Abstract
An oligonucleotide microarray hybridization system to differentiate microbial species was designed and tested. Seven microbial species were studied, including one Bacillus and six Pseudomonas strains. DNA sequences near the 5′ end of 16S rRNA genes were aligned and two contiguous regions of high variability, flanked by highly conserved sequences, were found. The conserved sequences were used to design PCR primers which efficiently amplified these polymorphic regions from all seven species. The amplicon sequences were used to design 88 9mer hybridization probes which were arrayed onto glass slides. Single-stranded, fluorescence-tagged PCR products were hybridized to the microarrays at 15°C. The experimental results were compared with the ΔG° values for all matched and mismatched duplexes possible between the synthetic probes and the 16S target sequences of the seven test species, calculated using a ‘virtual hybridization’ software program. Although the observed hybridization patterns differed significantly from patterns predicted solely on the basis of perfect sequence matches, a unique hybridization fingerprint was obtained for each of the species, including closely related Pseudomonas species, and there was a reasonable correlation between the intensity of observed hybridization signals and the calculated ΔG° values. The results suggest that both perfect and mismatched pairings can contribute to microbial identification by hybridization fingerprinting.
INTRODUCTION
Widespread environmental biomonitoring, involving analyses of very large numbers of biological samples, is not readily attainable with today’s technology (1). Analyses of DNA sequences and expression of genes are beginning to play key roles in molecular diagnostics and drug discovery. New processes (2) and devices (3) must be developed for collection, processing, and analyzing of very large numbers of biological samples in medical applications (4), and for industrial and ecological uses (5,6). This need has stimulated the development of numerous identification and typing techniques based on phenotypic and genetic characteristics. One of the emerging tools in molecular diagnostics is the genosensor, or DNA chip, a microarray of surface-tethered DNA sequences (probes) to which nucleic acid samples (target strands) are hybridized on a glass or silicon substrate (7). In the hybridization fingerprinting approach reported here, the sensitivity of PCR (8) and the specificity of oligonucleotide microarray hybridization are combined with a new virtual hybridization (VH) strategy to enable microbial identification through analysis of the 5′ region of prokaryotic 16S rRNA genes of different bacterial strains.
MATERIALS AND METHODS
Bacterial strains, media and growth conditions
The seven bacterial strains used in this study (Pseudomonas aeruginosa, Pseudomonas alcaligenes, Pseudomonas fluorescens, Pseudomonas veronii, Pseudomonas syringae, Pseudomonas putida and Bacillus pumillus) were isolated and identified as described in Wallace et al. (9). Cells were routinely cultured in complex media (Luria broth or agar) containing 1% (w/v) tryptone, 0.5% (w/v) yeast extract and 0.5% (w/v) NaCl. This medium was used to prepare bacterial cell stocks for DNA preparation. When required, liquid media were solidified by the addition of 1.5% (w/v) Bacto agar. Media were sterilized by autoclaving at 15 p.s.i. for 30 min. Liquid cultures of cells were grown at 28°C in Erlenmeyer flasks fitted with gas permeable tops, which contained culture volumes not exceeding one-tenth the nominal flask volume, and rotated at 240 r.p.m.
DNA isolation and quantitation
Genomic DNA was isolated as described by Goldberg and Ohman (10) and spectrophotometrically quantitated as previously described (11).
Synthesis and design of probes and primers
Oligodeoxyribonucleotide probes derivatized with a 3′- terminal aminopropanol function for attachment to glass (3,5) were acquired from Sigma Genosys (The Woodlands, TX). Except as noted otherwise, all other oligonucleotide probes and PCR primers were obtained from Integrated DNA Technologies (Coralville, IA). PCR forward primers were derivatized with a 5′-biotin group to facilitate single-stranded target DNA purification. PCR reverse primers were derivatized with a 5′-fluorescent label (CY3 or CY5). The sequences and names of primers were as follows. Region A: 5′-biotin forward primer (primer fa) 5′-CTCCTACGGGAGGCAGCAG-3′; 5′-CY3 reverse primer (primer ra1) or 5′-CY5 reverse primer (primer ra2) 5′-GTATTACCGCGGCTGCTGG-3′. Region B: 5′-biotin forward primer (primer fb) 5′-CCAGCAGCCGCGGTAATAC-3′; 5′-CY3 reverse primer (primer rb1) or 5′-CY5 reverse primer (primer rb2) 5′-GGCGTGGACTACCAGGGTATC-3′. For region AB 5′-biotin primer fa was used as the forward primer and 5′-CY3 primer rb1 or 5′-CY5 primer rb2 was used as the reverse primer.
For the design of the probes and primers the 16S rRNA gene sequences from the seven bacterial species (12–18) were aligned using Clustal-X version 1.80 (19) to identify regions with numerous sequence variations (for design of hybridization probes) flanked by highly conserved sequences (for design of PCR primers). Two flanking highly variable regions, designated ‘A’ and ‘B’, were selected near the 5′ end of the 16S rRNA gene sequences. The probes were designed using a newly developed program called Genosensor Probe Designer (GPD) (A.Méndez-Tenorio, K.L.Beattie, M.J.Doktycz, R.Maldonado-Rodríguez, A.Guerra-Trejo and A.Reyes-López, submitted for publication). Two sets of probes were designed, one group of 28 probes for region ‘A’ and another group of 62 probes for region ‘B’. The aligned 16S rRNA gene sequences (in regions ‘A’ and ‘B’) of all test species as well as the sequences and positions of all probes along the genes are included in Supplementary Material, while Table 1 lists each probe sequence, its calculated duplex Tm value, and the species to which it is specific. Tm values of perfectly matched oligonucleotide duplexes were calculated by the GPD program using formulae of SantaLucia (20), setting [Na+] = 50 mM and [oligonucleotide] = 100 µM. The Tm estimates are intended only to describe the relative stability of different oligonucleotides in a given hybridization reaction, since the absolute values will depend on variables of DNA and ionic concentrations. All selected probes had the same size (9mer) but different predicted Tm values, were specific (perfectly paired at a single site within the amplified 16S rRNA gene region) to one or two and occasionally three of the test species, and usually showed more than one base difference with the corresponding PCR target strand of the other bacterial strains. Probes 89 and 90 were controls, having the same sequences as probes 27 and 28 of region ‘A’, respectively, except for being two bases longer (11mer). Probes 60 and 63 were identical, as were probes 61 and 64.
Table 1. Listing of oligonucleotide probe sequences, calculated Tm and ΔG° values, and species to which each probe is specific (single perfect match).
| Probe | Sequence (5′→3′) | Tm (°C) | ΔG° (kcal/mol) | Targeted species (perfect sequence match with test strains) |
|---|---|---|---|---|
| 1 | AAGTCTGAC | 32.03 | –10.49 | B.pumilus |
| 2 | AGTCTGACG | 37.60 | –11.66 | B.pumilus |
| 3 | GGAGCAACG | 42.53 | –12.72 | B.pumilus |
| 4 | GTGAGTGAT | 32.18 | –10.54 | B.pumilus |
| 5 | TGAGTGATG | 32.22 | –10.55 | B.pumilus |
| 6 | GGAGAAGCC | 39.68 | –12.08 | – |
| 7 | GCAGTTACC | 35.95 | –11.27 | P.syringae, P.veronii |
| 8 | CAGTTACCT | 30.93 | –10.31 | P.syringae, P.veronii |
| 9 | CGAGAGTAA | 31.38 | –10.35 | B.pumilus |
| 10 | GAGAGTAAC | 28.35 | –9.62 | B.pumilus |
| 11 | GTAACTGCT | 33.02 | –10.71 | B.pumilus |
| 12 | ATACCTTGC | 32.77 | –10.71 | P.aeruginosa, P.alcaligenes |
| 13 | AACTGCTCG | 39.85 | –12.16 | B.pumilus |
| 14 | TACCTTGCT | 34.42 | –11.11 | P.aeruginosa, P.alcaligenes |
| 15 | TACGTTAGT | 28.85 | –9.93 | P.putida, P.fluorescens |
| 16 | CCTTGCTGT | 39.02 | –11.98 | P.aeruginosa, P.alcaligenes |
| 17 | CGTTAGTGT | 33.56 | –10.80 | P.putida, P.fluorescens |
| 18 | CGTGATTGT | 35.05 | –11.13 | P.syringae, P.veronii |
| 19 | GTTAGTGTT | 27.90 | –9.63 | P.putida, P.fluorescens |
| 20 | GCTGTTTGA | 35.24 | –11.16 | P.aeruginosa, P.alcaligenes |
| 21 | GCTGTTTTG | 34.18 | –10.86 | P.syringae |
| 22 | AGTGTTTCG | 34.90 | –11.08 | P.fluorescens |
| 23 | AGTGTTTTC | 29.30 | –9.91 | P.putida |
| 24 | GTTGGGATG | 35.50 | –11.20 | – |
| 25 | GTGTTTCGA | 35.04 | –11.10 | P.fluorescens |
| 26 | CCTTGACGG | 40.76 | –12.32 | B.pumilus |
| 27 | TTGACGTTA | 31.17 | –10.38 | P.syringae, P.aeruginosa, P.alcaligenes, P.veronii |
| 28 | GTACCTAAC | 28.08 | –9.60 | B.pumilus |
| 29 | GTGGTTCAG | 35.70 | –11.20 | P.aeruginosa, P.alcaligenes |
| 30 | TGGTTCAGC | 39.14 | –12.00 | P.aeruginosa, P.alcaligenes |
| 31 | GGTTCAGCA | 39.14 | –12.00 | P.aeruginosa, P.alcaligenes |
| 32 | GTTCAGCAA | 35.24 | –11.16 | P.aeruginosa, P.alcaligenes |
| 33 | TTCAGCAAG | 34.43 | –11.00 | P.aeruginosa |
| 34 | AGCAAGCTT | 37.74 | –11.77 | P.alcaligenes |
| 35 | AGCAAGTTG | 35.09 | –11.14 | P.aeruginosa |
| 36 | GCAAGCTTG | 39.04 | –11.94 | P.alcaligenes |
| 37 | GCAAGTTGG | 37.95 | –11.70 | P.aeruginosa |
| 38 | CAAGCTTGA | 34.43 | –11.00 | P.aeruginosa |
| 39 | AAGCTTGAT | 31.06 | –10.43 | – |
| 40 | AAGTCCGTT | 36.33 | –11.47 | B.pumilus |
| 41 | AAGTCTGAT | 28.56 | –9.93 | P.aeruginosa |
| 42 | AGCTTGATG | 33.72 | –10.88 | – |
| 43 | AGTCCGTTG | 38.76 | –11.92 | B.pumilus |
| 44 | AGTCTGATG | 31.34 | –10.38 | P.aeruginosa |
| 45 | GCTTGATGT | 34.54 | –11.04 | – |
| 46 | GTCCGTTGT | 39.53 | –12.08 | B.pumilus |
| 47 | GTCTGATGT | 32.18 | –10.54 | P.aeruginosa |
| 48 | CTTGATGTG | 31.25 | –10.25 | – |
| 49 | TCCGTTGTG | 39.56 | –12.09 | B.pumilus |
| 50 | TCTGATGTG | 32.22 | –10.55 | – |
| 51 | GTGGATACT | 30.35 | –10.21 | P.aeruginosa |
| 52 | AAGCTACTG | 32.18 | –10.55 | P.alcaligenes |
| 53 | AACTACTGA | 27.89 | –9.77 | P.fluorescens, P.veronii |
| 54 | AACTGACTG | 32.75 | –10.64 | P.aeruginosa |
| 55 | AGCTACTGA | 33.22 | –10.85 | P.alcaligenes |
| 56 | ACTACTGAG | 29.78 | –10.05 | P.fluorescens, P.veronii |
| 57 | ACTGACTGA | 33.81 | –10.94 | P.syringae, P.putida |
| 58 | ACTGGCAAG | 39.02 | –11.98 | P.aeruginosa |
| 59 | GCTACTGAG | 34.03 | –10.85 | P.aeruginosa, P.alcaligenes |
| 60 | CTACTGAGC | 34.03 | –10.85 | P.fluorescens, P.veronii |
| 61 | CTGACTGAC | 34.57 | –10.94 | P.syringae, P.putida |
| 62 | CTGGCAAGC | 42.84 | –12.78 | P.aeruginosa, P.alcaligenes |
| 63 | CTACTGAGC | 34.03 | –10.85 | P.fluorescens, P.veronii |
| 64 | CTGACTGAC | 34.57 | –10.94 | – |
| 65 | CTGGATGAC | 34.36 | –10.94 | P.fluorescens, P.veronii |
| 66 | GACTGACTA | 29.96 | –10.07 | P.syringae, P.putida |
| 67 | GGCAAGCTA | 38.60 | –11.91 | P.fluorescens, P.veronii |
| 68 | ACTGACTAG | 29.78 | –10.05 | P.syringae, P.putida |
| 69 | GCAAGCTAG | 36.30 | –11.35 | P.aeruginosa, P.alcaligenes |
| 70 | TGAGCTAGA | 32.52 | –10.71 | P.syringae, P.putida |
| 71 | CAAGCTAGA | 31.51 | –10.41 | – |
| 72 | CTAGAATGT | 25.55 | –9.21 | P.aeruginosa |
| 73 | GTAGAGGTG | 32.85 | –10.61 | P.aeruginosa |
| 74 | TAGAGGTGG | 34.23 | –11.01 | P.aeruginosa |
| 75 | AGAGGTGGT | 38.26 | –11.87 | P.aeruginosa |
| 76 | GAGGTGGTA | 35.07 | –11.17 | P.aeruginosa |
| 77 | AGGTGGTAG | 34.92 | –11.15 | P.aeruginosa |
| 78 | GGTGGTAGA | 35.07 | –11.17 | P.aeruginosa |
| 79 | TGGTAGAAT | 27.55 | –9.77 | P.fluorescens |
| 80 | GTGGTGAAG | 35.70 | –11.20 | B.pumilus |
| 81 | CTCTCTGGT | 35.15 | –11.17 | B.pumilus |
| 82 | TCTCTGGTC | 35.30 | –11.19 | B.pumilus |
| 83 | CTCTGGTCT | 35.15 | –11.17 | – |
| 84 | TGGACCAAC | 38.01 | –11.76 | P.aeruginosa |
| 85 | ACTGTACTG | 31.36 | –10.36 | – |
| 86 | ACCAACATT | 31.39 | –10.50 | P.aeruginosa |
| 87 | CTGTACTGA | 30.68 | –10.22 | – |
| 88 | CATTGACAC | 32.10 | –10.41 | – |
Probes 1–28 were designed for region A and probes 29–90 were designed for region B. Tm values of perfectly matched oligonucleotide duplexes were calculated by the GPD program using formulae of SantaLucia (20), setting [Na+] = 50 mM and [oligonucleotide] = 100 µM. The Tm estimates are intended only to describe the relative stability of different oligonucleotides in a given hybridization reaction, since the absolute values will depend on variables of DNA and ionic concentrations.
PCR amplifications
Two hundred nanograms of bacterial genomic DNA from each bacterial strain were used as a template for all the PCRs. For both regions ‘A’ and ‘B’ the PCR was performed in 1× PCR buffer (10 mM Tris–HCl, pH 8.3, 50 mM KCl) containing 1.0 µl of each 20 ng/µl primer stock, 1.0 µl of a stock solution containing 10 mM of each of the four dNTPs, 1.5 µl of 25 µM MgCl2, 0.4 µl of 5 U/µl AmpliTaq DNA polymerase (Perkin Elmer) and 200 ng of template, in a total volume of 25 µl. The following thermal profile was performed for both regions ‘A’ and ‘B’: 3 min at 94°C; 32 cycles of 40 s at 94°C, 30 s at 60.5°C, and 40 s at 72°C; finally, holding at 4°C. For the largest PCR product (region ‘AB’) the reaction was performed in 25 µl of total volume of 1× buffer (10 mM Tris–HCl, pH 8.3, 50 mM KCl) containing 1.0 µl of 20 ng/µl stocks of each primer, 1.0 µl of a stock containing 10 mM of each dNTP, 1.7 µl of 25 mM MgCl2, 0.4 µl of 5 U/µl AmpliTaq DNA polymerase (Perkin Elmer) and 200 ng of template in a 25 µl reaction volume. The mixture was subjected to amplification using the following thermal profile: 3 min at 94°C; 35 cycles of 40 s at 94°C, 30 s at 61.0°C and 40 s at 72°C; finally, holding at 4°C.
Ten microliters of the PCR products were loaded onto 2% agarose gels and purified by STE MIDI SELECT-D G-50 Microcentrifuge Spin Columns for the Purification of Radiolabeled DNA and RNA (5 Prime→3 Prime, Inc.).
Hybridization substrates
Glass microscope slides were prepared for probe attachment by soaking for 15 min in hexane, followed by three washes in water. The slides were then dried in an 80°C oven for several hours. If the attachment reaction was not to be performed immediately, the slides were kept dry in a dessicator under vacuum at room temperature until the oligonucleotide probes were attached.
Oligonucleotide probe attachment
Oligonucleotide probes derivatized at the 3′ end with aminopropanol function were dissolved in water. An aliquot (typically 10 nl) of each probe, diluted to 20 µM in water, was applied in duplicate to a clean, dry glass slide using a Microlab 2200 workstation (Hamilton Company, Reno, NV) equipped with a precision x–y substage and a solenoid valve-based ink jet using a sapphire dispense tip as described by Hicks et al. (21). The applied droplets were allowed to air dry (typically 5–15 min), then the slides were rinsed three times with distilled water at room temperature to remove unbound oligonucleotides. As discussed previously (3–5,22), the attachment of 3′-aminopropanol-derivatized oligonucleotides to plain, underivatized glass surfaces is rapid and convenient; the attachment reaction is completed during the few minutes required for robotically applied probe aliquots to dry on the glass slide.
Preparation of single-stranded target DNA
PCRs were conducted using forward primers labeled with biotin and reverse primers labeled with CY3 or CY5, yielding PCR products in which the target strand was fluorescently labeled at the 5′ end. As described previously (22,23), the PCR product was processed using a Millipore (Bedford, MA) Ultrafree spin-filter (30 000 mol. wt cutoff) to remove excess PCR primers, the retained material was applied to an AffiniTip Strep 20 column (Sigma Genosys), and the single-stranded target was eluted from the column, neutralized, desalted and concentrated by ethanol precipitation.
Hybridization and imaging
Hybridizations were performed in 5× standard saline, phosphate, EDTA (SSPE). The single-stranded target PCR product was dissolved in the hybridization solution at a concentration of 30–150 nM. A 50-µl aliquot of the target solution was applied to the area of the slide containing the array of attached probes, covered with a cover slip and incubated at 15°C for 12 h. The cover slips were removed and then slides were washed with 1× SSPE solution at 15°C for a minimum of 5 min. Each hybridization experiment (involving a given bacterial species and a given array of probes) was carried out with duplicate probes (per spotted array) and triplicate slides hybridized simultaneously. Thus, each hybridization result represented an average of six parallel determinations. Fluorescent hybridization images were acquired using a GeneTAC imaging system with GT Imaging & Review software (Genomic Solutions, Ann Arbor, MI). The GeneTAC imaging system has a dynamic range of 2–3 logs and displays detection linearity when the average pixel intensity is between 5000 and 50 000. Each slide was imaged for a period of time which yielded an average pixel intensity for the brightest spots of slightly below 50 000 (typically 30 000). For each hybridization result the average pixel intensities for the six determinations were averaged, and after subtraction of background signal (average pixel intensity typically <100) the hybridization result was assigned a qualitative intensity value of ‘strong’ (average pixel intensity 10 000–50 000), ‘weak’ (average pixel intensity <10 000 but typically >5000) or ‘absent/undetected’ (average pixel intensity <1000). To statistically compare the hybridization patterns obtained with the three spotted slides in each hybridization experiment, SigmaStat version 2.03 was used for Friedman repeated measures analysis of variance on ranks. This non-parametric test consistently yielded P-values of 1.000 for replicate hybridization intensities, thus the differences between experimental replicates were not statistically significant, supporting the conclusion that these hybridization experiments were highly reproducible.
Virtual hybridization
A VH module of the GPD software (A.Méndez-Tenorio, K.L.Beattie, M.J.Doktycz, R.Maldonado-Rodríguez, A.Guerra- Trejo and A.Reyes-López, submitted for publication) was used to predict the likelihood of hybridization of each probe with the 16S rDNA target of each species and to predict the binding strength at alternative hybridization sites along the target. GPD was written in Borland Delphi v. 5.0 (Borland International) and runs on Windows 95, 98, Millennium and NT operating systems. The GPD program is based on object-oriented programming methodology and includes classes for representing the user interface, molecules, criteria and interactions. The VH is based on calculation of the Gibbs free energy (ΔG°) of binding of any given probe when forming a duplex along the target strand (all positions throughout the entire target sequence are interrogated). To calculate the ΔG° of matched or mismatched oligonucleotide sequences the VH program can accept thermodynamic parameters representing either singlet or doublet data formats but in this study we employed SantaLucia’s extensive set of thermodynamic parameters for perfectly matched and mismatched oligonucleotide sequences, in the nearest neighbor (NN) doublet format in which the entire NN interaction is considered in a single parameter and thermodynamic values for each of the NN base pairs are used (24–28). Thermodynamic calculations for each probe aligned at all positions along the relevant target DNA sequence are performed and the ΔG° values over any desired negative free energy range are tabulated. SantaLucia’s thermodynamic parameters include all single base mismatches in different nearest neighbor sequence contexts. Tandem mismatches, which SantaLucia notes are not well correlated with the NN approach, are considered as internal loops. They are assigned positive free energy values using a function that gives a linear dependence of positive free energy value with loop size, as used in prediction of secondary structure. Terminal mismatches are assigned no energetic contribution other than the resultant decrease in duplex length.
RESULTS
Design of probes and PCR primers
Clustal-X alignment of the seven 16S rRNA gene sequences revealed a region near the 5′ end which contained two flanking stretches of variable sequence, named ‘A’ and ‘B’, flanked by conserved sequences. Conserved and variable sequences were used for the design of the PCR primers and probes, respectively. The PCR product from region ‘A’ is located between nucleotides 356 and 554, yielding a 197 bp fragment for P.aeruginosa and a 198 bp fragment for the six other species. The PCR product from region ‘B’ is positioned between nucleotides 533 and 830, forming a 293 bp fragment for P.aeruginosa and a 294 bp fragment for the remaining species. The distal pair of primers yields a larger ‘AB’ PCR product, encompassing regions ‘A’ and ‘B’, of length 471 bp fragment for P.aeruginosa and 473 bp for the other species. An additional computer search of the GenBank database suggested that the same primers can be used in numerous other bacteria. A list of the probes for each region, their Tms, and the species for which they were specific (perfectly matching) is included in Table 1.
PCR amplification and hybridization
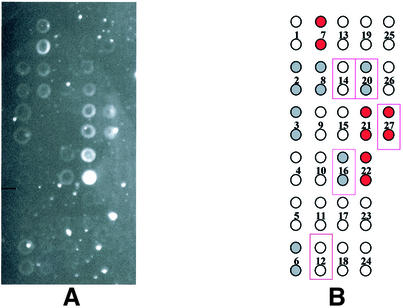
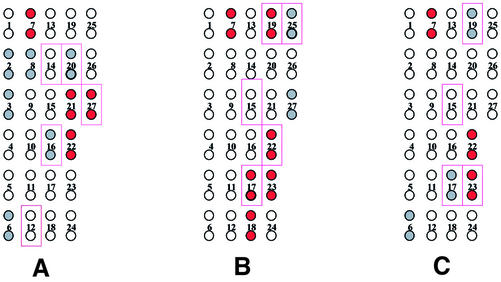
Using the above PCR primers, amplification of genomic DNA from all seven bacterial species tested yielded the expected single product in good yield as judged by gel electrophoresis, for regions ‘A’ and ‘B’ as well as ‘AB’ (Fig. 1). The hybridization pattern obtained with single-stranded fragment ‘A’ from P.aeruginosa is shown in Figure 2 as a representative example; similar experiments were performed for each species. Figure 2A shows the experimental hybridization signals and Figure 2B represents the location of each probe placed in duplicate across the genosensor and color-coded relative signal intensity, qualitatively assigned as strong (red circles), weak (gray circles) and absent (clear circles) hybridization. The positions of duplicate array elements corresponding to a perfect sequence match for P.aeruginosa are indicated by outlining. Of the five probes designed specifically for P.aeruginosa, only three yielded strong or weak hybridization signals. Additionally, unexpected hybridization signals of weak or strong intensity were seen for several probes on the array that were not designed for this species. This hybridization pattern, although significantly different from the originally anticipated pattern, was nevertheless consistently produced in multiple experiments performed with P.aeruginosa (data not shown). Similar analyses were performed for all seven bacterial species. Figure 3 represents the hybridization patterns produced in region ‘A’ of three Pseudomonas species (P.aeruginosa, P.fluorescens and P.putida). The patterns are different for each species. Again, not all of the perfectly matched probes yielded hybridization signals, and additional signals were seen, representing mismatched hybridizations.
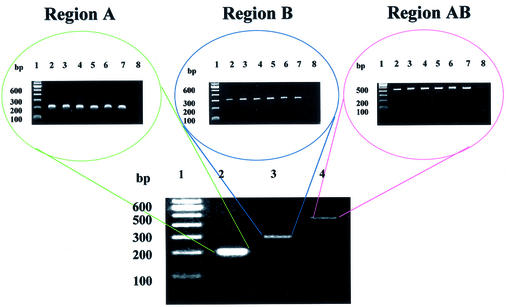
Figure 1.
Electrophoretic analysis of PCR products comprising regions A, B and AB. In the electrophoretic separations displayed at the top, the DNA sample was as follows: lane 1, 100 bp DNA ladder; lane 2, PCR product from P.aeruginosa; lane 3, PCR product from P.syringae; lane 4, PCR product from P.fluorescens; lane 5, PCR product from P.veronii; lane 6, PCR product from P.putida; lane 7, PCR product from B.pumillus; lane 8, material from a negative control PCR containing no template. In the electrophoretic separation shown at the bottom the DNA samples were as follows: lane 1, 100 bp DNA ladder; lane 2, PCR product derived from P.aeruginosa using primers designed for region A; lane 3, PCR product derived from P.aeruginosa using primers designed for region B; lane 4, PCR product derived from P.aeruginosa using primers designed for region AB.
Figure 2.
Hybridization pattern for region A of the P.aeruginosa 16S rRNA gene. Each probe was spotted in duplicate onto the glass slide. (A) The pattern of fluorescence and (B) a color-coded representation of the results. Red-filled circles represent strong experimental hybridization signals (average pixel intensity 10 000–50 000). Gray-filled circles represent weak experimental hybridization signals (average pixel intensity <10 000). Open circles represent absence of detectable hybridization signal (average pixel intensity <1000). Solid rectangular outlines represent positions of predicted hybridization based on the existence of a single perfect match within the target.
Figure 3.
Color-coded representation of hybridization results in region A of the 16S rRNA gene for three test species. (A) Pseudomonas aeruginosa, (B) P.fluorescens and (C) P.putida. Red-filled circles represent strong experimental hybridization signals (average pixel intensity 10 000–50 000). Gray-filled circles represent weak experimental hybridization signals (average pixel intensity <10 000). Open circles represent absence of detectable hybridization signal (average pixel intensity <1000). Solid rectangular outlines represent positions of predicted hybridization based on the existence of a single perfect match within the target.
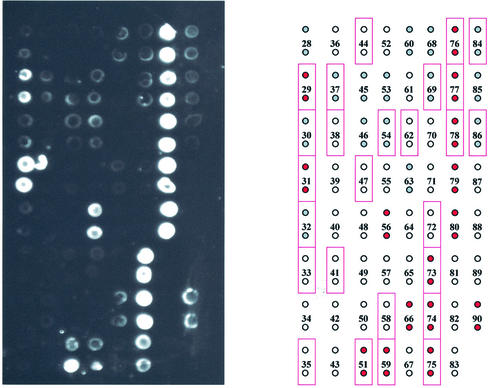
The hybridization pattern obtained for region ‘B’ of P.aeruginosa is shown in Figure 4. Again, the experimentally obtained image is shown on the left and a color-coded data summary (indicating qualitative/relative hybridzation intensities) is given on the right. In region ‘B’ 17 of the 26 predicted (perfectly matched) probes for P.aeruginosa gave a signal, 10 of them strong and seven weak. Thirteen additional probes produced unexpected hybridization signals, five strong and eight weak. The same pattern was repeatedly produced and distinct and reproducible patterns were also produced by each bacterial species (data not shown).
Figure 4.
Hybridization pattern for region B of the P.aeruginosa 16S rRNA gene. Each probe was spotted in duplicate onto the glass slide. The pattern of fluorescence is shown on the left and a color-coded representation of the results is shown on the right. Red-filled circles represent strong experimental hybridization signals (average pixel intensity 10 000–50 000). Gray-filled circles represent weak experimental hybridization signals (average pixel intensity <10 000). Open circles represent absence of detectable hybridization signal (average pixel intensity <1000). Solid rectangular outlines represent positions of predicted hybridization based on the existence of a single perfect match within the target.
Table 2 summarizes the total expected (perfect match), positive/expected, negative/expected and positive/unexpected (mismatched) hybridizations produced in both regions for all the analyses (except for region ‘A’ in B.pumillus and region ‘B’ in P.syringae which were not tested). A total of 49 of the 90 expected (perfectly paired) signals were experimentally seen, 41 expected signals were not seen, and 88 unexpected (mismatched) hybridization signals were obtained.
Table 2. Summary of all hybridization data, comparing expected and observed hybridization signals for each test species in regions A and B of the 16S rRNA gene.
| Test species | P.veronii | B.pumillus | P.aeruginosa | P.alcaligenes | P.fluorescens | P.putida | P.syringae | Total | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Region | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A + B |
| Total expected | 4 | 6 | (–) | 8 | 5 | 26 | 5 | 12 | 4 | 7 | 4 | 5 | 4 | (–) | 90 |
| Positive/expected | 1 | 0 | (–) | 5 | 3 | 17 | 2 | 4 | 4 | 4 | 3 | 3 | 3 | (–) | 49 |
| Negative/expected | 3 | 6 | (–) | 3 | 2 | 9 | 3 | 8 | 0 | 3 | 1 | 2 | 1 | (–) | 41 |
| Positive/unexpected | 3 | 5 | (–) | 8 | 7 | 13 | 3 | 11 | 4 | 10 | 3 | 17 | 4 | (–) | 88 |
Total expected signals are based on the existence of perfect sequence matches within the PCR targets. Positive/expected signals represent the detection of hybridization for probes with a perfect match within the target sequence. Negative/expected signals represent the lack of detectable hybridization for probes possessing a perfect match within the target sequence. Positive/unexpected signals represent the detection of hybridization signals for probes lacking a perfect match within the target sequence (mismatched hybridization). (–), untested.
Experimental versus virtual hybridization
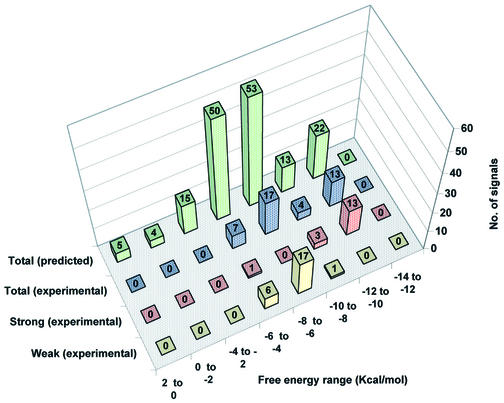
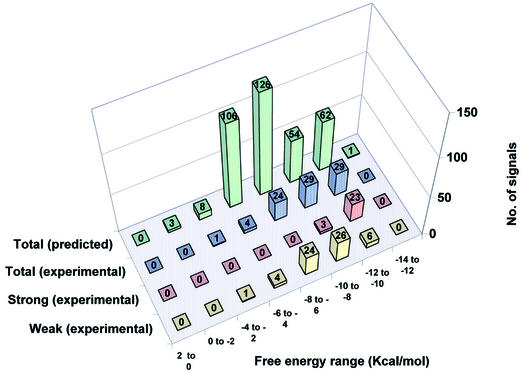
A VH analysis was conducted to predict hybridization signals, on the basis of ΔG° values obtained for all potential pairings of oligonucleotide probes along the PCR target sequences. These calculations utilized the thermodynamic parameters reported by SantaLucia (24–28) for perfectly matched and mismatched oligonucleotide hybrids, as detailed further in a separate manuscript (A.Méndez-Tenorio, K.L.Beattie, M.J.Doktycz, R.Maldonado-Rodríguez, A.Guerra-Trejo and A.Reyes-López, submitted for publication). The calculated ΔG° values for the 9mer probes arrayed for region ‘B’ hybridization are listed in Table 3, and the last column lists ΔG° values for each probe paired with its most stable site within P.aeruginosa region ‘B’. In Figures 5 and 6 the experimental hybridization results are compared with the ΔG° values predicted using the VH analysis for regions ‘A’ and ‘B’, respectively. For this analysis, the data were divided into groups at increments of 2 kcal/mol ΔG° value. For each incremental ΔG° group the rear column represents the total number of predicted cases in which any of the probes can form a matched or mismatched duplex anywhere within all the PCR target sequences (except for region ‘A’ in B.pumillus and region ‘B’ in P.syringae which were not tested). The next column shows the number of hybridization signals experimentally detected for probes predicted to bind, within each ΔG° increment. The next two columns divide the number of experimentally detected hybridization signals according to the intensity of the signal (strong and weak). Two important correlations are seen from these graphs. First, the proportion of predicted signals that were experimentally seen increases in direct relation with the binding energy (negative ΔG° value). Secondly, strong signals were generally produced only by hybrids with ΔG° values of –8 to –12 kcal/mol, while weak signals were produced by hybrids with ΔG° values of –6 to –10 kcal/mol.
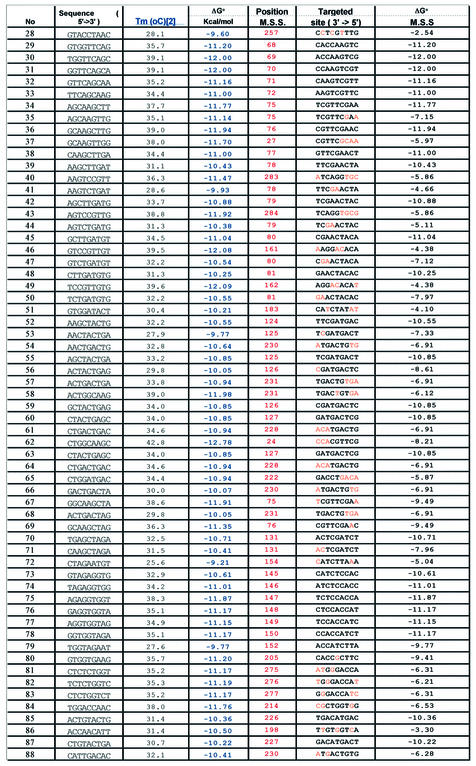
Table 3. Comparison of predicted versus experimentally observed hybridization in region B of P.aeruginosa.
The first column shows the 9mer probe number as given in Table 1. The second column displays the 9mer probe sequence (5′→3′ direction). The third column lists the calculated Tm values for each 9mer probe, paired with its complementary target sequence. Tm values of perfectly matched oligonucleotide duplexes were calculated by the GPD program using formulae of SantaLucia (20), setting [Na+] = 50 mM and [oligonucleotide] = 100 µM. The Tm estimates are intended only to describe the relative stability of different oligonucleotides in a given hybridization reaction, since the absolute values will depend on variables of DNA and ionic concentrations. The fourth column lists the binding energy (ΔG° in kcal/mol) calculated for each probe, paired with its complementary target sequence. The fifth column lists the first nucleotide position of the most stable site (MSS) for binding of each probe within region B of P.aeruginosa. The sixth column displays the target sequence (3′→5′ direction) for each MSS within region B of P.aeruginosa, with paired positions written in black and mispaired positions indicated in orange. The last (seventh) column lists the calculated binding energy (ΔG° in kcal/mol) for hybridization of each probe at its MSS within region B of P.aeruginosa.
Figure 5.
Distribution of predicted versus experimentally detected hybridization signals in region A of all test species grouped according to calculated binding energy. The data were divided into groups at increments of 2 kcal/mol ΔG° value. For each incremental ΔG° group the green columns represent the total number of predicted cases in which any of the probes can form a matched or mismatched duplex anywhere within all the PCR target sequences. The blue columns show the number of hybridization signals experimentally detected for probes predicted to bind, within each ΔG° increment. The red columns represent the number of experimentally detected strong hybridization signals (average pixel intensity 10 000–50 000) in each ΔG° increment, while the yellow columns represent the number of experimentally detected weak hybridization signals (average pixel intensity <10 000) in each ΔG° increment.
Figure 6.
Distribution of predicted versus experimentally detected hybridization signals in region B of all test species grouped according to calculated binding energy. The data were divided into groups at increments of 2 kcal/mol ΔG° value. For each incremental ΔG° group the green columns represent the total number of predicted cases in which any of the probes can form a matched or mismatched duplex anywhere within all the PCR target sequences. The blue columns show the number of hybridization signals experimentally detected for probes predicted to bind, within each ΔG° increment. The red columns represent the number of experimentally detected strong hybridization signals (average pixel intensity 10 000–50 000) in each ΔG° increment, while the yellow columns represent the number of experimentally detected weak hybridization signals (average pixel intensity <10 000) in each ΔG° increment.
DISCUSSION
The objective of this study was to implement oligonucleo tide array hybridization (genosensor chip technology) for microbial identification. For this work we selected a set of seven species, representing both divergent phylogeny (Gram-negative and Gram-positive species) and closely related species (six Pseudomonas species), and we selected as a gene target the 16S rRNA gene which is well characterized and known to contain considerable sequence polymorphism. A first step in the design of a genosensor chip for discrimination between different bacterial species is to find a genetic region that can be amplified using a single pair of PCR primers and also contains a reasonable degree of sequence variation within the amplicon. The 16S rRNA gene sequences of the seven bacterial species were aligned using Clustal-X version 1.8 (19) to search for an appropriate region. The Clustal-X alignment revealed that there is not sufficient sequence polymorphism in the 3′-end region of 16S rRNA genes to yield oligonucleotide probes that could distinguish the Pseudomonas species, although probes in this region have been used to distinguish many other species (8). The Clustal-X alignment did, however, reveal two contiguous regions with the required sequence variations, flanked by conserved primer sites, near the 5′ end of the 16S rRNA genes of all the bacterial species. The primers designed reproducibly yielded a single product in all the tested strains for both contiguous regions (‘A’ and ‘B’), as well as the combined product (‘AB’) using the distal primers. A further search of the GenBank database suggested that the same pairs of primers can probably generate the corresponding PCR products in more than 100 other bacterial species and in other organisms.
Some previous studies have focused on design of oligonucleotide probes of similar Tm values in order to avoid the well known effects of base composition and sequence on duplex stability and achieve uniform hybridization intensity across the array, under a single hybridization condition (29,30). Despite this design strategy, the experimental hybridization patterns have often contained missing or additional signals with respect to those expected. A missing signal may be due to the formation of secondary structure within the target DNA, while additional hybridization signals may be due to mismatched but nevertheless stable hybrids involving other sites within the analyte DNA molecules. In the case of bacterial strains derived from environmental or clinical samples, the precise target sequences are often unknown (6), since these strains typically possess some sequence differences compared with the available reference strains, making the interpretation of the data more difficult.
In the study reported here we designed 9mer oligonucleotide probes to interrogate sequence differences between the aligned 16S rRNA genes of seven species. The probes contained preferably no more than two identical bases in a row and preferably four or five [G+C], but at times three or six [G+C] out of nine bases. The probes were not designed to give uniform duplex stability across the array, displaying a significant predicted Tm variation (23–40°C) among the collection of probes. Each probe was designed to be specific (perfect match) for one or two and sometimes three of the bacterial species studied, while having (with few exceptions) no other binding sites containing fewer than two base mismatches within the full sequence of all DNA targets. It should be pointed out, however, that the probe design, as well as the evaluation of the stability of the matched and mismatched hybrids using the VH application of the GPD software, were based on the sequences of reference species reported in GenBank, whereas the actual environmentally isolated species may have differed slightly in target sequence from the reference species. Despite the range of duplex stability of probes and lack of certainty of target sequences, the probes and species utilized in this study were nevertheless useful to assess whether closely related species can be distinguished on the basis of experimental hybridization patterns and to assess whether the VH strategy can be useful in interpretation of microarray hybridization data, and ultimately in designing optimally specific sets of probes for a given target sequence. To maximize the probability that probes would yield a hybridization signal (even though probes of higher Tm values may yield hybridizations involving relatively stable mismatches) the hybridizations were performed at a relatively low temperature (15°C).
The experimental hybridization patterns showed, not surprisingly in light of the above discussion, significant differences from those initially predicted, as documented by the hybridization patterns for region ‘A’ of P.aeruginosa, P.fluorescens and P.putida (Figs 2 and 3). Table 2 summarizes the hybridization signals for each bacterial test species, categorized as total expected (perfect match), positive/expected, negative/expected and positive/unexpected (mismatched) hybridization signals. Of the 90 total hybridization signals expected (perfect match), 49 signals were detected and 41 were undetected, while 88 unexpected (mismatched) hybridization signals were seen. Although some of these results were unexpected, the hybridization patterns were nevertheless different for each species and some species-specific signals were seen.
To help explain the discrepancy between anticipated and observed results, the VH module of the GPD software was applied to the data. The VH program calculates the Gibbs free energy (ΔG°) of mismatched or matched probes when aligned to target DNA (Table 3). The GPD software runs each probe along the full target DNA sequence (including all test strains) and calculates binding energy as ΔG° for each position. The selection can be adjusted to search for pairings with high, medium or low probability to produce stable hybridization under a given hybridization condition. Figures 5 and 6 display the VH results for all 90 9mer probes in regions ‘A’ and ‘B’ for the seven bacteria tested. The results were divided into eight stability groups of increasingly negative ΔG° values (+2 to 0, 0 to –2, –2 to –4, –4 to –6, –6 to –8, –8 to –10, –10 to –12 and –12 to –14 kcal/mol). The green bars in the back row represent the total number of predicted hybridization signals for each stability group, including both perfectly matched and mismatched hybrids. The bars in the remaining rows represent total (blue), strong (red) and weak (yellow) experimentally observed signals. On top of each bar is indicated the total number of signals for all bacterial species that were predicted (green bars) or experimentally observed (other colors) in each stability group. One would expect a Gaussian distribution of predicted ΔG° values (green bars) for a random collection of probes of a given length. However, in the probe design, oligonucleotides having single mismatches at any site within the target sequences were generally excluded, and this restriction accounts for the reduced number of hybrids in the stability group with a ΔG° of –8 to –10 kcal/mol. For experimentally detected hybrids it can be seen that the proportion of signals detected generally decreased as the stability of the hybrid decreased. When the experimentally detected signals were divided into strong and weak signals (red and yellow bars, respectively), it could be seen that for region ‘A’ (Fig. 5), in the group with the highest ΔG° value (–10 to –12 kcal/mol) all signals were strong (100%). In the next stability group (–8 to –10 kcal/mol) most hybridization signals (75%) were strong and the remaining (25%) were weak. In the next group with even lower stability (–6 to –8 kcal/mol) all the signals (100%) were weak. A similar result was seen with the –4 to –6 kcal/mol group, where 85% of the signals were weak and 15% were strong. The same analysis was conducted for region ‘B’ with very similar results (Fig. 6). In general, these results suggest that the VH analysis helps to explain the hybridization results and that hybridization signals arise primarily from the most stable duplex structures found for each probe within the target (Table 3). Furthermore, it can be seen that the experimentally detected mismatched hybrids often contained mismatches positioned at their ends, where destabilization is minimal (30), and in many cases the mismatches were of a type known to have low destabilizing effect (22,28,30,31).
To provide additional confidence in the conclusion that the experimentally observed hybridization signals correlate with sites of stronger predicted binding energy (more negative ΔG° values), we carried out the following statistical tests. A series of Excel spreadsheets were created containing three columns of data. Column one contained the free energy values for the most stable sites (MSS) predicted for all 9mer probes along the target DNA, the second column contained free energy values for probes producing strong (S) hybridization signals, and the third column contained free energy values for probes producing weak (W) hybridization signals. The second and third columns could also be combined to represent all the observed hybridization signals (S + W). Individual spreadsheets were created for region A, region B and combined regions A + B. The Kolmogorov–Smirnov test was first performed to determine whether the predicted free energy values tabulated in a given column formed a normal distribution. The results indicated that the data were distributed normally (P < 0.05) only within MSS and S + W columns, which contained the greatest number of ΔG° values (individual S and W columns may not have contained sufficient data to reveal normal distributions). Next, parametric (Student’s 2-sample t-test) and non-parametric (Mann–Whitney test) statistical tests were then performed to evaluate whether the probability distributions were statistically different for total probes predicted to hybridize (MSS) versus probes yielding actual experimental hybridization (S, W, S + W). The t-test is traditionally used for normal distributions while the Mann–Whitney test is often used when normal distributions are not found. Using both parametric and non-parametric tests, small P-values indicated that in nearly all cases (with the exception of weak signals in region A) the free energy distributions for predicted versus observed hybridizations are significantly different, with the experimental hybridization signals occurring at significantly higher negative (more stable) free energy values. This result was also obtained when the statistical tests were repeated incorporating the new PCR sequence in region B of the P.aeruginosa sample. Thus, there appears to be a correlation between predictive free energy values and the presence of hybridization signals: the probability of observed hybridization is greatest when predicted binding energy is greatest. These statistical analyses are detailed fully in a separate manuscript which describes the GPD/VH software (A.Méndez-Tenorio, K.L.Beattie, M.J.Doktycz, R.Maldonado- Rodríguez, A.Guerra-Trejo and A.Reyes-López, submitted for publication).
In the case of pairings with a high negative ΔG° value that were not experimentally detected, these may be attributed to sequence differences between the samples and reference strains, or to the formation of secondary structure within the target (32). Certain hybridization signals that were observed but not predicted may also have been due to sequence differences between the environmental isolates and the reference species. In fact, after this manuscript was submitted, the actual sequence of the PCR product representing region B of the P.aeruginosa sample was sequenced (both strands), and it is clear that there are six single-base differences between the environmental isolate used in these studies and the published ‘reference sequence’ used to design the 9mer probes intended to represent P.aeruginosa. Furthermore, a new Clustal-X sequence alignment of region B including the sequenced PCR product, the reference strain (accession no. GI:45418), and 49 other published P.aeruginosa sequences (see Supplementary Material) revealed that the sequence of our environmental isolate was identical to the consensus sequence of the 49 published P.aeruginosa strains (which displayed a high degree of sequence conservation), whereas the ‘reference strain’ appeared to be rather atypical among the collection. The new sequence information resulted in a net loss of five sites with perfect match in region B (dropping from 26 in the reference strain to 21 in the sequenced environmental isolate). This sequence difference does not explain any of the ‘missing’ hybridization signals (at sites of perfect match) but does account for two of the ‘unexpected’ hybridization signals. In the case of probes 53 and 56, which yielded weak and strong hybridization signals, respectively, the PCR product sequencing revealed that these probes are actually perfectly paired with the target DNA. Furthermore, when the VH analysis was repeated using the PCR product sequence it was seen that for probes having the most stable hybridization sites of higher stability (–8 to –12 kcal/mol) in region B, the prediction of hybridization was somewhat better for the sequenced product than for the reference sequence. Another potential source of unpredicted positive hybridization could be mismatches that are unusually stable in certain sequence contexts. Unexpected hybridization signals, regardless of their origin, belong nevertheless to a specific target and, consequently, can contribute to the identification of bacterial species via hybridization fingerprinting.
Several other laboratories have recently reported the use of oligonucleotide probes targeted to 16S rRNA sequences to differentiate between microbial species (33–36). Bavykin et al. (33) and Liu et al. (34), employing arrays of 20mers immobilized within Mirzabekov’s gel pad microchip system, successfully differentiated closely related Bacillus species. Small et al. (35), utilizing an array of glass-immobilized 20mer capture probes in combination with labeled 20mer detector probes hybridized in tandem, directly identified several Geobacter and Desulfovibrio 16S rRNA sequences in bulk RNA extracted from soil samples. Wilson et al. (36) analyzed PCR amplicons derived from a diverse collection of cultured bacteria, using an Affymetrix GeneChip designed to interrogate sequence differences within an 82-bp region near the 3′ end of 16S rRNA genes. The GeneChip, containing 31 179 20mer probes selected from a subalignment of 1945 prokaryotes and 431 eukaryotes contained in version 5.0 of the Small Subunit Ribosomal Database Project, reliably identified nearly all microbial species tested. Although the studies cited above cannot be directly compared with those reported here due to significant differences in target sequences, probe length and other experimental conditions, the results further document the utility of 16S rRNA oligonucleotide fingerprinting in microbial identification.
A major and novel contribution emerging from the studies reported here is that the utility of oligonucleotide arrays can be extended beyond the current paradigm in which sequence recognition relies on single base mismatch discrimination. By using arrays of short oligonucleotide probes and hybridization conditions that would be considered ‘suboptimal’ in the above traditional approach, meaningful hybridization fingerprints involving mismatched hybrids in addition to perfect hybrids can be produced. Mismatch-containing hybridization fingerprints should be useful for a variety of microarray applica tions that examine sequence variations, including microbial identification and analysis of single nucleotide polymorphisms. The hybridization data obtained in this study, combined with available thermodynamic parameters for matched and mismatched duplexes, have provided the ‘working materials’ for initial development of a VH software tool that can assist in the prediction of hybridization patterns, a function which will become increasingly powerful as thermodynamic data become available for an expanded set of oligonucleotide interactions such as tandem mismatches, terminal mismatches, intrastrand secondary and tertiary structures, as well as interactions with other environmental components in the microarray assay such as solution additives and surfaces. As further detailed in a separate manuscript (A.Méndez-Tenorio, K.L.Beattie, M.J.Doktycz, R.Maldonado-Rodríguez, A.Guerra-Trejo and A.Reyes-López, submitted for publication), the VH approach can guide the selection of optimally specific sets of oligonucleotide probes for a given oligonucleotide microarray application.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Brent Harker of ORNL for arraying the probes onto glass slides, and thank Aaron Nagel of The University of Tennessee for providing valuable technical advice. The authors thank Engineer Armando Guerra-Trejo of the ENCB-IPN for substantial assistance with statistical analysis. Financial support for this work was provided by CONACYT and IPN at ENCB-IPN and by the US Department of Defense Strategic Environmental Research and Development Program (CU-1081) at ORNL.
REFERENCES
- 1.Ramsay G. (1998) DNA chips: state-of-the art. Nat. Biotechnol., 16, 40–44. [DOI] [PubMed] [Google Scholar]
- 2.Drmanac S., Kita,D., Labat,I., Hauser,B., Schmidt,C. and Burczak,J.D. (1998) Accurate sequencing by hybridization for DNA diagnostics and individual genomics. Nat. Biotechnol., 16, 54–58. [DOI] [PubMed] [Google Scholar]
- 3.Doktycz M.J. and Beattie,K.L. (1997) Genosensors and model hybridization studies. In Beugelsdiik,A. (ed.), Automation Technologies for Genome Characterization. John Wiley and Sons, New York, pp. 205–225.
- 4.Maldonado-Rodriguez R., Espinosa-Lara,M., Loyola-Avitia,P., Beattie,W.G. and Beattie,K.L. (1999) Mutation detection by stacking hybridization on genosensor arrays. Mol. Biotechnol., 11, 13–25. [DOI] [PubMed] [Google Scholar]
- 5.Beattie K.L. (1997) Analytical microsystems: emerging technologies for environmental biomonitoring. In Sayler,G.S., Sanseverino,J. and Davis,K.L. (eds), Biotechnology in the Sustainable Environment. Plenum Press, New York, pp. 249–260.
- 6.Fredrickson H.L., Perkins,E.J., Bridges,T.S., Tonucci,R.J., Fleming,J.K., Nagel,A., Diedrich,K., Mendez-Tenorio,A., Doktycz,M.J. and Beattie,K.L. (2001) Towards environmental toxicogenomics—development of a flow-through, high-density DNA hybridization array and its application to ecotoxicity assessment. Sci. Total Environ., 274, 137–149. [DOI] [PubMed] [Google Scholar]
- 7.Beattie K.L., Beattie,W.G., Meng,L., Turner,S., Bishop,C., Dao,D., Coral,R., Smith,D. and McIntyre,P. (1995) Advances in genosensors research. Clin. Chem., 41, 700–706. [PubMed] [Google Scholar]
- 8.Greisen K., Loeffelholz,M., Purohit,A. and Leong.D. (1994) PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J. Clin. Microbiol., 32, 335–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wallace W.H., Rice,J.F., White,D.C. and Sayler,G.S. (1994) Distribution of alginate genes in bacterial isolates from corroded metal surfaces. Microb. Ecol., 27, 213–223. [DOI] [PubMed] [Google Scholar]
- 10.Goldberg J.B. and Ohman,D.E. (1984) Cloning and expression in Pseudomonas aeruginosa of gene involved in the production of alginate. J. Bacteriol., 169, 1115–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sambrook J., Fritsh,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 12.Toschka H., Hoepfel,P., Ludwig,W., Schleifer,K.H., Ulbrich,N. and Erdmann,V.A. (1988) Nucleic Acids Res., DDBJ/EMBL/GenBank accession no. X06684. [Google Scholar]
- 13.Anzai Y., Kudo,Y. and Oyaizu,H. (1997) Int. J. Syst. Bacteriol., DDBJ/EMBL/GenBank accession no. D84006. [Google Scholar]
- 14.Moore E.R.B., Mau,M., Arnscheidt,A., Boettger,E.C., Hutson,R.A., Collins,M.D., Van de Peer,Y., De Wachter,R. and Timmis,K.N. (1996) Syst. Appl. Microbiol., DDBJ/EMBL/GenBank accession no. Z76662. [Google Scholar]
- 15.Elomari M., Coroler,L., Hoste,B., Gillis,M., Izard,D. and Leclerc,H. (1996) Int. J. Syst. Bacteriol., DDBJ/EMBL/GenBank accession no. AF064460. [DOI] [PubMed] [Google Scholar]
- 16.Sawada H., Takeuchi,T. and Matsuda,I. (1997) Appl. Environ. Microbiol., DDBJ/EMBL/GenBank accession no. AB001447. [Google Scholar]
- 17.Anzai Y., Kudo,Y. and Oyaizu,H. (1997) Int. J. Syst. Bacteriol., DDBJ/EMBL/GenBank accession no. D84020. [Google Scholar]
- 18.Tsukamoto T. (1998) Direct submission DDBJ/EMBL/GenBank accession no. AB020208.
- 19.Jeanmougin F., Thompson,J.D., Gouy,M., Higgins,D.G. and Gibson,T.J. (1998) Multiple sequence alignment with Clustal X. Trends Biochem Sci., 23, 403–405. [DOI] [PubMed] [Google Scholar]
- 20.SantaLucia J. (1998) A unified view of polymer, dumbbell and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA, 95, 1460–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hicks J.S., Harker,B.W., Beattie,K.L. and Doktycz,M.J. (2001) Modification of an automated liquid-handling system for reagent-jet, nanoliter-level dispensing. Biotechniques, 30, 878–885. [DOI] [PubMed] [Google Scholar]
- 22.Maldonado-Rodriguez R. and Beattie,K.L. (2001) Analysis of nucleic acids by tandem hybridization on oligonucleotide microarrays. In Rampal,J.B. (ed.), DNA Arrays. Methods and Protocols. Humana Press, Totowa, NJ, pp. 157–171. [DOI] [PubMed]
- 23.Maldonado-Rodriguez R., Espinosa-Lara,M., Calixto-Suárez,A., Beattie,W.G. and Beattie,K.L. (1999) Hybridization of glass-tethered oligonucleotide probes to target strands preannealed with labeled auxiliary oligonucleotides. Mol. Biotechnol., 11, 1–12. [DOI] [PubMed] [Google Scholar]
- 24.Allawi H.T. and SantaLucia,J. (1997) Thermodynamics and NMR of internal G-T mismatches in DNA. Biochemistry, 36, 10581–10594. [DOI] [PubMed] [Google Scholar]
- 25.Allawi H.T. and SantaLucia,J. (1998) Nearest neighbor thermodynamic parameters for internal G·A mismatches in DNA. Biochemistry, 37, 2170–2179. [DOI] [PubMed] [Google Scholar]
- 26.Allawi H.T. and SantaLucia,J. (1998b) Thermodynamics of internal C·T mismatches in DNA. Nucleic Acids Res., 26, 2694–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allawi H.T. and SantaLucia,J. (1998c) Nearest-neighbor thermodynamics of internal A-C mismatches in DNA. Sequence dependence and pH effects. Biochemistry, 37, 9435–9444. [DOI] [PubMed] [Google Scholar]
- 28.Peyret N., Ananda,S.P., Allawi,H.T. and SantaLucia,J.,Jr (1999) Nearest-neighbor thermodynamics and NMR of DNA sequences with internal A.A,C.C,G.G and T.T mismatches. Biochemistry, 38, 3468–3477. [DOI] [PubMed] [Google Scholar]
- 29.Beattie W.G., Meng,L., Turner,S., Varma,R.S. and Beattie,K.L. (1995) Hybridization of DNA targets to glass-tethered oligonucleotide probes. Mol. Biotechnol., 4, 213–225. [DOI] [PubMed] [Google Scholar]
- 30.Doktycz M.J., Morris,M.D., Dormady,S.J., Beattie,K.L. and Jacobson,K.B. (1995) Optical melting of 128 octamer DNA duplexes. Effects of base pair location and nearest neighbors on thermal stability. J. Biol. Chem., 270, 8439–8445. [DOI] [PubMed] [Google Scholar]
- 31.Yang M., McGovern,M.E. and Thompson,M. (1997) Genosensor technology and the detection of interfacial nucleic acid chemistry. Anal. Chim. Acta, 346, 259–275. [Google Scholar]
- 32.Southern E., Mir,K. and Shchepinov,M. (1999) Molecular interactions on microarrays. Nature Genet., 21 (suppl.), 5–9. [DOI] [PubMed] [Google Scholar]
- 33.Bavykin S.G., Akowski,J.P., Zakhariev,V.M., Barsky,V.E., Perov,A.N. and Mirzabekov,A.D. (2001) Portable system for microbial sample preparation and oligonucleotide microarray analysis. Appl. Environ. Microbiol., 67, 922–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu W.-T., Mirzabekov,A.D. and Stahl,D.A. (2001) Optimization of an oligonucleotide microchip for microbial identification studies: a non-equilibrium dissociation approach. Environ. Microbiol., 3, 619–629. [DOI] [PubMed] [Google Scholar]
- 35.Small J., Call,D.R., Brockman,F.J., Straub,T.M. and Chandler,D.P. (2001) Direct detection of 16S rRNA in soil extracts by using oligonucleotide microarrays. Appl. Environ. Microbiol., 67, 4708–4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson K.H., Wilson,W.J., Radosevich,J.L., DeSantis,T.Z., Viswanathan,V.S., Kuczmarski,T.A. and Andersen,G.L. (2002) High-density microarray of small-subunit ribosomal DNA probes. Appl. Environ. Microbiol., 68, 2535–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]