Abstract
The fission yeasts are members of the fungal order Schizosaccharomycetales, a candidate deep-diverging group within Ascomycota. Although a great deal of molecular information is available from Schizosaccharomyces pombe, a model eukaryote, very little is available from other members of this group. In order to better characterize mitochondrial genome evolution in this fungal lineage, the mitochondrial DNA (mtDNA) of two additional fission yeasts, Schizosaccharomyces octosporus and Schizosaccharomyces japonicus var. japonicus, was sequenced. Whereas the mtDNA of S.pombe is only 19 431 bp, the mtDNA of S.octosporus is 44 227 bp, and that of S.japonicus var. japonicus is over 80 kb. The size variation of these mtDNAs is due largely to non-coding regions. The gene content in the latter two mtDNAs is almost identical to that of the completely sequenced S.pombe mtDNA, which encodes 25 tRNA species, the large and small mitochondrial ribosomal RNAs (rnl and rns), the RNA component of mitochondrial RNaseP (rnpB), mitochondrial small subunit ribosomal protein 3 (rps3), cytochrome oxidase subunits 1, 2 and 3 (cox1, cox2 and cox3) and ATP-synthase subunits 6, 8 and 9 (atp6, atp8 and atp9). However, trnI2(cau) (C modified to lysidine) is absent in the S.octosporus mtDNA, as are corresponding ATA codons in its protein-coding genes, and rps3 and rnpB are not found in the mtDNA of S.japonicus var. japonicus. The mtDNA of S.octosporus contains five double hairpin elements, the first report of these elements in an ascomycete. This study provides further evidence in favor of the mobility of these elements, and supports their role in mitochondrial genome rearrangement. The results of our phylogenetic analysis support the monophyly of the Schizosaccharomycetales, but question their grouping within the Archiascomycota.
INTRODUCTION
The fission yeasts are a small group of unicellular, saprophytic organisms classified within the higher fungal division Ascomycota, order Schizosaccharomycetales (1,2), in the single genus Schizosaccharomyces. The placement of Schizosaccharomyces within Ascomycota is based on a set of fundamental features that delimit this division, including life cycle, mode of ascospore formation and non-centric mitosis (3). Furthermore, the morphology and development of the sexual sporangium of the fission yeasts is similar to the ascus of other ascomycetes (4,5). Phylogenetic studies including a broad range of species from all fungal lineages (6,7) consistently support this assignment.
Schizosaccharomyces pombe is an increasingly used organism for research, due largely to the many molecular genetic techniques available (8), and the availability of its complete nuclear genome sequence (9). Several features highlight the utility of this organism as an alternative model system to the budding yeast, Saccharomyces cerevisiae. For example, S.pombe has served as an important model organism in genetic and molecular studies of the cell cycle (10), as it displays features, such as a distinct G2 phase and visible chromosome condensation, typical of other eukaryotes but not found in budding yeasts (11). Of the many fission yeast species described since the initial description of S.pombe, most have been found to be conspecific with one of three species: S.pombe Lindner (12), Schizosaccharomyces octosporus Beijerinck (13) and Schizosaccharomyces japonicus Yukawa and Maki (14). Unfortunately, little molecular data are available from the latter two Schizosaccharomyces species. Comparative data from closely related organisms can allow the identification of, for example, regulatory elements or genes in the DNA sequence that would not be recognized by comparing more distantly related organisms (15–17).
The mitochondrial DNA (mtDNA) of S.pombe strain 50 h– has been completely sequenced (18,19); however, the only data available from the mitochondrial genomes of other fission yeasts are the sequence of the mitochondrial small ribosomal subunit from S.japonicus var. versatilis (GenBank accession number X72804). In order to provide comparative data from other members of Schizosaccharomyces to better define the mitochondrial genomes in this group, and to better resolve the fungal phylogeny, the mtDNAs of two additional fission yeasts, S.octosporus and S.japonicus var. japonicus, were sequenced.
The current taxonomic view of Ascomycota, based on molecular phylogenetic analyses of small subunit rRNA sequences (20,21), shows three major lineages: the euascomycetes (filamentous ascomycetes), the hemiascomycetes (budding yeasts) and the archiascomycetes (a heterogeneous group to which the genus Schizosaccharomyces is considered to belong; ‘Taphrinomycotina’, according to GenBank). The euascomycetes and hemiascomycetes form a monophyletic group, while the archiascomycetes are the most ancestral lineage of the ascomycetes. This basic topology of the phylum Ascomycota is generally consistent throughout most small subunit rRNA- and RPB2- (beta subunit of RNA polymerase II) (22) based analyses. However, there is no significant statistical support for this topology (23,24). In addition, certain phylogenies based on amino acid sequences (25,26) have placed S.pombe at the base of the budding yeasts, although again with poor support. In contrast, phylogenetic analyses based on multiple concatenated mitochondrial protein sequences have indicated that the position of S.pombe is at the base of the budding yeasts, often with good bootstrap support (6,24). Although both the budding yeasts and S.pombe form long branches in these analyses, maximum likelihood-based analyses that are known to minimize long-branch attraction artifacts (27) increase support for this topology (24). The present study further explores the position of Schizosaccharomyces within the Fungi.
MATERIALS AND METHODS
Strains, culture conditions and preparation of mtDNA
The strains used were S.octosporus (ATCC 2479) and S.japonicus var. japonicus (ATCC 10660). Cells were grown using Yeast Standard Medium, consisting of 1% yeast extract, 1 g/l KH2PO4 and 3% glycerol (for S.japonicus var. japonicus, 3% glucose was used). Cultures were grown for 24–48 h with shaking (100 r.p.m.) at 30°C. Purification of mtDNA was performed using 20–30 g (wet weight) of cells, harvested in the early stationary phase by centrifugation. After resuspension in a sorbitol buffer (0.6 M sorbitol, 5 mM EDTA, 50 mM Tris pH 7.4), the cells were broken mechanically by shaking with glass beads, and a crude mitochondrial fraction was isolated by differential centrifugation. The mitochondrial fraction was lysed in the presence of 1% SDS and 100 µg/ml proteinase K, at 50°C for 1 h. SDS was subsequently eliminated from the lysate by addition of 1 M NaCl, and after 1 h on ice, the precipitate (SDS–protein complex) was removed by centrifugation. The total nucleic acids were fractionated on a CsCl gradient (1.1 g/ml, 40 000 r.p.m. for 48 h) in the presence of 10 µg/ml bis-benzimide (Hoechst dye 33258). The upper band (A + T-rich DNA) was extracted and re-centrifuged in one or two subsequent CsCl gradients. Yields of 0.5–3 µg DNA were typical.
Cloning and sequencing of mtDNA
mtDNA was physically sheared by nebulization (28), and a size fraction of 500–3000 bp was recovered after agarose gel electrophoresis. DNAs were incubated in the presence of dNTPs, the Klenow fragment of DNA polymerase I and T7 DNA polymerase to generate blunt ends, and cloned into the SmaI site of a modified Bluescript II KS+ vector with a shortened multi-cloning site (pFBS). Recombinant plasmids containing mtDNA inserts were identified by colony hybridization using mtDNA as a probe. DNA sequencing was performed by the dideoxy chain termination method (29), using single-stranded DNA as template and [α-35S]dATP as label. Labeled DNA fragments were subjected to electrophoresis in 4% polyacrylamide gels, dried onto glass plates (30), and autoradiographed. Automated sequencing was performed on a LiCor 4000L apparatus, using an end-labeled primer and a cycle sequencing protocol (Amersham).
The mtDNA sequences of S.octosporus and S.japonicus var. japonicus have been deposited in GenBank (accession numbers AF275271 and AF547983, respectively).
Data analysis
Sequences were assembled using GAP (31) and custom-made command line interfaces, and stored in the MasterFile format (http://megasun.bch.umontreal.ca/ogmp/masterfile/ intro.html).
Sequence analysis was performed on SUN workstations, using software developed by the OGMP (Organelle Genome Megasequencing Project; see http://megasun.bch.umontreal. ca/ogmp/ogmpid.html) and others (such as the Staden sequence analysis package). The FASTA program (32) was used for searches of local databases. Sequence similarity searches were also performed at the National Center for Biotechnology Information (NCBI), using the BLAST network service (33).
RESULTS AND DISCUSSION
Genome size, gene content and gene order
The mtDNA of S.pombe (19) (Fig. 1A) is 19 431 bp, and codes for 25 tRNA species (capable of recognizing all codons, probably including the rarely used UGA at low efficiency; see below), the mitochondrial small and large ribosomal RNAs (rns and rnl, respectively), the RNA component of mitochondrial RNase P (rnpB; E.Seif and B.F.Lang, unpublished), the mitochondrial small subunit ribosomal protein 3 (rps3) (17), cytochrome oxidase subunits 1, 2 and 3 (cox1, cox2 and cox3), apocytochrome b (cob), and ATP-synthase subunits 6, 8 and 9 (atp6, atp8 and atp9) (Table 1). The A + T content of this genome is 70.0%, and all genes are expressed from the same DNA strand.
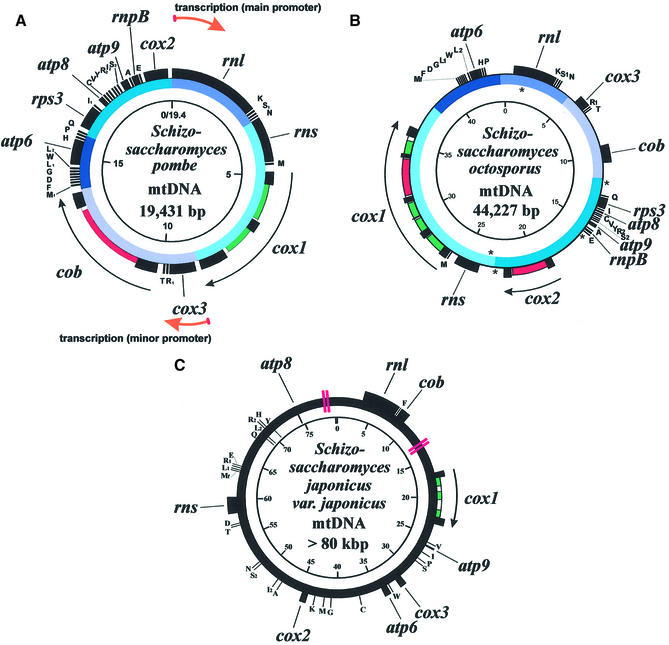
Figure 1.
The mtDNAs of (A) S.pombe, (B) S.octosporus and (C) S.japonicus var. japonicus. Inner circle gives scale in kb. Colors on the intermediate circles in the mtDNAs of S.pombe and S.octosporus show regions of gene order conservation. Outer circle indicates the location of genes, exons (black), introns (white) and intronic ORFs (group I, green; group II, magenta). Location of DHEs in the mtDNA of S.octosporus is indicated by asterisks. Double red lines in the mtDNA of S.japonicus var. japonicus indicate regions of ambiguous sequence assembly.
Table 1. Gene content and introns in Schizosaccharomyces mtDNAs.
| Genes | Schizosaccharomyces pombe | Schizosaccharomyces octosporus | Schizosaccharomyces japonicus var. japonicus |
|---|---|---|---|
| rns, rnl | ▪ | ▪ | ▪ |
| atp6, 8, 9 | ▪ | ▪ | ▪ |
| cob | ▪ | ▪ | ▪ |
| cox1, 2, 3 | ▪ | ▪ | ▪ |
| trnA-W | 25 | 24 | 25 |
| rnpB | ▪ | ▪ | □ |
| rps3 | ▪ | ▪ | □ |
| Group I introns | 2 | 4 | 2 |
| Group II introns | 1 | 2 | 0 |
Filled squares indicate the presence; open squares the absence of a gene or genes.
We have completely sequenced the mtDNA of S.octosporus (Fig. 1B). This genome is a circular-mapping molecule of 44 227 bp, i.e. more than twice the size of the mtDNA of S.pombe. The A + T content of this genome is 76.0%, and all genes are encoded on the same DNA strand, as in S.pombe. The only observed difference in gene content between the mtDNAs of S.pombe and S.octosporus is the absence of the trnI2(cau) gene (C residue modified to lysidine) in that of S.octosporus (discussed below). The gene order in this mtDNA is quite similar to that observed in S.pombe, with five blocks of gene order conservation comprising the entire genome [excluding the absent trnI2(cau) gene] (see color code in Fig. 1).
We have also sequenced the mtDNA of S.japonicus var. japonicus (Fig. 1C), a genome that is about four times as large as that of S.pombe. The sequence assembly of two regions of this mtDNA remains ambiguous, due to the presence of long repeat sequences (see Fig. 1C for locations; the version of the sequence assembly with the lowest number of conflicts is shown). The A + T content of this genome is 80.2% and, in contrast to the mtDNAs of S.pombe and S.octosporus, genes are encoded on both DNA strands. We have identified the same set of genes in this genome as in that of S.pombe, except that rps3 and rnpB are not present (which is not surprising, as these two genes have a patchy distribution in fungal mtDNAs) (17; E.Seif and B.F.Lang, unpublished). Extensive sequencing of clones from a random mtDNA library did not suggest the presence of additional genes within the repeat regions. The gene order in this mtDNA is completely dissimilar to that observed in the mtDNA of either S.pombe or S.octosporus. The gene order information derived from this study indicates that S.pombe and S.octosporus are more closely related to each other than either is to S.japonicus, as proposed by Sipiczki (3). The close relationship of S.pombe and S.octosporus is further reflected in sequence similarities of mitochondrial genes, and in phylogenetic analyses grouping them as sister species to the exclusion of S.japonicus (see Fig. 4).
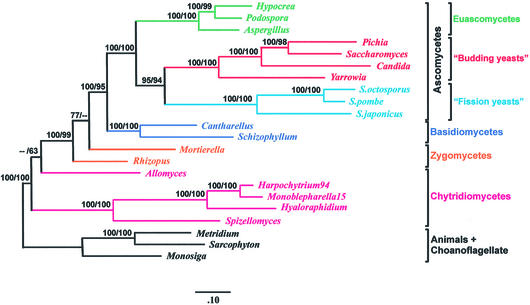
Figure 4.
Phylogenetic analysis based on concatenated mitochondrial proteins. The phylogenetic tree was constructed from unambiguously aligned portions of the concatenated protein sequences of Cox1, 2 and 3, Cob, Atp6 and 9, Nad1, 2, 3, 4, 4L and 5, using a total of 3010 amino acid positions. The topology shown was inferred using ProML (52) and the PMB model of protein evolution (E.Tillier, unpublished), and branch lengths were determined using CodeML, included in the PAML package (53). The alpha value used was 1.05, as determined by CodeML. Maximum likelihood-bootstrap support (percent, first number) was calculated from 100 replicates using SEQBOOT, provided with the PHYLIP package. Distance method bootstrap values (1000 replicates, percent, second number) were calculated using PUZZLE to generate maximum likelihood-distance tables, from which trees were inferred using Weighbor (54). Because distance methods are highly sensitive to lack of sequence information, while ML methods are not, we chose to use the full dataset of 3010 amino acid positions only for the inference with ProML. For the distance tree, we eliminated all Nad protein sequences from the alignment (the corresponding genes are not present in S.cerevisiae and S.pombe), as well as additional gaps in the alignment, retaining a total of 1464 amino acid positions. The tree topologies were similar with both methods, except that in the distance tree, the Zygomycota were found to be monophyletic, with high bootstrap support (95%), while they are paraphyletic (although without significant support) in the ML analysis. Trees inferred by both methods place Schizosaccharomyces at the base of the hemiascomycetes (‘budding yeasts’), with bootstrap support generally considered high (95% in maximum likelihood, 94% in distance). Sequences were obtained from GenBank: Schizophyllum commune (AF402141), Pichia canadensis (D31785), S.cerevisiae (AJ011856), Candida albicans (AF285261), Yarrowia lipolytica (AJ307410), S.octosporus (AF275271), S.pombe (X54421), S.japonicus (AF547983), Hypocrea jecorina (AF447590), Aspergillus nidulans (ODAS1, CAA33481, AAA99207, AAA31737, CAA25707, AAA31736, CAA23994, X15442, P15956, CAA23995, CAA33116, X00790, X15441, X06960, J01387, X01507), P.anserina (X55026), A.macrogynus (U41288), Hyaloraphidium curvatum (AF402142), Spizellomyces punctatus (AF402142), Metridium senile (AF000023), Sarcophyton glaucum (AF064823, AF063191). Protein sequences of Cantharellus cibarius, Monosiga brevicollis, Harpochytrium94, Monoblepharella15, Rhizopus stolonifer and Mortierella verticillata can be downloaded from http://megasun.bch.umontreal.ca/People/lang/FMGP/proteins/. The translations of mitochondrial proteins of the species C.albicans, P.canadensis and Y.lipolytica found in GenBank were found to be incorrect based on multiple alignments. It was found that these species, rather than using the S.cerevisiae mitochondrial genetic code, use the standard genetic code along with UGA-tryptophan. Consequently, we translated the proteins used in these analyses using this genetic code. In addition, intron annotation in H.jecorina, Y.lipolytica and C.albicans was corrected.
Mitochondria of S.japonicus var. japonicus (as well as S.japonicus var. versatilis) are highly reduced in most cytochromes, as determined by cytochrome spectroscopy (34). The data correlate with the inability of members of this species to grow on non-fermentable substrates (35). As all the expected (with respect to S.pombe) mtDNA-encoded genes coding for the translation machinery, respiratory chain and ATP-synthase complexes are present in the S.japonicus var. japonicus mtDNA, these phenotypes probably stem from an extra-mitochondrial source. Some nuclear pet mutants (i.e. deficient in growth on non-fermentable carbon sources) have been shown to have pleiotropic loss of cytochromes, but retain a wild-type mtDNA, in both S.cerevisiae (36) and S.pombe (37,38). It is probable that S.japonicus var. japonicus has acquired such a nuclear mutation.
Finally, most of the size variation (from 17.4 to 24.4 kb) between mitochondrial genomes of naturally occurring S.pombe strains is due to the presence or absence of introns (39). However, the large size variation (between 19 and >80 kb) between the mtDNAs of the three species compared here is due mostly to the size of highly A + T-rich (77.6, 80.7 and 82.0% in S.pombe, S.octosporus and S.japonicus var. japonicus, respectively) non-coding (intergenic) regions. Non-coding DNA accounts for only 11.1% of the mtDNA of S.pombe, whereas it accounts for 49.4% in S.octosporus and 76.5% in S.japonicus var. japonicus. The function of these non-coding sequences, if any, is unknown.
Intron content
The mtDNA of S.pombe contains two group I introns in the cox1 gene, and a single group II intron in the cob gene. Six introns are found in the mtDNA of S.octosporus, five located in the cox1 gene (four group I, one group II) and one group II in the cox2 gene. Two group I introns, both located in the cox1 gene, are found in the mtDNA of S.japonicus var. japonicus. All introns contained within these mtDNAs include open reading frames (ORFs) characteristic of the corresponding intron group; that is, two conserved dodecapeptide (LAGLIDADG) motifs in the group I intronic ORFs (40), and conserved reverse transcriptase sequence motifs in the group II intronic ORFs (41).
The ORFs encoded by both group II introns in the mtDNA of S.octosporus are significantly similar to that encoded by S.pombe cob-I1 (the first intron in the cob gene; FASTA opt scores of more than 400 in all cases). This may indicate the homology of the introns encoding these ORFs. Three sets of homologous group I introns were also found (set1, S.pombe cox1-I1 and S.octosporus cox1-I1; set2, S.octosporus cox1-I2 and S.japonicus var. japonicus cox1-I1; set3, S.pombe cox1-I2, S.octosporus cox1-I3 and S.japonicus var. japonicus cox1-I2). The introns in each set have identical insertion sites within the cox1 gene and encode highly similar ORFs. It is interesting to note that cox1-I2 of S.japonicus var. japonicus contains two ORFs, whereas its homologs (S.pombe cox1-I2 and S.octosporus cox1-I3) contain only one. The data suggest either the acquisition of a second ORF subsequent to the divergence of S.japonicus var. japonicus (mobility of intronic ORFs independent of the intron has been demonstrated in Podospora) (42), or the loss of this ORF from the introns in the other two species. Group I introns encoding two ORFs are uncommon, but have been described previously in the mtDNA of the filamentous ascomycete Podospora anserina (43) and the cellular slime mold Dictyostelium discoideum (44).
The only intron for which a homolog was not found in the other mtDNAs is S.octosporus cox1-I5. This intron is inserted in a unique location in Schizosaccharomyces mtDNAs, and the ORF that it encodes has no significant similarity to any other intronic ORF in the three Schizosaccharomyces species. BLAST searches of the polypeptide encoded by the intronic ORF, however, identified closely related ORFs encoded by P.anserina cox1-I15 (e = 4 × 10–27), D.discoideum cox1/2-I3 (e = 5 × 10–25) and Marchantia polymorpha cox1-I8 (e = 3 × 10–15). The introns containing these ORFs are inserted in identical positions within the cox1 gene (note that the cox1 and cox2 genes are fused in D.discoideum, therefore the delimitation of Cox1 is inferred by protein sequence alignment). The data suggest a common origin of these introns together with their intronic ORFs.
Genetic code
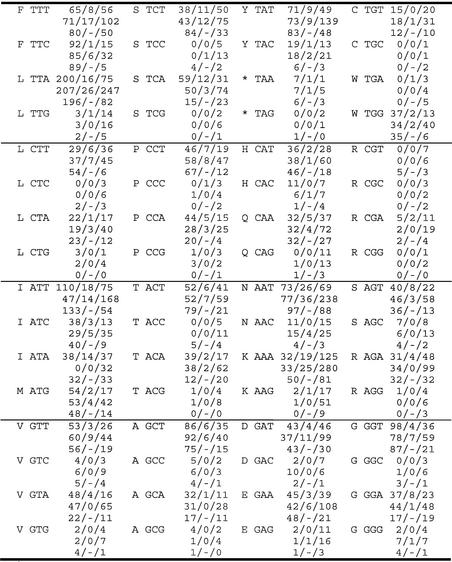
The universal translation code is used in the standard protein-coding mitochondrial genes (cox1, cox2, cox3, cob, atp6, atp8 and atp9) of S.pombe, S.octosporus and S.japonicus var. japonicus (Table 2). TGA codons are not found in these genes, nor is TGA used as a stop codon in these mtDNAs. However, TGA codons are found in rps3 of S.pombe (one of three trp codons is TGA, 33%) and in the intronic ORFs encoded in the mtDNAs of all three species (12 of 71 codons in intronic ORFs are TGA, 17%). If UGA was read as a stop codon during translation, this would result in truncated protein products, possibly compromising mitochondrial function. In fact, a truncated rps3 transcript has been shown to cause a respiratory-deficient phenotype in S.pombe (45). Alternatively, if the deviant genetic code in which UGA = Trp were used in these mtDNAs (as it is in S.cerevisiae), we would expect to find a single tryptophan tRNA (trnW) bearing the anticodon UCA (a modified U in the wobble position allows the recognition of both UGA and UGG by this anticodon). Strangely, the sole trnW in these mtDNAs is trnW(cca), which is able to recognize only UGG codons. We propose that UGA codons in this system are, albeit inefficiently, decoded as tryptophan by trnW(cca). A similar proposal has been made for the mtDNA of the basidiomycete Schizophyllum commune, which encodes only trnW(cca), although over 20% of total tryptophan codons are specified by UGA (6). It is possible that the C in the wobble position of the trnW(cca) anticodon is modified to permit the recognition of UGA (Trp) codons.
Table 2. Codon usage in the mtDNAs of S.pombe, S.octosporus and S.japonicus var. japonicusa.
aCognate amino acid is indicated in one-letter code. Asterix indicates a stop codon. Numbers indicate the total number of codons in standard protein-coding genes/rps3/intronic ORFs in S.pombe (upper numbers), S.octosporus (middle numbers) and S.japonicus var. japonicus (lower numbers).
Based on protein sequence alignments (not shown), ATA codes for isoleucine in S.pombe and S.japonicus var. japonicus (following the universal translation code) as opposed to methionine, as it does in animal and S.cerevisiae mitochondria. However, the mtDNA of S.octosporus lacks trnI2(cau) (C residue modified to lysidine), the tRNA that decodes AUA codons as isoleucine. The lack of this tRNA gene correlates with the absence of ATA codons in the standard protein-coding genes of this mtDNA as well as in the rps3 gene. It would appear that ATA codons and trnI2(cau) were eliminated from the S.octosporus mtDNA subsequent to the divergence of S.octosporus and S.pombe. It is interesting to note that intronic ORFs in the S.octosporus mtDNA do, in fact, contain ATA codons (32 of 235 Ile codons are ATA, 14%). It is possible that a tRNA is imported from the cytoplasm to recognize these codons, or that the intronic ORFs are neither translated nor required for intron splicing.
Finally, a strong tendency for A + T nucleotides is observed in the wobble position of codons in almost all protein-coding genes (including intronic ORFs), in the mtDNAs of all three species (see Table 2). Conversely, there is a tendency to use TTC codons to code for phenylalanine in standard mitochondrial protein-coding genes (266 of 482 Phe codons are TTC, 55%), whereas TTT codons are strongly preferred in rps3 and intronic ORFs (233 of 292 Phe codons are TTT, 80%). The tendency for a C in the wobble position of these codons in standard genes may serve to avoid frame-shifting at runs of U residues during translation, a phenomenon observed in yeast mitochondria (46).
Transcription and RNA transcript processing
The mitochondrial genome of S.pombe is transcribed in two units, from promoters situated immediately upstream of the rnl gene (5′-ATATATGTA-3′), and upstream of the cox3 gene (5′-ATATGTGA-3′) (6,47) (Fig. 1A). Similar sequence motifs have not been identified in either the S.octosporus or S.japonicus var. japonicus mtDNAs. Because of the large size of both of these genomes (from two to more than four times that of S.pombe), more than one promoter is probably required. In addition, since genes are encoded on both strands of the S.japonicus var. japonicus mtDNA, at least two promoters must be present. Capping experiments of the mitochondrial RNAs in these two fission yeast species should be undertaken in order to determine promoter locations.
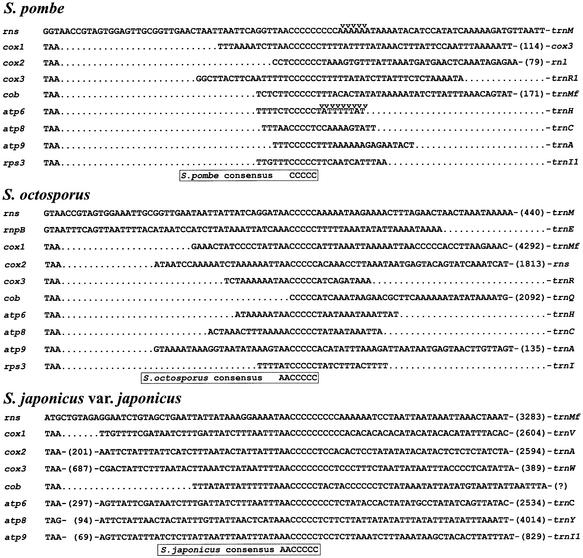
C-rich regions (Fig. 2) are present downstream of all mitochondrial protein-coding genes as well as downstream of rns in S.pombe. S1 nuclease protection signals are clustered immediately downstream of these regions in the atp6 (18) and rnl (48) transcripts, indicating that these C-rich regions are present in the mature transcripts of these genes. Similarly, C-rich regions are present downstream of all protein-coding genes and rns in the mtDNAs of both S.octosporus and S.japonicus var. japonicus, as well as downstream of rnpB in S.octosporus. The consensus sequence of these regions was found to be 5′-AACCCCC-3′ in these two genomes. When S.pombe is included in the comparison, the consensus is reduced to 5′-CCCCC-3′. The function of these sequences is unknown, but they may be involved in RNA tran script stability, protecting RNA 3′-ends from exonuclease degradation. Alternatively, they could act as sites for endonucleolytic cleavage.
Figure 2.
Conserved C-rich regions downstream of genes in the mtDNAs of S.pombe, S.octosporus and S.japonicus var. japonicus. Stop codons are indicated at the left of the alignment. S1 nuclease protection signals are indicated by arrows. Numbers in brackets indicate nucleotides not shown. ‘?’ indicates that the downstream gene is not known due to ambiguous sequence assembly.
Double hairpin elements (DHEs)
DHEs are potentially mobile, palindromic, repeat-containing sequences first described in the mitochondrial genome of the chytridiomycete fungus Allomyces macrogynus (49). These elements have the potential to form secondary structures consisting of two adjacent hairpins, of which one or both may contain up to two mismatches, and generally 3–5 nt loops. Extensive G-C pairing is often observed in at least one stem. DHEs have also been identified in the mitochondrial genomes of other members of the genus Allomyces, in members of the chytridiomycete order Monoblepharidales, and the chytridiomycete Spizellomyces punctatus. They are often found in intergenic regions, within intronic ORFs (often disrupting the reading frame), and sometimes within variable regions of ribosomal RNA genes. Their role in gene expression, if any, is unknown.
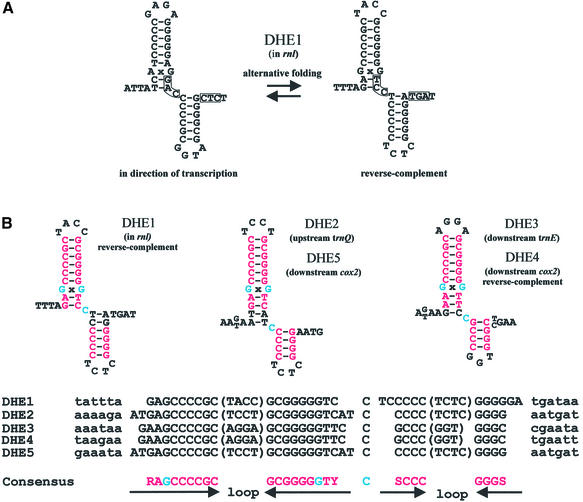
We have identified five DHEs in the mtDNA of S.octosporus (Fig. 3) that meet the consensus model for DHE secondary structure. This is the first such report in an ascomycete mtDNA. Four of these elements are located in intergenic regions (DHE2–5), and one is located in a variable region of the rnl gene (DHE1) (Fig. 1B). Their location is not suggestive of a role in gene expression. These five DHEs have the potential to form alternative structures (Fig. 3A). However, alignment of these elements (Fig. 3B) reveals a strong primary sequence consensus, which was subsequently used to model the secondary structure. The consensus sequence predicts two hairpins containing mostly G-C pairs, the longest of which includes a GxG mismatch. A cytosine separates these two hairpins in all five cases, and each hairpin contains a loop of 3–5 nt. The functional relevance of these features is unknown.
Figure 3.
DHEs in the mtDNA of S.octosporus. (A) Possible alternative secondary structures of DHE1. Nucleotides potentially involved in an alternative interaction are boxed. (B) Predicted secondary structures of S.octosporus DHEs based on a DNA sequence alignment. Universally conserved nucleotides are indicated in red, except the GxG mismatches and the C separating the two hairpins, which are indicated in blue. DHEs which are not found in the direction of transcription are labeled reverse-complement. R, purine; Y, pyrimidine; S, G or C.
Two sets of identical DHEs (DHE2 and 5, and DHE3 and 4) occur in distant locations in the S.octosporus mtDNA. This observation supports the mobility of DHEs, as was proposed for those found in the genus Allomyces (49). The mechanism for mobility has not yet been elucidated. The fact that some DHEs (DHE1 and 4) meet the overall consensus only when reverse-complemented (relative to the general transcription direction) suggests that it does not likely occur via an RNA intermediate. The absence of DHEs in the mtDNAs of S.pombe and S.japonicus var. japonicus (and in all other analyzed ascomycete mtDNAs) suggests that DHEs were acquired laterally in S.octosporus, rather than vertically from a common ancestor.
DHEs have been implicated in mtDNA rearrangements in Allomyces (49). Similarly, three of the five DHEs (DHE2, 4 and 5) in the mtDNA of S.octosporus are located near predicted sites of rearrangement with respect to the mtDNA of S.pombe (see Fig. 1A and B). Further, DHE3 is located downstream of trnE, a region of the mtDNA which is apparently the result of a duplication event involving the trnE gene and the 5′ section of the cox2 gene (not shown). This duplication has resulted in a cox2 pseudogene (as well as a tRNA that completely lacks a D-stem, showing high similarity to trnE, although it has the anticodon AAA) upstream of the intact cox2 gene. These results may indicate that the highly similar DHE sequences serve as preferential recombination sites, as do GC-rich palindromes in yeast (50).
Phylogenetic analysis
In order to determine whether additional data from the genus Schizosaccharomyces would add support for the placement of S.pombe with the hemiascomycetes (budding yeasts; e.g. S.cerevisiae), to the exclusion of the euascomycetes (e.g. P.anserina), we inferred phylogenetic trees using the amino acid sequences of 12 concatenated mitochondrial proteins (see legend of Fig. 4 for details). Our topology (Fig. 4) places Schizosaccharomyces at the base of the hemiascomycetes with a bootstrap support value of either 95% (with a maximum likelihood-based method) or 94% (with a distance-based method). Analysis of the same set of proteins and species, but excluding S.japonicus var. japonicus and S.octosporus produced bootstrap support values of either 94% (maximum likelihood) or 92% (distance method) for this placement of Schizosaccharomyces. Likelihood ratio tests were subsequently used for rigorous statistical testing of alternative topologies (Table 3). With the Approximately Unbiased test (AU) (51), the commonly accepted topology, in which Schizosaccharomyces branches at the base of the Ascomycota, had a P-value of 0.071, close to the accepted strict confidence level of 0.05 that is required to reject this topology. When only S.pombe is included in the dataset, the P-value for this standard topology was 0.158 using the AU test, far from permitting the rejection of this topology. Therefore, although addition of the data from these two additional Schizosaccharomyces species to mitochondrial protein-based phylogenetic analysis does lend more support to the placement of S.pombe at the base of the hemiascomycete branch, it is not possible to confidently reject the standard topology in which S.pombe diverges prior to the separation of the hemiascomycetes and the euascomycetes, with these data.
Table 3. Likelihood tests of alternate topologies.
| Tree topology | Dli | NP | AU | wKH | wSH |
|---|---|---|---|---|---|
| Best tree (Fig. 4; yeasts/schizos monophyletic) | –17.9 | 0.942 | 0.946 | 0.942 | 0.999 |
| (a) Yeasts/euascos monophyl, basidios below schizos | 17.9 | 0.056 | 0.071 | 0.058 | 0.228 |
| (b) Schizos/euascos monophyl, basidios below yeasts | 27.6 | 0.000 | 0.000 | 0.003 | 0.010 |
| (c) Yeasts/euascos monophyl, schizos below basidios | 46.9 | 0.001 | 0.002 | 0.005 | 0.030 |
| (d) Yeasts/euascos monophyl, schizos/basidios monophyl | 49.6 | 0.000 | 0.000 | 0.003 | 0.015 |
| (e) Yeasts/schizos monophyl, euascos below basidios | 82.3 | 0.000 | 0.000 | 0.000 | 0.000 |
| (f) Yeasts/schizos monophyl, euascos/basidios monophyl | 83.6 | 0.000 | 0.000 | 0.000 | 0.000 |
| (g) Yeasts/basidios monophyl, schizos/euascos monophyl | 116.9 | 0.000 | 0.000 | 0.000 | 0.000 |
| (h) Yeasts/basidios monophyl, euascos below schizos | 120.4 | 0.000 | 0.000 | 0.000 | 0.000 |
| (i) Schizos/euascos monophyl, yeasts below basidios | 123.1 | 0.000 | 0.000 | 0.000 | 0.000 |
| (j) Yeasts/basidios monophyl, schizos below euascos | 123.4 | 0.000 | 0.000 | 0.000 | 0.000 |
| (k) Schizos/basidios monophyl, euascos below yeasts | 124.5 | 0.000 | 0.000 | 0.000 | 0.000 |
| (l) Euascos/basidios monophyl, schizos below yeasts | 126.2 | 0.000 | 0.000 | 0.000 | 0.000 |
| (m) Euascos/basidios monophyl, yeasts below schizos | 127.8 | 0.000 | 0.000 | 0.000 | 0.000 |
| (n) Schizos/basidios monophyl, yeasts below euascos | 131.6 | 0.000 | 0.000 | 0.000 | 0.000 |
Statistical tests to evaluate phylogenetic placement of Schizosaccharomyces among the Fungi. The values represent, from left to right, the likelihood distance (Dli), the bootstrap probability of selection (NP), the P-value of the Approximately Unbiased test (AU), the weighted Kishino–Hasegawa test (wKH), and the weighted Shimodaira–Hasegawa test (wSH). The Dli indicates the maximum difference in absolute log-likelihood scores between the topology in question and other tested topologies. The remaining four tests are commonly used likelihood ratio tests, and are each subject to various forms of bias. The result is that the NP and wKH tests lead to overconfidence in the best tree, while wSH is overly conservative, and tends to include many trees in the confidence set. The AU test is adjusted for the bias of the NP and wKH tests, but is less conservative than wSH. Therefore, the likelihood ratio test currently recommended by Shimodaira (51) is AU. All tests were performed as described in Shimodaira (51), and evaluate all possible arrangements of the hemiascomycetes (‘yeasts’), euascomycetes (‘euascos’), basidiomycetes (‘basidios’) and the Schizosaccharomyces genus (‘schizos’). Using all four tests, both the best ML topology (shown in Fig. 4) and tree (a), in which Schizosaccharomyces is placed at the base of the ascomycetes (the topology produced in most SSU rRNA-based analyses) are within the confidence set (P = 0.05). More data from species considered to belong to the archiascomycete lineage will probably eliminate topology (a) at this strict confidence level. Note: ‘monophyl’ indicates that the taxa form a monophyletic group.
CONCLUSIONS
The comparative analysis presented here allows a more comprehensive understanding of the mitochondrial genomes found in the fungal genus Schizosaccharomyces. Current work is focused on the mtDNAs of other potentially deep-diverging members of Ascomycota (archiascomycetes, such Saitoella or members of the genus Taphrina), as well as members of Basidiomycota, in order to better characterize mitochondrial evolution in the higher fungi, and to test the hypothesis that archiascomycetes are a monophyletic lineage (21).
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs D. Spencer and A. Lohan for comments on the manuscript, and Z. Wang for excellent technical assistance. We also thank Dr M. Sipiczki for kindly providing the S.japonicus var. japonicus strain used in this study. This investigation was supported by the Canadian Institute of Health Research (grants MT-14028 and MOP 42475; BFL), the Canadian Institute for Advanced Research (CIAR; BFL), a generous academic equipment grant by SUN Microsystems (Palo Alto, CA; BFL), and the donation of an automatic sequencer by LiCor (Lincoln, NE; BFL).
REFERENCES
- 1.Prillinger H., Dörfer,C.H., Laaser,G., Eckerlein,B. and Lehle,L. (1990) Ein Beitrag zur Systematik und Entwicklungsbiologie der höheren Pilze: Hefe-Typen der Basidiomyceten. Teil I: Schizosaccharomycetales, Protomyces. Typ. Z. Mykol., 56, 219–250. [Google Scholar]
- 2.Erickson E., Svedskog,A. and Landvik,S. (1993) Molecular evidence for the evolutionary hiatus between Saccharomyces cerevisiae and Schizosaccharomyces pombe. Systema Ascomycetum, 11, 119–162. [Google Scholar]
- 3.Sipiczki M. (1995) Phylogenesis of fission yeasts. Contradictions surrounding the origin of a century old genus. Antonie Leeuwenhoek, 68, 119–149. [DOI] [PubMed] [Google Scholar]
- 4.Tanaka K. and Hirata,A. (1982) Ascospore development in the fission yeast Schizosaccharomyces pombe and S. japonicus. J. Cell Sci., 56, 263–279. [DOI] [PubMed] [Google Scholar]
- 5.Sipiczki M. (1983) Diploid protoplasts in Schizosaccharomyces pombe: formation, growth and sporulation. Can. J. Microbiol., 29, 593–595. [Google Scholar]
- 6.Paquin B., Laforest,M.-J., Forget,L., Roewer,I., Wang,Z., Longcore,J. and Lang,B.F. (1997) The fungal mitochondrial genome project: evolution of fungal mitochondrial genomes and their gene expression. Curr. Genet., 31, 380–395. [DOI] [PubMed] [Google Scholar]
- 7.Keeling P.J., Luker,M.A. and Palmer,J.D. (2000) Evidence from beta-tubulin phylogeny that Microsporidia evolved from within the fungi. Mol. Biol. Evol., 17, 23–31. [DOI] [PubMed] [Google Scholar]
- 8.Moreno S., Klair,A. and Nurse,P. (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol., 194, 795–823. [DOI] [PubMed] [Google Scholar]
- 9.Wood V., Gwilliam,R., Rajandream,M.A., Lyne,M., Lyne,R., Stewart,A., Sgouros,J., Peat,N., Hayles,J., Baker,S. et al. (2002) The genome sequence of Schizosaccharomyces pombe. Nature, 415, 871–880. [DOI] [PubMed] [Google Scholar]
- 10.Forsberg S.L. and Nurse,P. (1991) Cell cycle regulation in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe. Annu. Rev. Cell. Biol., 7, 227–256. [DOI] [PubMed] [Google Scholar]
- 11.Taylor J.W., Bowman,B., Berbee,M.L. and White,T.J. (1993) Fungal model organisms: phylogenetics of Saccharomyces, Aspergillus and Neurospora. Syst. Biol., 42, 440–457. [Google Scholar]
- 12.Lindner P. (1893) Schizosaccharomyces pombe n. sp., neuer Gärungserreger. Wochenschrift für Brauerei, 10, 1298–1300. [Google Scholar]
- 13.Beijerinck M.W. (1894) Schizosaccharomyces octosporus, eine achtsporige Alkoholhefe. Zentralblatt Bakteriologie Parasitenkunde, 16, 49–58. [Google Scholar]
- 14.Yukawa M. and Maki,T. (1931) Schizosaccharomyces japonicus nov. spec. La Bul. Sci. Falkultato Terkultura, Kjusu Imp. Univ., Fukuoka, Japan, 4, pp. 218–226.
- 15.Gray M.W., Lang,B.F., Cedergren,R., Golding,G.B., Lemieux,C., Sankoff,D., Turmel,M., Brossard,N., Delage,E., Littlejohn,T.G., Plante,I., Rioux,P., Saint-Louis,D., Zhu,Y. and Burger,G. (1998) Genome structure and gene content in protist mitochondrial DNAs. Nucleic Acids Res., 26, 865–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chesnick J.M., Goff,M., Graham,J., Ocampo,C., Lang,B.F., Seif,E. and Burger,G. (2000) The mitochondrial genome of the stramenopile alga Chrysodidymus synuroideus. Complete sequence, gene content and genome organization. Nucleic Acids Res., 28, 2512–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bullerwell C.E., Burger,G. and Lang,B.F. (2000) A novel motif for identifying Rps3 homologs in fungal mitochondrial genomes. Trends Biochem. Sci., 25, 363–365. [DOI] [PubMed] [Google Scholar]
- 18.Lang B.F., Ahne,F., Distler,S., Trinkl,H., Kaudewitz,F. and Wolf,K. (1983) Sequence of the mitochondrial DNA, arrangement of genes and processing of their transcripts in Schizosaccharomyces pombe. In Nasim,A., Young,P. and Johnston,B.F. (eds), Molecular Biology of the Fission Yeast. Academic Press, San Diego, pp. 313–329.
- 19.Lang B.F. (1993) The mitochondrial genome of Schizosaccharomyces pombe. In O’Brien,S.J. (ed.), Genetic Maps, 6th Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, pp. 3118–3119.
- 20.Nishida H. and Sugiyama,J. (1993) Phylogenetic relationships among Taphrina, Saitoella and other higher fungi. Mol. Biol. Evol., 10, 431–436. [DOI] [PubMed] [Google Scholar]
- 21.Nishida H. and Sugiyama,J. (1994) Archiascomycetes: detection of a major new lineage within the Ascomycota. Mycoscience, 35, 361–366. [Google Scholar]
- 22.Liu Y.J., Whelen,S. and Hall,B.D. (1999) Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Mol. Biol. Evol., 16, 1799–1808. [DOI] [PubMed] [Google Scholar]
- 23.Berbee M.L., Carmean,D.A. and Winka,K. (2000) Ribosomal DNA and resolution of branching order among the Ascomycota: how many nucleotides are enough? Mol. Phylogenet. Evol., 17, 337–344. [DOI] [PubMed] [Google Scholar]
- 24.Leigh J., Seif,E., Rodriguez,N., Jacob,Y. and Lang,B.F. (2003) Fungal evolution meets fungal genomics. In Arora,D. (ed), Handbook of Fungal Biotechnology, 2nd Edn. Marcel Dekker Inc., New York, in press.
- 25.Baldauf S.L., Roger,A.J., Wenk-Siefert,I. and Doolittle,W.F. (2000) A kingdom-level phylogeny of eukaryotes based on combined protein data. Science, 290, 972–977. [DOI] [PubMed] [Google Scholar]
- 26.Landvik S., Eriksson,O.E. and Berbee,M.L. (2001) Neolecta—a fungal dinosaur? Evidence from beta-tubulin amino acid sequences. Mycologia, 93, 1151–1163. [Google Scholar]
- 27.Felsenstein J. (1978) Cases in which parsimony and compatibility methods will be positively misleading. Syst. Zool., 27, 27–33. [Google Scholar]
- 28.Okpodu C.M., Robertson,D., Boss,W.F., Togasaki,R.K. and Surzycki,S.J. (1994) Rapid isolation of nuclei from carrot suspension culture cells using a Bionebulizer. Biotechniques, 16, 154–159. [PubMed] [Google Scholar]
- 29.Sanger F., Nicklen,S. and Coulson,A.R. (1977) DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA, 74, 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lang B.F. and Burger,G (1990) A rapid, high resolution DNA sequencing gel system. Anal. Biochem., 188, 176–180. [DOI] [PubMed] [Google Scholar]
- 31.Dear S. and Staden,R. (1991) A sequence assembly and editing program for efficient management of large projects. Nucleic Acids Res., 19, 3907–3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pearson W.R. (1990) Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol., 183, 63–98. [DOI] [PubMed] [Google Scholar]
- 33.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 34.Sipiczki M., Kucsera,J., Ulaszewski,S. and Zsolt,J. (1982) Hybridization studies by crossing and protoplast fusion within the genus Schizosaccharomyces Lindner. J. Gen. Microbiol., 128, 1989–2000. [Google Scholar]
- 35.Bulder E.A. (1964) Induction on petite mutation and inhibition of synthesis of respiratory enzymes in various yeasts. Antonie van Leeuwenhoek, 30, 1–9. [DOI] [PubMed] [Google Scholar]
- 36.Tzagoloff A. and Dieckman,C.L. (1990) PET genes of Saccharomyces cerevisiae. Microbiol. Rev., 54, 211–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolf K., Sebald-Althaus,M., Schweyen,R.J. and Kaudewitz,F. (1971) Nuclear mutations causing pleiotropic loss of cytochrome c activity. Mol. Gen. Genet., 110, 101–109. [DOI] [PubMed] [Google Scholar]
- 38.Goffeau A., Landry,Y., Foury,F. and Briquet,M. (1973) Oligomycin resistance of mitochondrial adenosine triphosphatase in a pleiotropic chromosomal mutant of a ‘petite-negative’ yeast, Schizosaccharomyces pombe. J. Biol. Chem., 248, 7097–7105. [PubMed] [Google Scholar]
- 39.Zimmer M., Welser,F., Oraler,G. and Wolf,K. (1987) Distribution of mitochondrial introns in the species Schizosaccharomyces pombe and the origin of the group II intron in the gene encoding apocytochrome b. Curr. Genet., 12, 329–336. [DOI] [PubMed] [Google Scholar]
- 40.Michel F., Jacquier,A. and Dujon,B. (1982) Comparisons of fungal mitochondrial introns reveals extensive homologies in RNA secondary structure. Biochimie, 64, 867–881. [DOI] [PubMed] [Google Scholar]
- 41.Michel F. and Lang,B.F. (1985) Mitochondrial class II introns encode proteins related to the reverse transcriptases of retroviruses. Nature, 316, 641–643. [DOI] [PubMed] [Google Scholar]
- 42.Sellem C.H. and Belcour,L. (1997) Intron open reading frames as mobile elements and evolution of a group I intron. Mol. Biol. Evol., 14, 518–526. [DOI] [PubMed] [Google Scholar]
- 43.Cummings D.J., Michel,F. and McNally,K.L. (1989) DNA sequence analysis of the 24.5 kilobase pair cytochrome oxidase subunit I mitochondrial gene from Podospora anserina: a gene with sixteen introns. Curr. Genet., 16, 381–406. [DOI] [PubMed] [Google Scholar]
- 44.Ogawa S., Matsuo,K., Angata,K., Yanagisawa,K. and Tanaka,Y. (1997) Group-I introns in the cytochrome c oxidase genes of Dictyostelium discoideum: two related ORFs in one loop of a group-I intron, a cox1/2 hybrid gene and an unusually large cox3 gene. Curr. Genet., 31, 80–88. [DOI] [PubMed] [Google Scholar]
- 45.Neu R., Goffart,S., Wolf,K. and Schäfer,B. (1998) Relocation of urf a from the mitochondrion to the nucleus cures the mitochondrial mutator phenotype in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet., 258, 389–396. [DOI] [PubMed] [Google Scholar]
- 46.Fox T.D. and Weiss-Brummer,B. (1980) Leaky +1 and –1 frameshift mutations at the same site in a yeast mitochondrial gene. Nature, 288, 60–63. [DOI] [PubMed] [Google Scholar]
- 47.Lang B.F., Cedergren,R. and Gray,M. (1987) The mitochondrial genome of the fission yeast, Schizosaccharomyces pombe: sequence of the large-subunit ribosomal RNA gene, comparison of potential secondary structure in fungal mitochondrial large-subunit rRNAs and evolutionary considerations. Eur. J. Biochem., 169, 527–537. [DOI] [PubMed] [Google Scholar]
- 48.Trinkl H., Lang,B.F. and Wolf,K. (1989) Nucleotide sequence of the gene encoding the small ribosomal RNA in the mitochondrial genome of the fission yeast Schizosaccharomyces pombe. Nucleic Acids Res., 17, 6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paquin B., Laforest,M.J. and Lang,B.F. (2000) Double-hairpin elements in the mitochondrial DNA of Allomyces: evidence for mobility. Mol. Biol. Evol., 17, 1760–1768. [DOI] [PubMed] [Google Scholar]
- 50.Dieckman C.L. and Gandy,B. (1987) Preferential recombination between GC clusters in yeast mitochondrial DNA. EMBO J., 6, 4197–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shimodaira H. (2002) An approximately unbiased test of phylogenetic tree selection. Syst. Biol., 51, 492–508. [DOI] [PubMed] [Google Scholar]
- 52.Felsenstein J. (2002) Phylip (Phylogeny Inference Package) Version 3.6 a 2.1. Distributed by the author. (University of Washington, Seattle).
- 53.Yang Z. (1997) PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci., 13, 555–556. [DOI] [PubMed] [Google Scholar]
- 54.Bruno W.J., Socci,N.D. and Halpern,A.L. (2000) Weighted neighbor joining: a likelihood-based approach to distance-based phylogeny reconstruction. Mol. Biol. Evol., 17, 189–197. [DOI] [PubMed] [Google Scholar]