutant appears to be completely unstructured. A moderate aminoacylation defect is related to this structural disruption, and other aspects of tRNA function more highly dependent on the anticodon stem may be even more significantly affected.
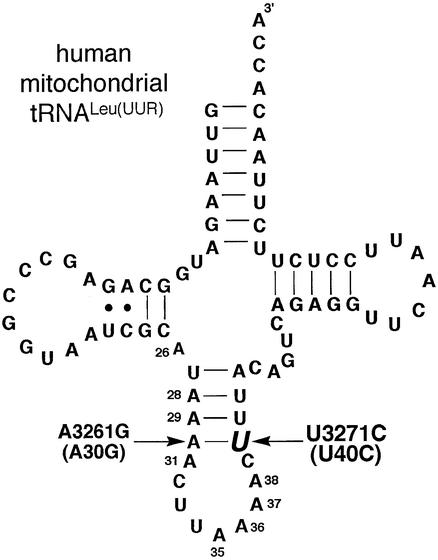
Figure 1.
Predicted cloverleaf secondary structure of hs mt tRNALeu(UUR). The pathogenic U3271C (U40C) mutation is highlighted. The location of the A3261G (A30G) compensatory mutation is also indicated. The mutations will be referred to by their four-digit genomic numbers except when the structural references are made; in these cases, the two-digit number indicating their location within the tRNA will be used.
MATERIALS AND METHODS
Cloning and preparation of tRNA constructs
DNA templates for in vitro transcription were created by ligating overlapping oligonucleotides into pUC18 digested with BamHI and PstI. The hs mt tRNALeu(UUR) genes were flanked by a T7 polymerase promoter sequence and a BstNI cleavage site. Plasmids were harvested in milligram quantities from Escherichia coli DH5α and cleaved with BstNI to generate the 3′-CCA end. In vitro transcription reactions contained template DNA (100–200 µg in 1–2 ml), T7 RNA polymerase (overexpressed in E.coli), 40 mM Tris–HCl (pH 8), 10 mM NaCl, 2 mM spermidine, 20 mM MgCl2, 4 mM NTPs and 5 mM dithiothreitol. Transcription reactions were incubated for 3 h at 37°C. Plasmid DNA was digested by the addition of DNase I (Takara). Reactions were extracted with 5:1 phenol (pH 4.7)/chloroform, ethanol precipitated, and purified by 12% denaturing polyacrylamide gel electrophoresis (PAGE) using a 0.5× TBE buffer (45 mM Tris base/45 mM boric acid/1 mM EDTA) on a 16.5 cm (w) × 26 cm (h) × 3 mm (d) gel for 12 h. tRNA samples were recovered by electroelution, ethanol precipitated and resuspended in 0.5× TE [5 mM Tris–HCl (pH 8), 0.5 mM EDTA].
Concentrations of tRNA solutions were determined by quantitating the absorbance at 260 nm and applying an extinction coefficient of 895 000 M–1 (mononucleotide) cm–1 (http://www.scripps.edu/mb/gottesfeld/ExtCoeff.html). Charging plateaus performed with an excess of hs mt leucyl-tRNA synthetase (LeuRS) confirmed the correspondence of concentrations determined spectrophotometrically with levels of chargable tRNA (5), but absorbance-based measurements were used to determine tRNA concentrations, as these values yielded more reproducible and accurate results. tRNA samples were annealed by incubation at 70°C for 5 min in 0.5× TE, addition of MgCl2 (10 mM), and immediate cooling on ice.
Preparation of hs mt LeuRS
Hs mt LeuRS was cloned and purified as described (5,18). SDS–PAGE was used to confirm the purity of the protein, and a Bradford assay was used to determine protein concentration. The activity of the enzyme was confirmed using a pyrophosphate exchange assay.
Preparation of 5′-32P-labeled hs mt tRNALeu(UUR)
Prior to labeling, tRNA samples were dephosphorylated using calf intestinal alkaline phosphatase (Takara). Dephos phorylated tRNA (100 pmol) was 5′ radiolabeled in a reaction containing 100 pmols of [γ-32P]ATP (ICN Biomedicals), 100 U of T4 polynucleotide kinase (New England Biolabs), 70 mM Tris–HCl (pH 7.6), 10 mM MgCl2 and 5 mM dithiothreitol. The samples were incubated at 37°C for 30 min and purified using G-25 columns (Amersham Pharmacia). Further purification of labeled samples was performed using a 12% denaturing PAGE with a 0.5× TBE buffer. Labeled tRNAs were isolated, electroeluted and ethanol precipitated. Samples were resuspended in 0.5× TE unless otherwise noted.
Aminoacylation assays
Prior to the aminoacylation assay, tRNA samples were annealed as described above. Assays were performed at 37°C in reaction mixtures containing 50 mM HEPES (pH 7.6), 100 µM spermine, 25 mM KCl, 0.2 mg/ml bovine serum albumin, 2.5 mM ATP, 100 µM leucine, 4.6 µM [3,4,5-3H] leucine, 7 mM MgCl2, 40 or 50 nM enzyme, and 400 nM to 20 µM tRNA. Assays were otherwise executed and analyzed as described (19). Km and kcat values were extracted from a non-linear fit of initial rate values (measured with at least six different tRNA concentrations) to the following adaptation of the Michaelis–Menten equation: νo = Vmax[S] / Km + [S].
Chemical probing
Experiments conducted under non-denaturing conditions [approximating the native environment of hs mt tRNALeu(UUR)] (20) were performed with tRNAs prepared in 50 mM sodium cacodylate (pH 7), 25 mM NaCl and 10 mM MgCl2. Reaction solutions were heated at 70°C for 5 min and cooled on ice for 20 min. Neat diethylpyrocarbonate (DEPC) (4 µl) was added and the samples were incubated for 1 h at 37°C. Reactions were halted by precipitation with 0.3 M sodium acetate (pH 5) and 2.5 vol of ethanol followed by incubation at –80°C. For comparison (data not shown), chemical probing was also carried out under denaturing conditions, which used buffer conditions that differed in the omission of 25 mM NaCl and 10 mM MgCl2 and inclusion of 1 mM EDTA.
Strand scission of carbethoxylated hs mt tRNAs
To achieve strand scission at modified positions each sample was resuspended in 20 µl of 1 M aniline (pH 4.5 in concentrated glacial acetic acid) (21). Samples were incubated in the dark for 20 min at 60°C. To halt the reaction, samples were flash frozen in liquid nitrogen and dried. The samples were resuspended in 20 µl of H2O, flash frozen and dried. After final drying, each sample was resuspended in 10 µl of denaturing PAGE loading buffer (containing 8 M urea).
Sequencing reactions
RNase T1 digestion was performed to create a G ladder. Wild-type (WT) hs mt tRNALeu(UUR) (4 µM) in 50 mM Tris base and 2 mM EDTA was incubated at 70°C for 5 min and cooled on ice (22). The reaction solution (5 µl) was incubated at 37°C for 20 min with 0.02 U of RNase T1 (Fermentas). WT hs mt tRNALeu(UUR) in 10 mM NaHCO3 and 2 mM EDTA (5 µl) was heated for 8 min at 90°C to form an alkaline ladder (22). Both reactions were stopped through the addition of 5 µl of denaturing PAGE loading dye and cooling on ice.
Gel electrophoresis of DEPC-treated tRNAs
Samples were loaded onto a 21 cm (w) × 40 cm (h) × 0.4 mm (d) 15% polyacrylamide gel (19:1) containing 8 M urea buffered with 0.5× TBE. Gels were typically electrophoresed for 1.25 h at 50 mA and then exposed to a phosphorimager plate overnight.
RESULTS AND DISCUSSION
To investigate how the U3271C mutation affected the structure of hs mt tRNALeu(UUR), we monitored the reactivity of anticodon stem nucleotides with the chemical probe DEPC under non-denaturing conditions. Three tRNAs were used in these studies: the WT hs mt tRNALeu(UUR), a tRNA construct containing the U3271C mutation, and a tRNA construct containing the U3271C mutation and a compensatory A3261G mutation reintroducing a Watson–Crick pair at the site of the pathogenic substitution (Fig. 1). DEPC was used for the chemical probing because this reagent reacts preferentially with adenines and the region of interest was the AU-rich anticodon stem (20,21).
PAGE analysis of WT, U3271C and A3261G/U3271C hs mt tRNALeu(UUR) treated with DEPC illustrated striking structural differences among these constructs (Fig. 2A). The cleavage ratios at the various adenine positions were quantitated and normalized, yielding protection values for each adenine position within the folded structures of the mutants in comparison with WT hs mt tRNALeu(UUR) (Fig. 2B). The protection values clearly establish that the structural integrity of the anticodon stem is lost in the presence of the U3271C substitution, as A28, A29, A30 and A31 are heavily cleaved by DEPC. The WT hs mt tRNALeu(UUR) undergoes an intermediate level of cleavage at these positions, and the A3261G/U3271C mutant exhibits strong protection from DEPC. It appears that the stabilizing mutation significantly strengthens the anticodon stem by introducing a GC pair into this domain that is otherwise composed of AU pairs in WT hs mt tRNALeu(UUR). It is noteworthy that the native tRNALeu(UUR) contains a pseudouridine residue at position 27 which may impart a slight increase in the stability of this stem compared with the unmodified constructs studied here.
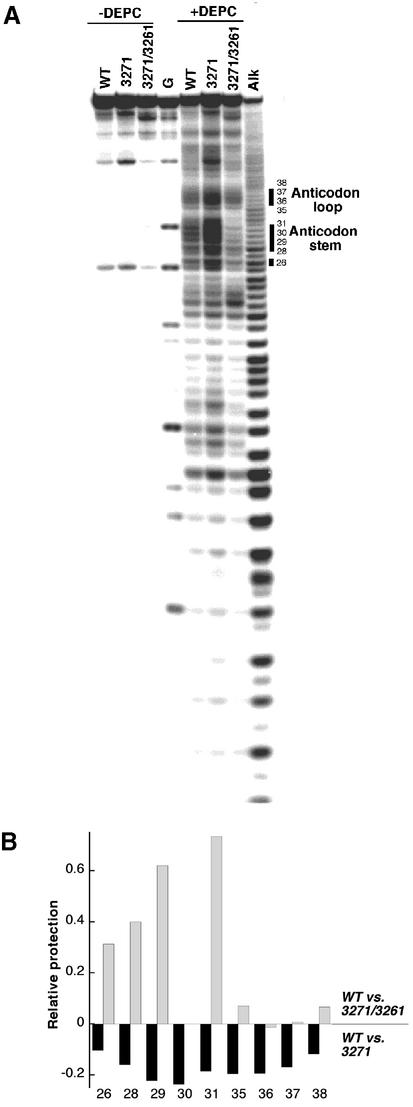
Figure 2.
(A) PAGE autoradiogram for DEPC chemical probing experiments with WT, U3271C and A3261G/U3271C hs mt tRNALeu(UUR). Lanes 1–3 (labeled WT, U3271C and A3261G/U3271C) are control lanes for the 5′-radiolabeled hs mt tRNALeu(UUR) samples. Lane G is a T1 digest of denatured WT hs mt tRNALeu(UUR). Lanes 5–7 are chemical probing reactions performed under native conditions. Samples were analyzed by 15% PAGE. The numbering of the bands corresponds to the positions of adenosine within the hs mt tRNALeu(UUR) sequence. Increased nuclease susceptibility was observed reproducibly for the U3271C mutant, as evidenced by the larger intensities of the bands in the control lane for this sample. (B) Analysis of chemical probing. The relative percent cleavage at the various adenine positions was quantitated (Image Quant) by first normalizing the values against the uncleaved tRNA within each lane to adjust the values for loading variations. The cleavage intensities at each adenine position for the mutant tRNAs were compared with those of the WT sample. Position 30 within the A3261G/U3271C mutant sequence is not an adenine. Data were obtained from greater than three independent trials; protection values from different trials varied ±0.02.
The same protection trends are observed for A26 (Fig. 2A and B). This nucleotide is typically part of the anticodon domain (23), but is more distant from the site of the pathogenic mutation than the nucleotides discussed above. Interestingly, this position is less protected for the U3271C mutant than WT hs mt tRNALeu(UUR), but appears less exposed (as judged from protection values) than the anticodon stem nucleotides. This indicates that the structural disruption may be less severe in the upper portion of this domain.
Different levels of cleavage at anticodon loop nucleotides are also observed for the different tRNA constructs. A35, A36, A37 and A38 exhibit similar reactivities for the WT and A3261G/U3271C tRNAs, but appear significantly more accessible in the U3271C mutant (Fig. 2A). Nucleotides within the anticodon loop, despite not being involved directly in interactions stabilizing secondary or tertiary structure, commonly exhibit less reactivity with chemical probes in folded tRNAs where stacking may occur (20). The pronounced accessibility of these residues in the pathogenic mutant relative to the other constructs studied [and the fact that the intensities of cleavage at these positions are comparable under non-denaturing and denaturing conditions (data not shown)] offers another piece of evidence that the anticodon domain is completely unstructured.
The effects of the U3271C mutation on more distal domains of the structure of hs mt tRNALeu(UUR) can also be evaluated from the DEPC cleavage data. A number of D-stem adenines (e.g. A12 and A14) are cleaved with higher frequency when the U3271C mutation is present, indicating that the conformational perturbation introduced by the mutation in the anticodon stem does affect the adjacent domain. However, the adenines of the acceptor stem display only slight increases in cleavage intensities for the U3271C construct, suggesting that the mutant tRNA is not completely denatured.
To explore how the structural effects of the U3271C mutation impacted an aspect of the function of hs mt tRNALeu(UUR), we monitored aminoacylation, an essential step in protein synthesis. Recombinant hs mt LeuRS expressed in E.coli was used to test the reactivity of the transcripts of both WT hs mt tRNALeu(UUR) and the U3271C mutant [previous studies indicated that hs mt tRNALeu(UUR) transcripts are suitable substrates for this enzyme (5)]. This experiment revealed that the rate of aminoacylation for the U3271C mutant was reduced (Fig. 3) relative to WT hs mt tRNALeu(UUR).
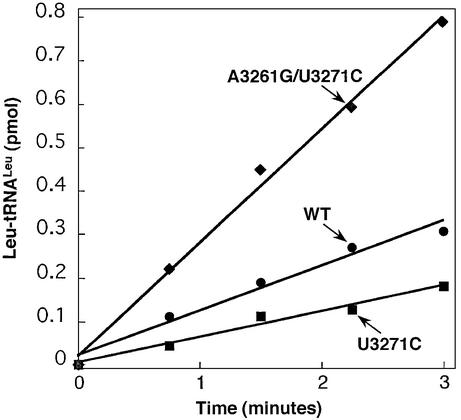
Figure 3.
Aminoacylation of WT (circles), U3271C (squares) and A3261G/U3271C (diamonds) hs mt tRNALeu(UUR) by hs mt LeuRS. Assays were performed at pH 7.6 at 37°C with 5 µM tRNA and 40 nM hs mt LeuRS.
The presence of a binding defect for U3271C hs mt tRNALeu(UUR) was elucidated through the measurement of kinetic parameters for the aminoacylation reaction. The U3271C substitution increased the apparent binding affinity (Km), but did not affect the catalytic rate constant (kcat) for aminoacylation (Table 1). The apparent Km of the U3271C hs mt tRNALeu(UUR) mutant was almost 5-fold higher (9.7 µM) than WT (2.3 µM). The kcat values for WT hs mt tRNALeu(UUR) and the U3271C mutant were comparable (0.020 and 0.024 s–1, respectively).
Table 1. Kinetic parameters for the aminoacylation of hs mt tRNALeu(UUR) constructs.
| Km (µM) | kcat (s–1) | kcat / Km (relative) | |
|---|---|---|---|
| WT | 2.3 ± 0.5 | 0.020 ± 0.002 | 1 |
| U3271C | 9.7 ± 0.9 | 0.024 ± 0.001 | 0.3 |
| A3261G/U3271C | 2.3 ± 0.4 | 0.047 ± 0.004 | 2.3 |
To determine whether the structural perturbation detected in chemical probing experiments was the source of the Km defect, we also investigated the aminoacylation of the construct containing both the pathogenic U3271C and compensatory A3261G mutations. Indeed, the A3261G/U3271C mutant exhibited markedly increased aminoacylation activity compared with the U3271C mutant, and was even more efficiently aminoacylated than the WT substrate (Fig. 3). The restored activity of the A3261G/U3271C mutant indicates that a loss of secondary structure in the anticodon stem causes the reduced aminoacylation activity for the U3271C mutant. Measurement of the kinetic parameters revealed that Km for the A3261G/U3271C mutant was identical to WT hs mt tRNALeu(UUR) (Table 1). Interestingly, the increase in the rate of aminoacylation reflected a 2-fold enhancement in kcat (0.047 s–1) above WT.
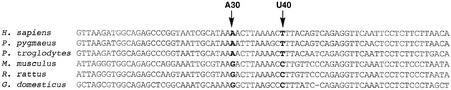
The 3261/3271 (30/40) base pair in the anticodon stem of hs mt tRNALeu(UUR) is not an A-U pair in all higher eukaryotes. Rattus rattus, Gallus domesticus and Mus musculus have a terminal G30/C40 base pair in the anticodon stem; however, other species besides Homo sapiens (e.g. Pongo pygmaeus and Pan troglodytes) have an A30/U40 base pair (Fig. 4) (24). As described here, the presence of the G30/C40 base pair in hs mt tRNALeu(UUR) increased the rate of aminoacylation compared with the WT sequence containing the AU base pair. It is interesting that mt tRNALeu(UUR) in some mammals contains this stabilizing feature in the anticodon stem, although species most closely related to humans retain the weaker AU pair.
Figure 4.
Alignment of mitochondrial tRNALeu(UUR) sequences. Sequences shown were obtained from the tRNA compilation cited in Sprinzl et al. (24).
The lowered aminoacylation activity of the U3271C hs mt tRNALeu(UUR) mutant, reduced only by a factor of ∼3, may not be the primary cause of the cellular defect manifested at the physiological level as MELAS or MIDD. Previous studies have detected losses of in vivo aminoacylation of a similar magnitude, but also provided evidence that steady-state levels of this tRNA are severely reduced when the mutation is present (9). Consistent with this observation, other experiments revealed that tRNA processing is severely impaired by the U3271C mutation (8). Indeed, the loss of structural integrity caused by this mutation may interfere with other aspects of tRNA function more dependent on the anticodon stem, and the more loosely folded structure may be susceptible to degradation. Likely, the structural instability of the U3271C hs mt tRNALeu(UUR) mutant, as revealed in these studies, interferes with many of its functional roles apart from aminoacylation.
Losses in function resulting from the disruption of fragile structural elements by pathogenic mutations were detected previously for hs mt tRNAIle (6,7). In this tRNA, mutations producing CA mispairs in the D-stem and anticodon stem of hs mt tRNAIle drastically decreased aminoacylation activities because of large kcat decreases (6). The fact that the aminoacylation defects were related to structural defects was confirmed by the introduction of compensatory mutations that restored base pairing and activity (6). The parallel observations described here for hs mt tRNALeu(UUR), with a mismatch-producing mutation impacting function, provide further evidence that the weak structures of hs mt tRNAs promote susceptibility to functional deactivation.
The patterns of reactivity for mutants of hs mt tRNALeu(UUR) and tRNAIle with structural disruptions within the anticodon domain also highlight interesting differences between the two systems. For hs mt tRNAIle, the losses in aminoacylation were severe (up to 103-fold) and resulted from kcat defects (6), while for hs mt tRNALeu(UUR), aminoacylation activity was affected to a lesser degree and was lowered due to an increase in Km. Studies of bacterial IleRS and LeuRS indicate that these two enzymes rely on the anticodon domain of their cognate tRNAs for aminoacylation efficiency to different extents (25–27). While aminoacylation of tRNAIle by IleRS is strongly affected by alterations in the anticodon stem and loop (25,26), the aminoacylation of tRNALeu by LeuRS is much less sensitive to changes in this domain (27). Indeed, recent studies indicate that an E.coli tRNALeu substrate completely lacking the anticodon stem and loop is aminoacylated, albeit with 10-fold lower efficiency than for the intact tRNA (27). However, E.coli tRNAIle substrates lacking the anticodon, or with point mutations in the anticodon stem are very poor substrates for IleRS, with activities reduced by ∼106 (25,26). These observations, made during studies exploring the bacterial analogs of tRNAIle and tRNALeu, are mirrored in the behavior of hs mt tRNAs containing pathogenic mutations in the anticodon stem. Although the structural disruptions that disease-related mutations introduce into hs mt tRNAs may be similar in different molecules, their effects on tRNA-specific enzymes, e.g. the aminoacyl-tRNA synthetases, likely differ because of the idiosyncrasies of tRNA recognition.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported in part by the Research Corporation, the Dreyfus Foundation NIH (GM63890) and Boston College.
REFERENCES
- 1.Anderson S., Bankier,A.T., Barrell,B.G., de Bruijn,M.H., Coulson,A.R., Drouin,J., Eperon,I.C., Nierlich,D.P., Roe,B.A., Sanger,F. et al. (1981) Sequence and organization of the human mitochondrial genome. Nature, 290, 457–465. [DOI] [PubMed] [Google Scholar]
- 2.Taanman J.W. (1999) The mitochondrial genome: structure, transcription, translation and replication. Biochim. Biophys. Acta., 1410, 103–123. [DOI] [PubMed] [Google Scholar]
- 3.Martin N. (1995) Organellar tRNAs: biosynthesis and function. In Söll,D. and RajBhandary,U. (eds), tRNA: Structure, Biosynthesis and Function. American Society for Microbiology, Washington, DC, pp. 127–140.
- 4.Helm M., Brule,H., Friede,D., Giegé,R., Putz,D. and Florentz,C. (2000) Search for characteristic structural features of mammalian mitochondrial tRNAs. RNA, 6, 1356–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wittenhagen L.M. and Kelley,S.O. (2002) Dimerization of a pathogenic human mitochondrial tRNA. Nature Struct. Biol., 9, 586–590. [DOI] [PubMed] [Google Scholar]
- 6.Kelley S.O., Steinberg,S.V. and Schimmel,P. (2000) Functional defects of pathogenic human mitochondrial tRNAs related to structural fragility. Nature Struct. Biol., 7, 862–865. [DOI] [PubMed] [Google Scholar]
- 7.Kelley S.O., Steinberg,S.V. and Schimmel,P. (2001) Fragile T-stem in disease-associated human mitochondrial tRNA sensitizes structure to local and distant mutations. J. Biol. Chem., 276, 10607–10611. [DOI] [PubMed] [Google Scholar]
- 8.Rossmanith W. and Karwan,R.M. (1998) Impairment of tRNA processing by point mutations in mitochondrial tRNALeu(UUR) associated with mitochondrial diseases. FEBS Lett., 433, 269–274. [DOI] [PubMed] [Google Scholar]
- 9.Yasukawa T., Suzuki,T., Ueda,T., Ohta,S. and Watanabe,K. (2000) Modification defect at anticodon wobble nucleotide of mitochondrial tRNAsLeu(UUR) with pathogenic mutations of mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes. J. Biol. Chem., 275, 4251–4257. [DOI] [PubMed] [Google Scholar]
- 10.Levinger L., Jacobs,O. and James,M. (2001) In vitro 3′-end endonucleolytic processing defect in a human mitochondrial tRNASer(UCN) precursor with the U7445C substitution, which causes non-syndromic deafness. Nucleic Acids Res., 29, 4334–4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chomyn A., Enriquez,J.A., Micol,V., Fernandez-Silva,P. and Attardi,G. (2000) The mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episode syndrome-associated human mitochondrial tRNALeu(UUR) mutation causes aminoacylation deficiency and concomitant reduced association of mRNA with ribosomes. J. Biol. Chem., 275, 19198–19209. [DOI] [PubMed] [Google Scholar]
- 12.Degoul F., Brule,H., Cepanec,C., Helm,M., Marsac,C., Leroux,J., Giegé,R. and Florentz,C. (1998) Isoleucylation properties of native human mitochondrial tRNAIle and tRNAIle transcripts. Implications for cardiomyopathy-related point mutations (4269, 4317) in the tRNAIle gene. Hum. Mol. Genet., 7, 347–354. [DOI] [PubMed] [Google Scholar]
- 13.Helm M., Florentz,C., Chomyn,A. and Attardi,G. (1999) Search for differences in post-transcriptional modification patterns of mitochondrial DNA-encoded wild-type and mutant human tRNALys and tRNALeu(UUR). Nucleic Acids Res., 27, 756–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yasukawa T., Hino,N., Suzuki,T., Watanabe,K., Ueda,T. and Ohta,S. (2000) A pathogenic point mutation reduces stability of mitochondrial mutant tRNAIle. Nucleic Acids Res., 28, 3779–3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yasukawa T., Suzuki,T., Ishii,N., Ohta,S. and Watanabe,K. (2001) Wobble modification defect in tRNA disturbs codon–anticodon interaction in a mitochondrial disease. EMBO J., 20, 4794–4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goto Y., Nonaka,I. and Horai,S. (1991) A new mtDNA mutation associated with mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes (MELAS). Biochim. Biophys. Acta, 1097, 238–240. [DOI] [PubMed] [Google Scholar]
- 17.Tsukuda K., Suzuki,Y., Kameoka,K., Osawa,N., Goto,Y., Katagiri,H., Asano,T., Yazaki,Y. and Oka,Y. (1997) Screening of patients with maternally transmitted diabetes for mitochondrial gene mutations in the tRNALeu(UUR) region. Diabet. Med., 14, 1032–1037. [DOI] [PubMed] [Google Scholar]
- 18.Bullard J.M., Cai,Y.C. and Spremulli,L.L. (2000) Expression and characterization of the human mitochondrial leucyl-tRNA synthetase. Biochim. Biophys. Acta, 1490, 245–258. [DOI] [PubMed] [Google Scholar]
- 19.Shepard A., Shiba,K. and Schimmel,P. (1992) RNA binding determinant in some class I tRNA synthetases identified by alignment-guided mutagenesis. Proc. Natl Acad. Sci. USA, 89, 9964–9968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peattie D.A. and Gilbert,W. (1980) Chemical probes for higher-order structure in RNA. Proc. Natl Acad. Sci. USA, 77, 4679–4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peattie D.A. (1979) Direct chemical method for sequencing RNA. Proc. Natl Acad. Sci. USA, 76, 1760–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Helm M., Brule,H., Degoul,F., Cepanec,C., Leroux,J.P., Giegé,R. and Florentz,C. (1998) The presence of modified nucleotides is required for cloverleaf folding of a human mitochondrial tRNA. Nucleic Acids Res., 26, 1636–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim S. (1979) Crystal structure of yeast tRNAPhe and general structural features of other tRNAs. In Schimmel,P., Söll,D. and Abelson,J. (eds), Transfer RNA: Structure, roperties and Recognition. Cold Spring Harbor Laboratory, USA, pp. 83–100.
- 24.Sprinzl M., Horn,C., Brown,M., Ioudovitch,A. and Steinberg,S. (1998) Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res., 26, 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nureki O., Niimi,T., Muramatsu,T., Kanno,H., Kohno,T., Florentz,C., Giege,R. and Yokoyama,S. (1994) Molecular recognition of the identity-determinant set of isoleucine transfer RNA from Escherichia coli. J. Mol. Biol., 236, 710–724. [DOI] [PubMed] [Google Scholar]
- 26.Nordin B. and Schimmel,P. (1999) RNA determinants for translational editing. J. Biol. Chem., 274, 6835–6838. [DOI] [PubMed] [Google Scholar]
- 27.Larkin D.C., Williams,A.M., Martinis,S.A. and Fox,G.E. (2002) Identification of essential domains for Escherichia coli tRNALeu aminoacylation and amino acid editing using minimalist RNA molecules. Nucleic Acids Res., 30, 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]