Abstract
The six minichromosome maintenance proteins (Mcm2–7) are required for both the initiation and elongation of chromosomal DNA, ensuring that DNA replication takes place once, and only once, during the S phase. Here we report on the cloning of a new human Mcm gene (hMcm8) and on characterisation of its protein product. The hMcm8 gene contains the central Mcm domain conserved in the Mcm2–7 gene family, and is expressed in a range of cell lines and human tissues. hMcm8 mRNA accumulates during G1/S phase, while hMcm8 protein is detectable throughout the cell cycle. Immunoprecipitation-based studies did not reveal any participation of hMcm8 in the Mcm3/5 and Mcm2/4/6/7 subcomplexes. hMcm8 localises to the nucleus, although it is devoid of a nuclear localisation signal, suggesting that it binds to a nuclear protein. In the nucleus, the hMcm8 structure-bound fraction is detectable in S, but not in G2/M, phase, as for hMcm3. However, unlike hMcm3, the hMcm8 structure-bound fraction is not detectable in G1 phase. Overall, our data identify a new Mcm protein, which does not form part of the Mcm2–7 complex and which is only structure-bound during S phase, thus suggesting its specific role in DNA replication.
INTRODUCTION
DNA replication is an organised and strictly coordinated biological process, which ensures the continuity and preservation of genetic material. This requires that DNA replication takes place once and only once per cell cyle in the S phase and that a given DNA fragment is not amplified more than once (1).
The strategy of eukaryotic cells to restrict DNA replication to one round per cell cycle can be viewed as a concerted effort to regulate the many activities of the Mcm complex (2). The key to this strategy is the periodic recruitment and discharge of the Mcm complex at replication origins, and the temporal separation of pre-replication complex assembly from initiation of DNA synthesis (3). In fact, the current view of replication initiation is that the origins are licensed for firing during G1, but are only activated under cellular conditions that preclude their relicensing (4).
A series of limiting steps has been described, which renders chromatin competent for replication (reviewed in 1,3,5). Formation of the pre-replication complex during the G1 phase of the cell cycle results from the sequential loading of: (i) six origin recognition complex (ORC) proteins, which act as a replication ‘landing pad’ (6–8); (ii) CDC6p and Cdt1 (9,10), which interact with the ORC and are required for loading of minichromosome maintenance (Mcm) proteins; (iii) Mcm proteins on specific DNA regions called ‘origins of replication’ (11,12). Recruitement of the Mcm complex is restricted to the period of time between the exit from mitosis and the initiation of DNA synthesis. It occurs indiscriminately to all replication origins, potentially licensing them for replication, although not all licensed origins are activated at the same time. In fact, the initiation of DNA synthesis appears to be controlled locally, the earliest events taking place at the G1 to S phase transition and later events occurring throughout the S phase.
All six Mcm proteins contain a central region of approximately 200 amino acids of extensive similarity. One element similar to the A motif of the Walker-type NTP-binding sequence and the Mcm signature motif IDEFDKM (13) are found in this region. In addition, an N-terminal zinc finger-type motif CX2CX18/19CX2/4C is present in four of the Mcm proteins (Mcm2, Mcm4, Mcm6 and Mcm7). Homologies in the N- and C-terminals are interspersed. The overall identity between the different Mcm proteins reaches ∼30% and the similarity ∼50% (11). The divergency between the primary structure of the Mcm proteins highlights the specialised functions of each member of the Mcm family. Indeed, inactivation or loss of a member inhibits DNA replication, both in vitro (14–16) and in vivo (17–19).
Cell extracts contain subcomplexes of Mcm proteins, the Mcm3/5 dimer and the Mcm2/4/6/7 tetramer. Subcomplex Mcm4/6/7, but not the six-membered Mcm complex, has exhibited limited helicase activity, only unwinding very short duplex DNA (20,21). However, further support for the hypothesis of Mcm helicase function was provided by the finding that the archeal hexameric Mcm complex behaves like a processive DNA helicase (22–24) and, more recently, by the demonstration that the processive DNA helicase activity of the Mcm4/6/7 complex requires forked DNA structures (25). Mcm proteins have been reported to be involved in transcriptional control through their interaction with RNA II polymerase holoenzyme (26), and to activate transcription through interaction with the transcription activating domain of STAT1 (27). Furthermore, recent evidence has highlighted the role of the Mcm complex in the elongation phase of DNA replication (17).
To date, all human Mcm genes have been cloned as the human counterparts of yeast Mcm genes. In a screening program aimed at isolating cancer-related genes (28), we have analysed the hepatitis B virus (HBV) DNA integration sites in the cellular DNA extracted from human hepatocellular carcinomas. In one tumor, the HBV genome was found to be integrated into a cellular sequence identical to part of a Mcm gene-like sequence on chromosome 20p12.3–13. We show here that the protein encoded by this gene is a new member of the Mcm2/7 protein family which, although it has a central conserved Mcm domain, does not participate in the Mcm3/5 and Mcm2/4/6/7 subcomplexes and only binds to chromatin in the S phase.
MATERIALS AND METHODS
Cloning of a new human Mcm cDNA
Total RNA was extracted from human normal liver using TRIzol (Life Technologies, Gibco BRL, Bethesda, MD). Poly(A)+ RNA was obtained using Dynabeads oligo(dT) (Dynal, France). Double-stranded cDNA was synthetized with the poly(T) primer according to the Marathon Kit instructions (Clontech, Palo Alto, CA). Amplification of the hMcm8 coding sequence was achieved using 5′ATG (5′-ATGAATGGAGAGTATAGAGGC-3′) and 3′STOP (5′-TTACATAGTTTGAAGCTGGTAA-3′) specific primers (ATG and STOP sequences are in bold), designed on the Mcm2/Mcm3/Mcm5 gene-like genomic sequence (accession no. AL035461). Non-coding hMcm8 sequences (5′ and 3′) were obtained using RACE–PCR with primers close to the 5′ATG and 3′STOP primers (5′-CACATTTACCATTGTTTCTCCATCA-3′ and 5′-CTGATGAATTTGGGAACCTAGATTTTGA-3′, respectively) and AP1 adaptor Marathon Kit primers, according to the manufacturer’s instructions. PCR bands were purified using the QiaQuick spin PCR purification kit (Qiagen, Hilden, Germany), cloned into the pGEM-T vector (Boehringer Mannheim) and sequenced to select the longest clones overlapping with the coding sequence. The hMcm8 coding sequence was subcloned into the pcDNA3.1/Myc-His eukaryotic expression vector (Invitrogen, NV Leek, The Netherlands) containing the CMV promoter and a C-terminal Myc epitope (hMcm8-Myc) or into the pET21b prokaryotic expression vector (Novagen, Madison, WI).
Sequence comparisons and phylogenetic analysis
Sequence comparisons and phylogenetic analysis of Mcm family proteins were performed as described elsewhere, using Dbclustal software with manual adjustment of the final alignment (29).
Cell culture, cell synchronisation and transient transfection
The following cell lines were used: Hs27 cells (CRL1634, American Type Culture Collection) derived from human newborn foreskin primary fibroblasts; HeLa cells (CCL-2, American Type Culture Collection, ATCC) derived from human cervix epithelioid carcinoma; HEK-293 (CRL-1573, ATCC) derived from a human embryonic kidney cell line; CCL13 cells (ATCC) derived from liver tumor cells; HuH7 cells derived from a human hepatocellular carcinoma. The cells were grown in DMEM medium supplemented with 10% foetal calf serum (FCS) containing 10 U/µl penicillin and 0.1 mg/ml streptomycin (Life Technologies). Hs27 cells were synchronised in the G0 phase by growth to confluence in DMEM medium supplemented with 10% FCS. Cells were then released into the cell cycle by splitting them in a 1:3 ratio. HeLa cells were blocked in mitosis by exposure for 20 h to 50 ng/ml nocodazole (Sigma). Mitotic cells were detached by gentle pipetting and then re-seeded in drug-free DMEM medium with 10% FCS after three washes in phosphate-buffered saline (PBS). The synchronisation of HeLa cells before entry into the S phase was achieved by adding mimosine (0.5 mM final concentration) (Sigma) for 25 h. Aphidicoline (10 µM final concentration) (Sigma) was added to the medium for 16 h to synchronise cells at early S phase. The synchronisation in the cell cycle was checked by propidium iodide staining of the cells followed by flow cytometry using FACScan analysis (Becton Dickinson) and Lysis II software. Data are presented as histograms showing relative DNA content (x-axis) and cell number (y-axis). Transient transfections were carried out using the Ca2PO4 co-precipitation method (20 µg/10 cm dish).
Northern blot analysis
Northern blot analysis was performed using poly(A)+ RNA, as described elsewhere (30). A PCR fragment corresponding to nucleotides 1–856 of the hMcm8 cDNA sequence or to full-lenght hMcm4 was labelled radioactively with [32P]dCTP using the Megaprime labelling kit (Amersham Pharmacia, UK) and used as a probe.
Production of anti-hMcm8 antibody and recombinant protein expression in Escherichia coli
Polyclonal antibody was raised against a synthetic hMcm8 N-terminal oligopeptide (amino acids 2–17: NGEYRGRGFGRGRFQS) derived from the hMcm8 predicted protein sequence. The 16 amino acid peptide was synthesised and coupled to keyhole limpet haemocyanin. An aliquot of 250 µg of peptide was emulsified in Freund’s complete adjuvant (for the first injection) and subsequently in incomplete adjuvant (for the three other injections) and injected into New Zealand rabbits at 14 day intervals. The specific antibody was purified by affinity chromatography (Hi-Trap Sepharose NHS column).
BL21/Lys E.coli were transformed using the hMcm8-pET21b construct. Expression of the recombinant protein was induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 37°C for 3 h. His-tagged recombinant protein (His-hMcm8) was then purified by chromatography on a NTA–agarose column (Qiagen), according to the manufacturer’s instructions.
Immunoprecipitation and western blotting
For immunoprecipitation, 5 × 106 exponentially growing HeLa cells were washed three times with PBS, scraped and resuspended in ice-cold hypotonic buffer A (1 mM HEPES, pH 7.3, with a mixture of protease inhibitors). After 20 min incubation on a wheel at 4°C and homogenisation with a Dounce homogeniser (100 strokes), cells were centrifuged at 20 000 g for 30 min at 4°C. The cell extract was then cleared by two more centrifugations (20 000 g at 4°C for 30 min). The supernatant was converted to buffer B (10 mM HEPES, pH 7.3, 50 mM NaCl, 10% glycerol) and incubated at 4°C for 2 h with protein A–Sepharose beads (Boehringer Mannheim) and anti-hMcm4 (0.2 mg/ml) or anti-hMcm3 (0.24 mg/ml) antibody (31). After five washes with 1 ml buffer B, 2× SDS–Laemmli buffer was added and the samples were vortexed, boiled for 5 min at 100°C and used for western blotting.
Lysates were loaded at an equal protein concentration, subjected to 8% SDS–PAGE and transferred onto Immobilon-P+ membranes (Millipore, Bedford, MA). The filter was blocked with TBS–Tween buffer (10 mM Tris–HCl, pH 7.5, 100 mM NaCl, 0.2% Tween 20) containing 5% non-fat dried milk, incubated for 1 h with the primary antibody (anti-hMcm8, 1:500; anti-Mcm2–7 antibodies, 1:1000 dilution) and washed three times with TBS–Tween buffer. After incubation for 1 h with the secondary antibody coupled to horseradish peroxidase (1:2000 dilution; Amersham), three TBS–Tween washes were performed. The blots were revealed using the ECL Kit (Amersham Pharmacia) and exposed to Biomax-ML film (Kodak).
Immunofluorescence
HeLa cells were grown on coverslips, fixed with 3.7% paraformaldehyde for 10 min and permeabilised with 0.2% Triton X-100 for 2 min. Non-specific binding sites were blocked with 10% goat serum (Sigma). Immunostaining was performed with anti-hMcm8 antibody (diluted 1:50) for 1 h. After three PBS washes, immunodetection was carried out using secondary antibody conjugated to fluorescein isothiocyanate (FITC) diluted 1:200. After washing three times with PBS, the nuclei were stained with 7-amino-actinomycin D (7-AAD) (Sigma) and the coverslips were mounted and analysed using fluorescence microscopy (Olympus BX60) and confocal laser scanning microscopy (Zeiss LSM 510).
Chromatin binding assay
HeLa cells (7 × 105) were harvested during the exponential phase of growth and incubated for 30 min on ice in a permeabilisation buffer (10 mM HEPES–KOH, pH 7.5, 7 mM MgCl2, 1.5 mM KCl, 1 mM DTT and 1% Triton X-100) with a mixture of protease inhibitors (Boehringer). After centrifugation (2000 g for 30 min at 4°C) the pellet was resuspended in the same volume of permeabilisation buffer with 0, 0.1, 0.2, 0.5, 1 or 1.5 M NaCl and then incubated on ice for 30 min. After centrifugation at 20 000 g for 30 min at 4°C, identical volumes of the supernatant were blotted.
RESULTS
Cloning of the cDNA encoding the hMcm8 protein
In a human hepatocellular carcinoma tissue, we identified the integration of HBV DNA next to a 485 bp sequence identical to a part of the GenBank genomic clone HS967N21 on chromosome 20p12.3–13 (28). FGENES, GENSCAN, CpG island and poly(A) feature analysis of this genomic clone by the Sanger Centre chromosome 20 mapping group showed that this part of the sequence could correspond to a hypothetical gene encoding for a novel Mcm2/Mcm3/Mcm5 family member.
We cloned the cDNA sequence (ATG–STOP) of hMcm8 from human normal liver poly(A)+ RNA using RT–PCR and obtained the 5′ and 3′ non-coding regions by RACE–PCR. The total length of hMcm8 cDNA is 2523 bases (ATG–STOP). Analysis of the hMcm8 cDNA and its amino acid sequence showed a central Mcm conserved domain (Mcm box) containing the Mcm signature IDEFDKM and the Walker A-type NTP-binding site GXXGXGKS (Supplementary Material, Fig. S1). hMcm8 protein also contains a zinc finger motif CX2CX18CX4C, but neither a classical SV40 large T antigen type of nuclear localisation signal (NLS) nor a bipartite type of NLS. The overall content of the basic residues is 12.3%. We have made the mRNA sequence of hMcm8 available in the EMBL Nucleotide Sequence Database (accession no. AJ439063).
The longest open reading frame encodes an 840 amino acid protein highly homologous with P1/Mcm2–7 protein family members (Fig. S1). The homology between hMcm8 protein and other hMcm is 61.1% (hMcm4), 59.3% (hMcm2), 55.7% (hMcm3), 55.0% (hMcm5), 54.5% (hMcm6) and 52.1% (hMcm7). The identity between hMcm8 and other hMcm is 26.5% (hMcm2), 26.4% (hMcm4), 22.6% (hMcm3), 24.0% (hMcm5), 21.7% (hMcm6) and 21.3% (hMcm7). The highest homology between hMcm8 and other hMcms was found in the central region of 210 amino acids (residues 401–611), which contains the Walker A-type NTP-binding site ‘GXXGXGKS’ and the Mcm signature sequence IDEFDKM.
hMcm8 is conserved during evolution
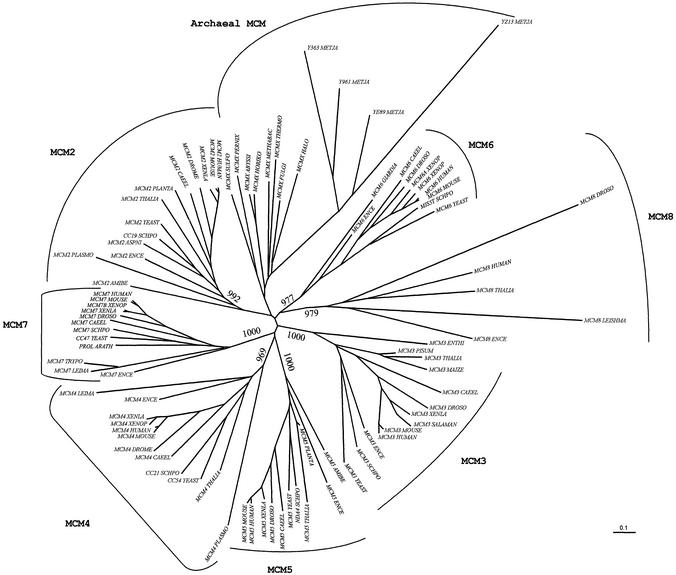
Sequence comparison and phylogenetic analysis of the Mcm protein family and its new member, hMcm8, was performed using Dbclustal software with manual adjustment of the final alignment (29). The tree shown in Figure 1 is based on full-length protein sequences. A reduced version of the multiple alignment excluding the variable N- and C-termini resulted in a similar tree (data not shown). Homologues of hMcm8 protein were found in Encephalitozoon cuniculi, Leishmania major, Arabidopsis thaliana and Drosophila. Furthermore, we were able to identify an EST sequence of Xenopus laevis and a peptide sequence of Mus musculus highly homologous to the hMcm8 sequence. We could not find any yeast or Caenorhabditis elegans homologues of the hMcm8 protein. The accession numbers of the Mcm sequences used for this analysis and the sequence alignments are available on the web site www-igbmc.u-strasbg.fr/BioInfo/Mcm.
Figure 1.
Phylogenetic tree of Mcm protein sequences. The bootstrapped non-rooted tree was created from the NJ algorithm implemented in the Clustal software with manual adjustment of the final alignment. The bootstrap values of the branch-root of each Mcm sub-family are shown when these values are higher than 800 over the 1000 bootstraps performed.
Characterisation of anti-hMcm8 antibodies
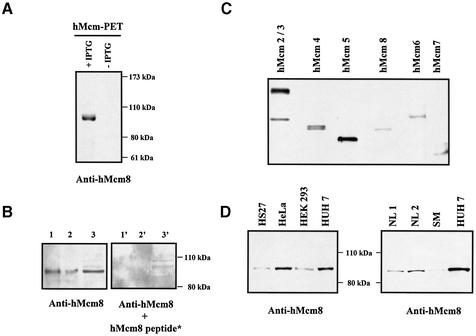
In order to detect the protein expression of hMcm8, we produced a polyclonal antibody raised against a 16 amino acid synthetic peptide located in the N-terminal part of the protein (amino acids 2–17). This antibody was then characterised by subcloning the hMcm8 cDNA sequence into a prokaryotic and a eukaryotic expression vector. The anti-hMcm8 antibody recognised the recombinant hMcm8 protein produced in bacteria (Fig. 2A) or overexpressed as a myc-tagged protein in the CCL13 cell line (data not shown).
Figure 2.
Characterisation of the anti-hMcm8 antibody. (A) Immunoblotting with anti-hMcm8 antibody of E.coli whole cell lysates harbouring the hMcm8 full-length prokaryotic expression vector after (+IPTG) or before (–IPTG) induction with IPTG. (B) Immunoblotting of the E.coli whole cell lysate expressing the hMcm8 recombinant protein (lanes 1, 1′, 2 and 2′) and Hs27 whole cell lysate (30 µg) (lanes 3 and 3′) with the anti-Mcm8 antibody. hMcm8 protein detection without (lanes 1, 2 and 3) or with (lanes 1′, 2′ and 3′) adsorption of the antibody using the 16 amino acid synthetic hMcm8 peptide. (C) Immunoblotting of the HeLa whole cell lysates (30 µg/lane) with human Mcm specific antibodies: anti-hMcm2 (top band) and anti-hMcm3, anti-hMcm4, anti-hMcm5, anti-hMcm8, anti-hMcm6 and anti-hMcm7. (D) Immunoblot analysis of whole cell lysates of ∼1 × 105 cells (Hs27, HeLa, HEK-293 and Huh7 cell lines) or 50 µg human tissue protein extracts (NL, human normal liver; SM, human skeletal muscle) with anti-hMcm8 antibody.
To rule out any cross-reaction between the anti-hMcm8 antibody and human Mcm2–7 proteins, we performed a western blot analysis using HeLa total cell extract, and then hybridised the stripes corresponding to parallel wells using anti-hMcm8 antibody or antibodies against other known Mcm proteins. The anti-hMcm8 antibody detected a single 95 kDa band (Fig. 2C), and no other band corresponding to the size of the other hMcm was detected after longer exposures (data not shown). The hMcm of a most similar size was hMcm4, with a molecular weight of 97 kDa; however, the hMcm4 band appeared as a double band corresponding to the differentially phosphorylated forms of hMcm4 (32), and the unique size of hMcm8 was slightly smaller than the lower band of hMcm4. Furthermore, the adsorption of anti-hMcm8 antibody with the 16 amino acid synthetic peptide (used to raise the antibody) abolished detection of both the recombinant and endogenous hMcm8 proteins (Fig. 2B). We therefore concluded that the polyclonal anti-hMcm8 antibody specifically recognises the hMcm8 protein.
Using this antibody, we detected a unique, endogenous 95 kDa band in fibroblasts (Hs27) and tumor-derived human cell lines (HeLa, HEK-293 and Huh7) as well as in normal human liver and skeletal muscle (Fig. 2D), which could be abolished by the adsorption test (data not shown). It is noteworthy that the accumulation of hMcm8 protein was higher in a proliferative, liver-derived, tumor cell line (HuH7) than in the corresponding non-proliferative tissue (normal liver).
hMcm8 mRNA is expressed during late G1 to S phase
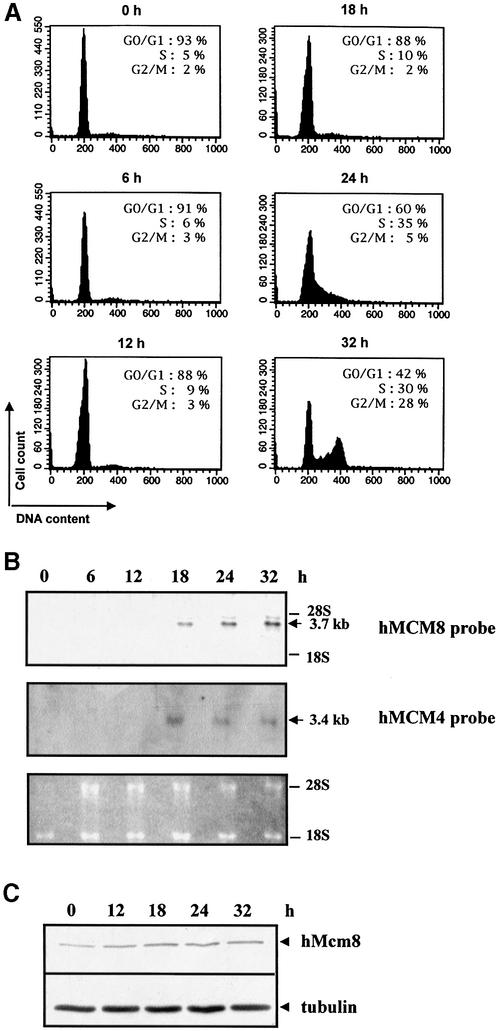
Cells from the human fibroblast cell line Hs27 were density arrested in G0, trypsinised when no mitosis was apparent and then released by re-seeding cells at a lower density. Flow cytometric analysis revealed that 93% of the cells were arrested in G0. Under these conditions, 35% of cells entered the S phase 24 h after release and 28% reached the G2/M phase 32 h later (Fig. 3A). The mRNA of both hMcm4, used as a control, and hMcm8 were absent in cells analysed 0, 6 and 12 h after release (Fig. 3A and B). hMcm8 and hMcm4 mRNA accumulation were first observed 18 h after release and continued to increase during the S and G2/M phases (Fig. 3B). Since it is known that hMcm4 accumulation occurs in G1 (and not in G0) and since the accumulation patterns of hMcm8 and hMcm4 were identical, we conclude that, like for hMcm4, hMcm8 mRNA accumulation begins in G1 and continues into the S and G2/M phases of the cell cycle (Fig. 3B). This result also indicates that, in our experimental conditions, synchronised Hs27 cells are in G0 phase at 0, 6 and 12 h after release, and in G1 phase 18 h after release.
Figure 3.
Expression of the hMcm8 mRNA and protein during the cell cycle. Hs27 human fibroblast cells were arrested in G0 by confluence and released by splitting cells in a 1:3 ratio. (A) Cell cycle pattern analysed by flow cytometry. Cells were harvested at the indicated times. (B) Northern blot of total RNA (20 µg) probed with a 32P-labelled hMcm8 or hMcm4 probe and ethidium bromide staining of the RNA before blotting (bottom). Positions of 28S and 18S rRNA are indicated on the right. (C) Whole cell lysates (15 µg) of the same Hs27 population were analysed by immunoblotting with the anti-hMcm8 antibody. The bottom part of the same blot was revealed with anti-tubulin antibody used as reference for protein loading.
hMcm8 protein levels remain stable during the cell cycle
We then evaluated the level of hMcm8 protein during the cell cycle. All known mammalian Mcm proteins exhibit only minor variations in their protein levels during the cell cycle. This is probably because of their stability and low turnover (33,34), the exception being the hMcm6 protein, the level of which rises dramatically during G1/S phase (35).
Using western blots, we analysed the total protein extracts of the same synchronised Hs27 cells used for northern blot analysis, and re-hybridised the same western blot using anti-tubulin antibody as a reference for protein loading (Fig. 3D). hMcm8 protein expression is very low in G0 phase (0 and 12 h after release) while its level increases as cells enter G1 and remains stable during the S and G2/M phases.
hMcm8 is a nuclear protein
Mammalian Mcm2–7 proteins are nuclear throughout the cell cycle. However, only Mcm2 and Mcm3 proteins contain a NLS. In fact, the murine homologues of Mcm4–Mcm7 are mainly cytoplasmic when overexpressed in COS cells, while their co-expression with Mcm2 or Mcm3 leads to a shift in their localisation from the cytoplasm to the nucleus (36). Furthermore, the association of Mcm proteins in a hexameric complex is a prerequisite for their nuclear localisation in yeast (37). It is thus plausible that the Mcm2 and Mcm3 NLS is involved in translocating the complex from the cytoplasm to the nucleus.
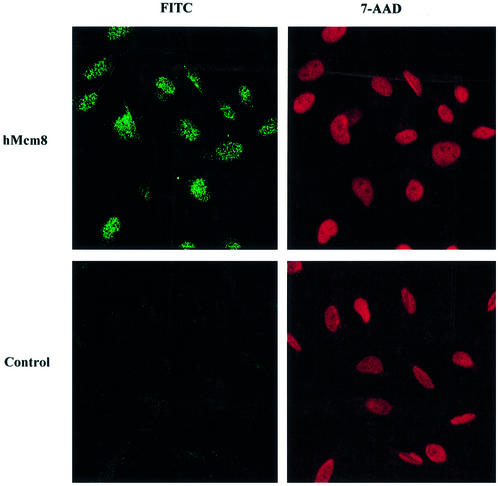
The subcellular localisation of hMcm8 was determined by laser confocal microscopy. Non-transfected Hs27 or HeLa cells were stained by immunofluorescence (FITC) with the anti-hMcm8 antibody. The nuclei were counterstained with 7-AAD. As shown in Figure 4, the anti-hMcm8 signal was mainly nuclear, while no specific staining was observed using the preimmune serum (negative control). The absence of a NLS in the hMcm8 protein sequence suggests that this protein could be carried to the nucleus as a result of its interaction with other nuclear proteins. The variable pattern of hMcm8 punctate nuclear staining could be related to differences in the expression level of hMcm8 and/or to changes in the proportion of structure-bound versus nucleosolic hMCM8 in different phases of the cell cycle.
Figure 4.
hMcm8 protein localises to the nucleus. Paraformaldehyde-fixed asynchronous HeLa cells were permeabilised with 0.2% Triton X-100, and incubated with either the anti-hMcm8 antibody or the preimmune serum (as control) followed by incubation with FITC-conjugated donkey anti-rabbit IgG antibodies. After immunostaining, the nuclei were stained with 7-AAD and the samples were analysed by confocal microscopy.
hMcm8 protein has a structure-bound and a nucleosolic fraction
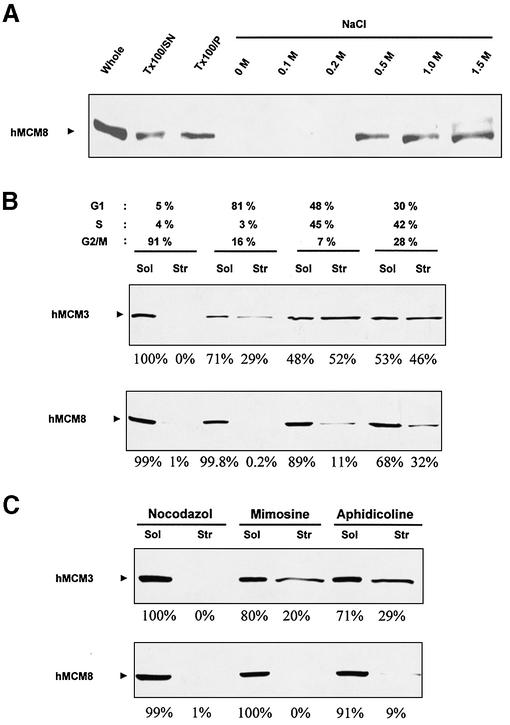
It has been shown that Mcm proteins comprise two nuclear fractions, a nucleosolic soluble fraction, which is released by permeabilising the nuclei with non-ionic detergents, and a chromatin-bound fraction which can be extracted by increasing the salt concentration (38). In order to determine whether this is also the case for hMcm8, HeLa cells in the exponential phase of growth were treated with a hypotonic buffer containing Triton X-100. After centrifugation, we obtained the soluble hMcm8 nucleosolic fraction in the supernatant and the structure-bound fraction in the pellet. As indicated in Figure 5A, a fraction of total hMcm8 is structure-bound. Subsequent extractions of this fraction using a buffer containing increasing salt concentrations dissociated hMcm8 from the chromatin (Fig. 5A, NaCl). Therefore, hMcm8 localises to the nucleus, where it exists as soluble, nucleosolic and structure-bound protein.
Figure 5.
Binding to chromatin of hMcm8 protein occurs in S phase. (A) The Triton X-100-extractable fraction (Tx100/SN) of HeLa cells was obtained as described (see Materials and Methods). The remaining pellet fraction (Tx100/P) was extracted with permeabilization buffer adjusted to the indicated NaCl concentrations. Immunoblot analysis with anti-hMcm8 antibodies of equal amounts of each fraction is shown. (B) HeLa cells were synchronised with nocodazole and then released to enter the cell cycle. At time points 0, 5, 10 and 20 h after release cells were harvested and equal amounts of nucleosolic (Sol) and chromatin-bound (Str) fractions were blotted. The relative proportions of nucleosolic and structure-bound hMcm3 and hMcm8 protein were assessed by image analysis of specific bands obtained by western blot. (C) HeLa cells were synchronised with nocodazole (block in G2/M), mimosine (block in late G1) or aphidicoline (block in early S) and equal amounts of nucleosolic (Sol) and chromatin-bound (Str) fractions were blotted. Quantifications were performed as described in (B).
hMcm8 protein is structure-bound in the S phase, after the establishment of DNA replication forks
Chromatin binding of Mcm2–Mcm7 proteins is tightly regulated during the cell cycle (1). In order to analyse the behaviour of hMcm8 during the cell cycle, HeLa cells were synchronised with nocodazole and then released to enter the cell cycle. At 0, 5, 10 and 20 h after release, cells were harvested and equal amounts of nucleosolic and structure-bound fractions were blotted (Fig. 5B). The relative proportions of nucleosolic and structure-bound hMcm3 and hMcm8 protein were assessed by image analysis of specific bands obtained by western blot. The majority of hMcm3 protein was nucleosolic in cells arrested in the G2/M phase by nocodazole. An increasing fraction of hMcm3 protein was structure-bound starting from G1 phase (29% of total protein), reaching 52% at S phase. The hMcm8 protein was also nucleosolic in nocodazole-arrested cells. However, unlike hMcm3, the structure-bound fraction of hMcm8 in G1 phase was barely detectable (0.2%). A fraction (11%) of hMcm8 protein started to be structure-bound in S phase, then reaching 32% of the total protein. In order to consider this point in greater depth, we repeated the same experiment using HeLa cells treated with 0.5 mM mimosine for 25 h or with aphidicoline (Fig. 5C). Under these conditions, mimosine blocks the cells in the late G1 phase (thus before the establishment of active replication forks) (39). Aphidicoline is an inhibitor of DNA polymerase α and it suppresses the migration of the replication fork, but it does not prevent the initiation of replication. Therefore, aphidicoline causes a cell cycle arrest in the beginning of the S phase. In mimosine-arrested cells, an important fraction (20%) of hMcm3 is structure-bound. Strikingly, all the hMcm8 protein is nucleosolic in these cells, indicating that, unlike other Mcm proteins, hMcm8 is not structure-bound in cells arrested in late G1 phase. In cells arrested at the beginning of S phase with aphidicoline, 29% of hMcm3 protein is structure-bound, while only a small fraction (9%) of hMcm8 protein is structure-bound. These data are consistent with the view that, at variance with the other members of the Mcm protein family, hMcm8 is structure-bound in the early S phase after the establishment of replication forks, and not in G1 phase.
hMcm8 protein does not co-immunoprecipitate with the Mcm2–7 complex
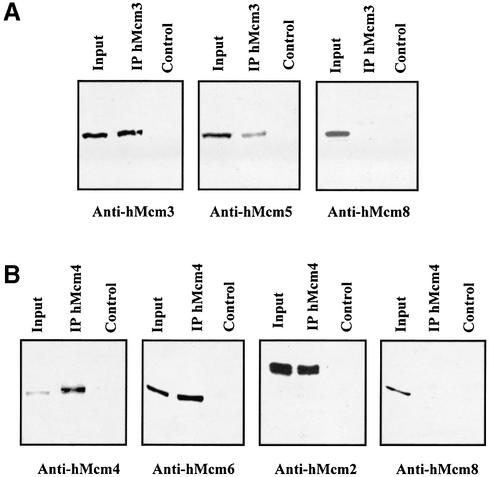
Mcm2–7 proteins are found as a heterohexameric complex in HeLa cells. In addition to the Mcm2–7 complex (31), Mcm2/4/6/7, Mcm4/6/7 and Mcm3/5 subcomplexes have also been identified (21,40). To investigate whether hMcm8 can associate with one of these Mcm subcomplexes, we performed immunoprecipitation and western blot analyses using exponentially growing, asynchronous HeLa cell extracts and polyclonal antibodies specific to hMcm3 or hMcm4. As shown in Figure 6A, anti-hMcm3 antibody immunoprecipitated both hMcm3 and hMcm5, but not hMcm8. Using the anti-hMcm4 antibody, we were able to immunoprecipite hMcm4, hMcm6 and hMcm2, but not hMcm8 (Fig. 6B). Although we do not rule out any possible interaction between hMcm8 and other Mcm proteins, hMcm8 does not seem to be involved in the Mcm2–7 complex.
Figure 6.
hMcm8 protein does not immunoprecipitate with the P1/Mcm2–7 protein complexes. The extracts of exponentially growing HeLa cells were prepared as described. An aliquot of one-tenth of the extract was loaded as input. The extracts were immunoprecipitated with anti-hMcm3 or anti-hMcm4 antibodies or with rabbit immunoglobin as control (control). (A) Immunoprecipitation using anti-hMcm3 antibody. The blots were revealed with anti-hMcm3, anti-hMcm5 or anti-hMcm8 antibodies. (B) Immunoprecipitation using anti-hMcm4 antibody. The blots were revealed with anti-hMcm4, anti-hMcm6, anti-hMcm2 or anti-hMcm-8 antibodies.
DISCUSSION
Our understanding of the mechanisms involved in eukaryotic replication origins has been greatly improved in the last few years and several key proteins have been identified. These studies have revealed a sophisticated programme determining that initiation occurs only once at every cell division and fixing the time at which individual origins fire during the S phase (1,3,5). However, this finely tuned process has not yet been completely elucidated, and the identification of new players may throw further light on its complexity.
Mcm genes were first discovered in budding and fission yeast as genes required to enable progression through the cell division cycle or to maintain minichromosomes containing autonomously replicating sequences (ARS). Characterisation of the Saccharomyces cerevisiae genes Mcm2 (41), Mcm3 (42) and Cdc46/Mcm5 (43,44) suggested their involvement in DNA replication. The protein family grew rapidly with cloning of the Schizosaccharomyces pombe genes cdc21/Mcm4 (45) and mis5/Mcm6 (46) and of the S.cerevisiae Cdc47/Mcm7 genes (47). The evolutionarily preserved homologues of the six yeast Mcm proteins were rapidly characterized in fruit fly, frog, mouse and humans. No other homologues of these genes have been identified in the complete genome of S.cerevisiae. Therefore, since the replication initiation mechanisms seem to be highly conserved from yeast to humans, it has been hypothesised that the six members of the Mcm family did not diversify any further (11).
In contrast with this view, we describe here a new member of the human Mcm protein family, hMcm8. It contains the typical domains and functional motifs of the Mcm2–7 protein family. hMcm8 mRNA accumulates during the G1/S–G2/M phase, while its protein product is detectable throughout the normal cell cycle. Its nuclear localisation and association with chromatin in a cell cycle-regulated manner suggest its role in DNA replication. Database searches have revealed that the Mcm8 protein is evolutionarily conserved, thus allowing the prediction of its potentially relevant biological function. However, the precise role of hMcm8 remains unknown. Our results show that hMcm8 does not participate in the Mcm3–5 and Mcm2/4/6/7 subcomplexes, and that its binding to chromatin is restricted to the S phase. The nuclear localisation of hMcm8 protein in the absence of a NLS suggests that hMcm8 may have a nuclear protein partner(s) which could cause its translocation to the nucleus. However, the data we obtained in HeLa cells show that this partner is not a known member of the Mcm family. At present, we can only speculate as to the biological function of Mcm8. It may be involved in conformational changes to the Mcm complex during the S phase and/or in the melting of origin DNA. Mcm proteins have also been shown to be involved in transcriptional control (26) and activation (27). Furthermore, the role of the Mcm complex in the elongation phase of DNA replication has recently been demonstrated (17). hMcm8 may participate in one or several of these activities, and further study is required to explore its function.
It has recently been shown that the down-regulation of Mcm proteins is a common downstream mechanism causing a loss of proliferative capacity in human cells (48). In fact, a loss of replication licensing coincides with the commitment of precursor cells to a specific lineage of differentiation or with cell quiescence. Mcm proteins have been proposed as new biomarkers capable of detecting G1 cells ‘with growth potential’. These properties of Mcm proteins highlight their potential usefulness in cancer biology as tumor markers (49,50), and in studies focusing on stem cell differentiation. Our preliminary data show that hMcm8 expression is higher in a proliferative cell line than in the corresponding (liver) non-proliferative tissue. However, these results need to be extended to a wider range of tissues and completed by cell cycle analyses.
In this context, it is also noteworthy that we recently reported on an in vivo hMcm8 gene mutation occurring selectively in a tumorous tissue and not in adjacent, non-tumorous cells, suggesting that hMcm8 mutations may play a role in cell transformation (see the mutated hMcm8 protein expressed in the tumor in Fig. 3C) (28).
In conclusion, we have described a new member of the Mcm protein family, evolutionarily conserved, which, unlike the other known Mcm proteins, displays restriction of nuclear structure binding to the S phase and does not participate in the Mcm complex. Although further work is required to elucidate the biological function of this new gene, its mutation in clonally expanded human tumor cells suggests its potential impact on the control of cell proliferation.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by grants from INSERM (Institut National de Santé et Recherche Médicale), LNC (Ligue Nationale contre le Cancer) and ARC (Association pour la Recherche contre le Cancer).
DDBJ/EMBL/GenBank accession no. AJ439063
REFERENCES
- 1.Ritzi M. and Knippers,R. (2000) Initiation of genome replication: assembly and disassembly of replication-competent chromatin. Gene, 245, 13–20. [DOI] [PubMed] [Google Scholar]
- 2.Tye B.K. (2000) Insights into DNA replication from the third domain of life. Proc. Natl Acad. Sci. USA, 97, 2399–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lei M. and Tye,B.K. (2001) Initiating DNA synthesis: from recruiting to activating the MCM complex. J. Cell Sci., 114, 1447–1454. [DOI] [PubMed] [Google Scholar]
- 4.Donaldson A.D. and Blow,J.J. (1999) The regulation of replication origin activation. Curr. Opin. Genet. Dev., 9, 62–68. [DOI] [PubMed] [Google Scholar]
- 5.Tye B.K. (1999) MCM proteins in DNA replication. Annu. Rev. Biochem., 68, 649–686. [DOI] [PubMed] [Google Scholar]
- 6.Coleman T.R., Carpenter,P.B. and Dunphy,W.G. (1996) The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell, 87, 53–63. [DOI] [PubMed] [Google Scholar]
- 7.Rowles A., Chong,J.P., Brown,L., Howell,M., Evan,G.I. and Blow,J.J. (1996) Interaction between the origin recognition complex and the replication licensing system in Xenopus. Cell, 87, 287–296. [DOI] [PubMed] [Google Scholar]
- 8.Romanowski P., Madine,M.A., Rowles,A., Blow,J.J. and Laskey,R.A. (1996) The Xenopus origin recognition complex is essential for DNA replication and MCM binding to chromatin. Curr. Biol., 6, 1416–1425. [DOI] [PubMed] [Google Scholar]
- 9.Maiorano D., Moreau,J. and Mechali,M. (2000) XCDT1 is required for the assembly of pre-replicative complexes in Xenopus laevis. Nature, 404, 622–625. [DOI] [PubMed] [Google Scholar]
- 10.Nishitani H., Lygerou,Z., Nishimoto,T. and Nurse,P. (2000) The Cdt1 protein is required to license DNA for replication in fission yeast. Nature, 404, 625–628. [DOI] [PubMed] [Google Scholar]
- 11.Kearsey S.E. and Labib,K. (1998) MCM proteins: evolution, properties and role in DNA replication. Biochim. Biophys. Acta, 1398, 113–136. [DOI] [PubMed] [Google Scholar]
- 12.Maiorano D., Lemaitre,J.M. and Mechali,M. (2000) Stepwise regulated chromatin assembly of MCM2-7 proteins. J. Biol. Chem., 275, 8426–8431. [DOI] [PubMed] [Google Scholar]
- 13.Koonin E.V. (1993) A common set of conserved motifs in a vast variety of putative nucleic acid-dependent ATPases including MCM proteins involved in the initiation of eukaryotic DNA replication. Nucleic Acids Res., 21, 2541–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chong J.P., Mahbubani,H.M., Khoo,C.Y. and Blow,J.J. (1995) Purification of an MCM-containing complex as a component of the DNA replication licensing system. Nature, 375, 418–421. [DOI] [PubMed] [Google Scholar]
- 15.Kubota Y., Mimura,S., Nishimoto,S., Takisawa,H. and Nojima,H. (1995) Identification of the yeast MCM3-related protein as a component of Xenopus DNA replication licensing factor. Cell, 81, 601–609. [DOI] [PubMed] [Google Scholar]
- 16.Madine M.A., Khoo,C.Y., Mills,A.D. and Laskey,R.A. (1995) MCM3 complex required for cell cycle regulation of DNA replication in vertebrate cells. Nature, 375, 421–424. [DOI] [PubMed] [Google Scholar]
- 17.Labib K., Tercero,J.A. and Diffley,J.F. (2000) Uninterrupted MCM2-7 function required for DNA replication fork progression. Science, 288, 1643–1647. [DOI] [PubMed] [Google Scholar]
- 18.Todorov I.T., Attaran,A. and Kearsey,S.E. (1995) BM28, a human member of the MCM2-3-5 family, is displaced from chromatin during DNA replication. J. Cell Biol., 129, 1433–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujita M., Kiyono,T., Hayashi,Y. and Ishibashi,M. (1996) Inhibition of S-phase entry of human fibroblasts by an antisense oligomer against hCDC47. Biochem. Biophys. Res. Commun., 219, 604–607. [DOI] [PubMed] [Google Scholar]
- 20.Lee J.K. and Hurwitz,J. (2000) Isolation and characterization of various complexes of the minichromosome maintenance proteins of Schizosaccharomyces pombe. J. Biol. Chem., 275, 18871–18878. [DOI] [PubMed] [Google Scholar]
- 21.Ishimi Y. (1997) A DNA helicase activity is associated with an MCM4, -6 and -7 protein complex. J. Biol. Chem., 272, 24508–24513. [DOI] [PubMed] [Google Scholar]
- 22.Kelman Z., Lee,J.K. and Hurwitz,J. (1999) The single minichromosome maintenance protein of Methanobacterium thermoautotrophicum DeltaH contains DNA helicase activity. Proc. Natl Acad. Sci. USA, 96, 14783–14788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chong J.P., Hayashi,M.K., Simon,M.N., Xu,R.M. and Stillman,B. (2000) A double-hexamer archaeal minichromosome maintenance protein is an ATP-dependent DNA helicase. Proc. Natl Acad. Sci. USA, 97, 1530–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shechter D.F., Ying,C.Y. and Gautier,J. (2000) The intrinsic DNA helicase activity of Methanobacterium thermoautotrophicum delta H minichromosome maintenance protein. J. Biol. Chem., 275, 15049–15059. [DOI] [PubMed] [Google Scholar]
- 25.Lee J.K. and Hurwitz,J. (2001) Processive DNA helicase activity of the minichromosome maintenance proteins 4, 6 and 7 complex requires forked DNA structures. Proc. Natl Acad. Sci. USA, 98, 54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yankulov K., Todorov,I., Romanowski,P., Licatalosi,D., Cilli,K., McCracken,S., Laskey,R. and Bentley,D.L. (1999) MCM proteins are associated with RNA polymerase II holoenzyme. Mol. Cell. Biol., 19, 6154–6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DaFonseca C.J., Shu,F. and Zhang,J.J. (2001) Identification of two residues in MCM5 critical for the assembly of MCM complexes and Stat1-mediated transcription activation in response to IFN-gamma. Proc. Natl Acad. Sci. USA, 98, 3034–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gozuacik D., Murakami,Y., Saigo,K., Chami,M., Mugnier,C., Lagorce,D., Okanoue,T., Urashima,T., Brechot,C. and Paterlini-Brechot,P. (2001) Identification of human cancer-related genes by naturally occurring Hepatitis B Virus DNA tagging. Oncogene, 20, 6233–6240. [DOI] [PubMed] [Google Scholar]
- 29.Thompson J.D., Plewniak,F., Thierry,J. and Poch,O. (2000) DbClustal: rapid and reliable global multiple alignments of protein sequences detected by database searches. Nucleic Acids Res., 28, 2919–2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Church G.M. and Gilbert,W. (1984) Genomic sequencing. Proc. Natl Acad. Sci. USA, 81, 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richter A. and Knippers,R. (1997) High-molecular-mass complexes of human minichromosome-maintenance proteins in mitotic cells. Eur. J. Biochem., 247, 136–141. [DOI] [PubMed] [Google Scholar]
- 32.Musahl C., Schulte,D., Burkhart,R. and Knippers,R. (1995) A human homologue of the yeast replication protein Cdc21. Interactions with other Mcm proteins. Eur. J. Biochem., 230, 1096–1101. [DOI] [PubMed] [Google Scholar]
- 33.Schulte D., Burkhart,R., Musahl,C., Hu,B., Schlatterer,C., Hameister,H. and Knippers,R. (1995) Expression, phosphorylation and nuclear localization of the human P1 protein, a homologue of the yeast Mcm 3 replication protein. J. Cell Sci., 108, 1381–1389. [DOI] [PubMed] [Google Scholar]
- 34.Musahl C., Holthoff,H.P., Lesch,R. and Knippers,R. (1998) Stability of the replicative Mcm3 protein in proliferating and differentiating human cells. Exp. Cell Res., 241, 260–264. [DOI] [PubMed] [Google Scholar]
- 35.Tsuruga H., Yabuta,N., Hosoya,S., Tamura,K., Endo,Y. and Nojima,H. (1997) HsMCM6: a new member of the human MCM/P1 family encodes a protein homologous to fission yeast Mis5. Genes Cells, 2, 381–399. [DOI] [PubMed] [Google Scholar]
- 36.Kimura H., Ohtomo,T., Yamaguchi,M., Ishii,A. and Sugimoto,K. (1996) Mouse MCM proteins: complex formation and transportation to the nucleus. Genes Cells, 1, 977–993. [DOI] [PubMed] [Google Scholar]
- 37.Pasion S.G. and Forsburg,S.L. (1999) Nuclear localization of Schizosaccharomyces pombe Mcm2/Cdc19p requires MCM complex assembly. Mol. Biol. Cell, 10, 4043–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kimura H., Nozaki,N. and Sugimoto,K. (1994) DNA polymerase alpha associated protein P1, a murine homolog of yeast MCM3, changes its intranuclear distribution during the DNA synthetic period. EMBO J., 13, 4311–4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krude T. (1999) Mimosine arrests proliferating human cells before onset of DNA replication in a dose-dependent manner. Exp. Cell Res., 247, 148–159. [DOI] [PubMed] [Google Scholar]
- 40.Burkhart R., Schulte,D., Hu,D., Musahl,C., Gohring,F. and Knippers,R. (1995) Interactions of human nuclear proteins P1Mcm3 and P1Cdc46. Eur. J. Biochem., 228, 431–438. [PubMed] [Google Scholar]
- 41.Yan H., Gibson,S. and Tye,B.K. (1991) Mcm2 and Mcm3, two proteins important for ARS activity, are related in structure and function. Genes Dev., 5, 944–957. [DOI] [PubMed] [Google Scholar]
- 42.Gibson S.I., Surosky,R.T. and Tye,B.K. (1990) The phenotype of the minichromosome maintenance mutant mcm3 is characteristic of mutants defective in DNA replication. Mol. Cell. Biol., 10, 5707–5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hennessy K.M., Clark,C.D. and Botstein,D. (1990) Subcellular localization of yeast CDC46 varies with the cell cycle. Genes Dev., 4, 2252–2263. [DOI] [PubMed] [Google Scholar]
- 44.Hennessy K.M., Lee,A., Chen,E. and Botstein,D. (1991) A group of interacting yeast DNA replication genes. Genes Dev., 5, 958–969. [DOI] [PubMed] [Google Scholar]
- 45.Coxon A., Maundrell,K. and Kearsey,S.E. (1992) Fission yeast cdc21+ belongs to a family of proteins involved in an early step of chromosome replication. Nucleic Acids Res., 20, 5571–5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takahashi K., Yamada,H. and Yanagida,M. (1994) Fission yeast minichromosome loss mutants mis cause lethal aneuploidy and replication abnormality. Mol. Biol. Cell, 5, 1145–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dalton S. and Whitbread,L. (1995) Cell cycle-regulated nuclear import and export of Cdc47, a protein essential for initiation of DNA replication in budding yeast. Proc. Natl Acad. Sci. USA, 92, 2514–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stoeber K., Tlsty,T.D., Happerfield,L., Thomas,G.A., Romanov,S., Bobrow,L., Williams,E.D. and Williams,G.H. (2001) DNA replication licensing and human cell proliferation. J. Cell Sci., 114, 2027–2041. [DOI] [PubMed] [Google Scholar]
- 49.Freeman A., Morris,L.S., Mills,A.D., Stoeber,K., Laskey,R.A., Williams,G.H. and Coleman,N. (1999) Minichromosome maintenance proteins as biological markers of dysplasia and malignancy. Clin. Cancer Res., 5, 2121–2132. [PubMed] [Google Scholar]
- 50.Todorov I.T., Werness,B.A., Wang,H.Q., Buddharaju,L.N., Todorova,P.D., Slocum,H.K., Brooks,J.S. and Huberman,J.A. (1998) HsMCM2/BM28: a novel proliferation marker for human tumors and normal tissues. Lab. Invest., 78, 73–78. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.