Abstract
Shunting is a mechanism that permits translational initiation at internal codons positioned in proximity to a ribosome acceptor sequence. Sendai virus exploits shunting to express a series of proteins that initiate at the fourth and fifth start sites on the P/C mRNA (namely, the Y1 and Y2 proteins, respectively). Shunt-mediated initiation at these sites is codon independent. In an attempt to characterise the acceptor site, an extensive deletion analysis was performed spanning the entire C ORF. Only mutants flanking the Y1/Y2 start sites exhibited altered shunt phenotypes. Some of these significantly enhanced shunting efficiency to the point where the Y1/Y2 proteins were the major translational products of the mRNA. Additionally, removal of a short region just downstream of the Y2 start codon (referred to as Δ10) ablated all Y protein initiation via shunting but had no effect on Y expression when the AUG codons were viewed by a scanning ribosome. Point mutations introduced into this Δ10 sequence severely perturbed shunt-mediated initiation. We also provide evidence that changes in this region of the P/C mRNA may be used to modulate Y protein expression levels in different viral strains.
INTRODUCTION
In the majority of eukaryotic mRNAs, translation begins with the binding of the eukaryotic initiation factor 4F (eIF4F) to the capped 5′ end, an interaction that is mediated via its eIF4E subunit. The eIF4F cap-binding complex is also composed of eIF4A, a member of the ‘DEAD Box’ family of RNA helicases, and eIF4G. Binding of eIF4F to the mRNA leads to recruitment of the ribosomal 43S pre-initiation complex. The 40S ribosome, and associated factors, is then thought to linearly scan the RNA unwinding intermolecular structure in an ATP-dependent fashion until an AUG codon is encountered and a 48S initiation complex forms. Codon–anticodon base-pairing in the ribosomal P site is then thought to trigger hydrolysis of the eIF2-bound GTP, and joining of the 60S ribosomal subunit (1–3). This multicomponent, coordinated series of events is strictly regulated and responds to both intra- and extracellular signals (4–6).
An increasingly important alternative strategy employed by both cellular mRNAs and viruses, is the direct recruitment of the ribosome to the start codon via an internal ribosome entry site (IRES) (7,8). These elements, which generally consist of complexed RNA secondary/tertiary structural elements within the 5′ UTR, effectively by-pass the requirement for cap recognition, and hence eIF4E. This has important physiological consequences because eIF4E [being a factor whose concentration is rate limiting during initiation (9,10)] is an important target for translational regulation (reviewed in 6). In addition to being eIF4E independent, certain viral IRESs, in particular hepatitis C virus (HCV) and classical swine fever virus (CSFV), are able to recruit the 43S ribosome to the start site independent of any of the eIF4 factors in a manner reminiscent of the prokaryotic Shine and Dalgarno element (11).
A second alternative strategy is ribosomal shunting or discontinuous scanning. Shunting combines features of linear scanning and IRESs. Ribosomes are loaded via the 5′ cap, start to scan, and then at a defined donor site translocate to an internal acceptor site located close to the start codon (12,13). The most studied shunts are those on the cauliflower mosaic virus (CaMV) 35S RNA (14,15) and the adenovirus late mRNAs (16,17). Shunting has also been reported to occur on the cellular hsp70 mRNA (17), suggesting that this is a generalised mode of translational initiation within eukaryotes. Both the CaMV and adenovirus shunts are characterised by the presence of stable secondary structural elements within the 5′ UTR that effectively delineate the donor and acceptor sites. These structural elements are a critical feature in both shunts. In CaMV it probably serves to spatially co-localise the donor and acceptor sites (18), whereas the adenovirus tripartite leader may recruit cellular factors required for ribosomal translocation (16). In the case of CaMV, a short ORF precedes the donor site, and translation of this ORF is required for shunting (19–21). Therefore, the CaMV shunt represents a very particular form of translational re-initiation (22). However, in the adenovirus tripartite leader, short ORFs within the 5′ UTR do not play a role in shunting. Rather, it appears that shunting is enhanced as a result of base-pairing between 18S rRNA and sequence elements within the 5′ UTR (17). Both the CaMV and adenovirus shunts are modulated by trans-acting viral factors. The CaMV transactivator TAV enhances shunt mediated initiation 2–3-fold (19,23) by facilitating translational re-initiation. Adenovirus expresses a 100 kDa protein late in infection. This protein interacts with eIF4G displacing Mnk1 kinase whose loss from the eIF4F complex leads to an accumulation of hypophosphorylated eIF4E (24). This compromises scanning-dependent initiation but not that mediated by shunting in which ribosomal loading at the 5′ cap appears to be less sensitive to the phosphorylation status of eIF4E (16;17). Therefore, at least in the case of adenovirus, shunting is a mechanism that ensures the expression of a subset of mRNAs under a particular physiological condition.
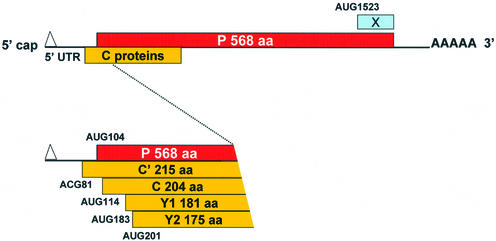
The Sendai virus (SeV) P/C mRNA has become a paradigm for translational flexibility expressing six polypeptides by ribosomal choice (25). Five of these polypeptides are derived from initiation sites located at the 5′ end (Fig. 1). The second start site (AUG104) gives rise to the 568 amino acid long P protein, an essential cofactor for the viral polymerase (26). The first (ACG81), third (AUG114), fourth (AUG183) and fifth (AUG201) start sites generate a nested set of four C proteins (termed C′, C, Y1 and Y2, respectively) with a common C-terminus. The first three start sites are accessed by leaky scanning, whereas Y1/Y2 are initiated via a shunt that bypasses the three upstream initiation codons (27,28). This shunt is characterised by the apparent absence of a precise donor site element within the 5′ UTR (28), and stable RNA structural elements around the Y1/Y2 start codons. However, shunt-dependent initiation from Y1/Y2 occurs efficiently from non-AUG codons, suggesting that the acceptor site may position the initiation complex directly at these start sites. Positioning of the ribosome in this manner was also proposed to explain the shunt-mediated non-AUG initiation for the badnavirus RTBV (29), and the IRES-mediated non-AUG initiation on HCV genomic RNA (30). The sixth translation product of the P/C mRNA, the X protein, represents the C-terminal 95 amino acids of the P protein, and initiates from an AUG codon more than 1500 nt from the 5′ cap (31). Curiously, like the Y proteins, the X protein is initiated by shunting, suggesting that SeV has exploited more than once this mode of translational initiation to expand its coding repertoire.
Figure 1.
Schematic representation illustrating the translational initiation sites on the SeV P/C mRNA. The 1943 nt long SeV P/C mRNA is shown as a horizontal line with the various ORFs as boxes. The P ORF is shown above the line, and the C ORF (+1 relative to P, encoding the C′, C, Y1, Y2) is below the line. The 81 nt 5′ UTR is also indicated. Numbers refer to nucleotide positions relative to the 5′ end of the P/C mRNA.
In this current work, we have identified a cis-acting sequence element just downstream of the Y2 start codon (referred to as Δ10) that is essential for shunt-mediated Y1/Y2 expression on the P/C mRNA. In addition, a number of deletion mutants upstream of the Y2 start codon significantly enhance shunting efficiency. In these latter constructs, the Y proteins are major translation products of the mRNA even when their start codons are non-AUG, thereby demonstrating that under certain conditions this type of shunt can give rise to efficient initiation. We have also identified a strain of SeV in which shunt-mediated initiation is impaired due to changes within the Δ10 region.
MATERIALS AND METHODS
Cell culture, transient transfection and metabolic labelling
HeLa and A549 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) supplemented with l-glutamine, penicillin, streptomycin and 5% fetal calf serum at 37°C in a 5% CO2 atmosphere.
Infection and transfection were performed when the cells were 70–80% confluent. Cells were infected at 2–5 p.f.u./cell with a vaccinia virus recombinant expressing T7 RNA polymerase (vTF7-3, referred to in the text as vaccinia-T7) (32). Transfection mixes containing 0.5 ml of DMEM medium, 16 µl of transfectASE (33) and 3 µg of plasmid DNA were added to the cells 30 min after infection.
For protein labelling, cells were starved for 30 min in DMEM minus methionine and cysteine (Gibco). This was then replaced with the same medium containing 120 µCi/ml 35S-translabel for 2 h. Cytoplasmic extracts were prepared by solubilising the monolayer in 150 mM NaCl, 50 mM Tris–HCl pH 7.4, 10 mM EDTA, 0.6% NP-40, 1 mM AEBSF (Boehringer) and 1% aprotinin (Sigma). Nuclei were removed by pelleting at 20 000 g for 5 min. Cytoplasmic extracts were immunoprecipitated overnight at 4°C with 2.5 vol of RIPA buffer (150 mM NaCl, 1% deoxycholate, 1% Triton X-100, 0.1% SDS, 10 mM Tris pH 7.8) containing a monoclonal antibody to the HA-tag (16b12; Eurogentec) or a polyclonal antibody to the C/C′ proteins raised in rabbits against a bacterially expressed His-tagged protein. The immune complexes were recovered on protein-A Sepharose (Pharmacia) for 5 h at 4°C, washed once with 0.5 M LiCl, 1% β-mercaptoethanol, and twice with RIPA buffer. Samples were boiled with sample buffer (10% glycerol, 5% β-mercaptoethanol, 2.5% SDS, 60 mM Tris–HCl pH 6.8, bromophenol blue) and proteins were resolved by 17.5% SDS–PAGE.
Immunoblotting of the cytoplasmic extracts was performed as outlined in Curran and Kolakofsky (31) using the CDP-star chemiluminescent substrate (Boehringer).
Plasmid construction
Standard procedures were used for restriction nuclease digestion, plasmid DNA construction and purification. The AUG81 clone (27) was transferred into the vector pBlueScript SK (pBS) as a KpnI/PstI fragment. This made a unique NarI site within the cDNA clone at position 155. Mutations around the Y1/Y2 start sites (Δ8, Δ9, Δ9NsiI, Δ10, Δ11, Δ12, Δ13, Δ14, Δ9NsiΔ10, Δ9NsiΔ10Y2AUG) were introduced into the pBS AUG81 by PCR using a positive sense oligonucleotide that carried the change and which spanned the NarI site, and a negative sense oligonucleotide downstream of a unique XbaI site (position 1027 in the P/C gene). These constructs and the mutations Δ15, Δ16, Δ17, Δ18, Δ9Y1UGUY2UGU, AUG81MVC, AUG81H*, Δ9AUG81H* and AUG81Y2ACG (see Figs 2A, 4A and 7A) were introduced into a pBS vector which carries a triple HA-tag between the XbaI and NotI sites. Inserts were generated by PCR using either the T7 primer as the positive sense oligonucleotide or a positive sense oligonucleotide that carried the change and spanned the NarI site or SalI site (Δ15, Δ16, Δ17, Δ18), and a negative sense oligonucleotide with a XbaI site which overlapped the stop codon of the C ORF. The AUG81MVC construct was made by PCR from the pTM1-CMVC-GFP (34). The Δ5′ deletions were generated by removal of a fragment spanning the Acc65I to NarI sites (Fig. 3A). Point mutations within the Δ10 region were generated by PCR using an oligonucleotide spanning the NarI site, and were introduced into a P/Cwt clone (35).
Figure 2.
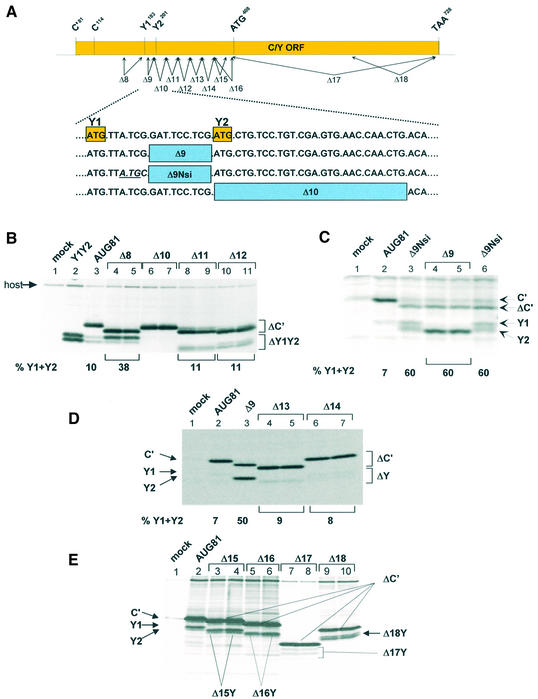
Deletion analysis reveals the presence of sequences downstream of Y1/Y2 required for shunt-mediated initiation. (A) Schematic representation of the deletion series used to map the acceptor site. The solid rectangle in the upper panel corresponds to the C ORF with the position of the start sites indicated above. The arrows below represent the approximate positions of the deletion series generated. All clones were introduced into an AUG81 background. The lower panel gives the sequence immediately downstream of the Y1 start codon (written as DNA) and the exact position of three of the deletion mutants. (B–E) A549 cells (B and C) and HeLa cells (D and E) infected with vaccinia-T7 (vTF7-3) were transfected with the plasmid clones as indicated above each panel. Two independent clones of each deletion were examined. In the clone Y1Y2 (B, lane 2), the Y1 AUG start codon was positioned just downstream of the encephalomyocarditis virus IRES. This provided positional markers for the Y1 and Y2 proteins. As a mock control, cells were infected but not transfected. Cells were metabolically labelled with 35S-translabel. Cytoplasmic extracts were prepared by scraping the cells into lysis buffer and proteins were immunoprecipitated with either a polyclonal anti-C antiserum (B and C), or a monoclonal anti-HA antibody (D and E). Immunoselected proteins were resolved on a 17.5% SDS–polyacrylamide gel and visualised by fluorography. The numbers below the panels give the percentage of Y1/Y2 proteins expressed relative to the total translation products of the C ORF (i.e. C′ + Y1 + Y2) based upon a densitometric scan of the gel films (Molecular Dynamics).
Figure 4.
The Y2 AUG codon is not a critical element of the Δ10 phenotype. (A) Sequence of the region immediately downstream of the Y1 start codon (written as DNA) indicating the position of the deletions analysed below. (B) A549 cells were transfected with the plasmid clones as indicated, and metabolically labelled with 35S-translabel at 18–20 h.p.i. Cytoplasmic extracts from these cells were immunoprecipitated with anti-C antiserum and analysed on a 17.5% SDS–polyacrylamide gel (see Fig. 2). The numbers below the panels give the percentage of Y1/Y2 proteins expressed relative to the total translation products of the C ORF based upon a densitometric scan of the film.
Figure 7.
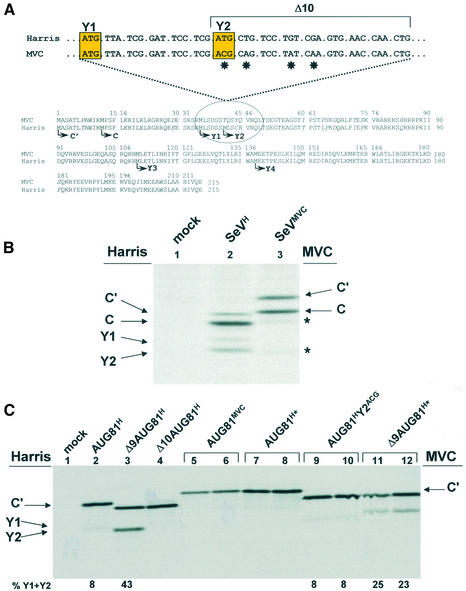
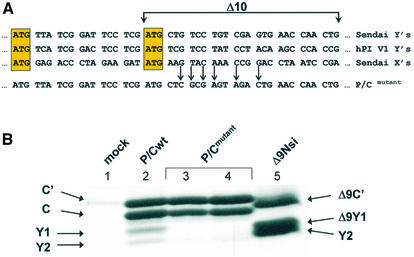
Shunting in the SeV MVC strain is impaired due to mutations within the Δ10 region. (A) The lower panel shows an alignment of the C ORF primary sequence between the SeV MVC (SeVMVC) and Harris strains (SeVH). Differences are underlined and in italics. Arrows indicate initiation sites. The upper panel aligns the nucleotide sequences from a region immediately downstream of the Y1 initiation codon (and including the Δ10 region) and stars indicate changes. (B) HeLa cells were infected with SeVH, SeVMVC or mock infected and metabolically labelled at 18–20 h.p.i. Cytoplasmic extracts were then prepared and products of the C ORF immunoselected with anti-C polyclonal antiserum. Proteins were resolved on a 17.5% SDS–polyacrylamide gel and visualised by fluorography. The stars in lane 3 indicate the presence of selected proteins smaller than the C protein. (C) Vaccinia-T7 infected HeLa cells were transfected with AUG81H (Harris strain, lane 2), Δ9AUG81H (Harris strain, lane 3), Δ10AUG81H (Harris strain, lane 4), AUG81MVC (MVC strain, lanes 5 and 6), AUG81H* (an AUG81H clone into which had been introduced the four changes found in the Δ10 region of SeVMVC; lanes 7 and 8), AUG81HY2ACG (an AUG81H clone in which the Y2 start codon has been changed to ACG; lanes 9 and 10) and Δ9AUG81H* (a Δ9AUG81H clone into which had been introduced the four changes found in the Δ10 region of SeVMVC; lanes 11 and 12). All clones carried the 3HA tag at the C-terminus. A mock transfection provided the negative control (lane 1). Transfected cells were metabolically labelled at 18–20 h.p.i. Cytoplasmic extracts were immunoprecipitated with the anti-HA monoclonal antibody and immunoselected proteins were analysed as outlined in the legend to Figure 2. The numbers below the panels give the percentage of Y1/Y2 proteins expressed relative to the total translation products of the C ORF obtained from a phosphorimager analysis of the gel (BioRad).
Figure 3.
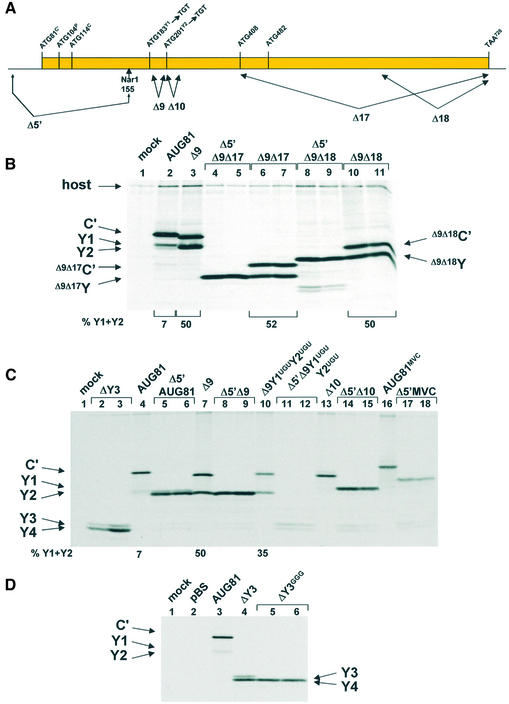
Scanning dependent initiation at the Y start codons: Δ10 perturbs specifically shunt-mediated initiation. (A) The solid rectangle represents the C ORF with the position of the initiation sites and stop codon (written as DNA) indicated above. Below the line is depicted the position of the Δ9, Δ17, Δ18 and Δ5′ deletions. The latter was designed to promote linear scanning through the Y1 and Y2 start sites by positioning them close to the 5′ end of the mRNA. It was generated by deleting all sequences upstream of the NarI site indicated. (B) The Δ17 and Δ18 deletions were introduced into the Δ9AUG81 clone carrying a triple HA tag at the C-terminus. To provide positional markers for the truncated Y proteins these clones were also constructed with the Δ5′ deletion. HeLa cells were transfected with the plasmids as indicated above the panel. Cells were metabolically labelled at 18–20 h.p.i., Cytoplasmic extracts were immunoprecipitated with the anti-HA monoclonal antibody. Immunoselected proteins were analysed as outlined in the legend to Figure 2. (C) The Δ5′ deletion was introduced into AUG81, Δ9AUG81, Δ10AUG81, Δ9Y1UGUY2UGUAUG81 (a clone in which the Y start codons had been changed to UGU) and AUG81MVC (a cDNA clone from the MVC strain of SeV). All clones carried a 3HA tag at the C-terminus. HeLa cells were transfected with the plasmids as indicated above the panel and processed as indicated in (B). The numbers below (B) and (C) give the percentage of Y1/Y2 proteins expressed relative to the total translation products of the C ORF based upon a densitometric scan of the films. (D) Sequences upstream of AUG408 (the putative start site for an Y3 protein) were deleted from AUG81 carrying the C-terminal 3HA tag to generate ΔY3. The AUG408 codon in this clone was then mutated to GGG to create ΔY3GGG. These constructs, plus the parent AUG81 and a pBS plasmid control, were transfected into HeLa cells and metabolically labelled at 18–20 h.p.i. Proteins were immunoselected with the anti-HA antibody, resolved on a 17.5% SDS–polyacrylamide gel and visualised by fluorography.
The sag465HA plasmid [gift from B. Conrad (36)] contains an unique AvrII site, at nucleotide 162 from the AUG start site, which served to introduce the region Δ10 in the positive and negative orientation.
RESULTS
Nucleotides flanking the Y1 and Y2 start sites modulate shunt-mediated initiation
The seminal observation in this work was that changing the C′ ACG start codon to AUG (referred to as AUG81) ablated expression of the P and C proteins (the second and third start sites) but not Y1/Y2 (the fourth and fifth start sites). The absence of P and C precluded leaky scanning past the AUG81 codon, suggesting that the Y proteins were initiated via a ribosomal shunt (28). The amount of Y proteins expressed in this background was between 5 and 10% the levels of C′, and since the proteins have similar stabilities this in turn indicates that only a fraction of ribosomes recruited to the 5′ cap actually shunt. We proposed that the shunt acceptor site might flank the Y1/Y2 initiation codons (28). In an attempt to locate this region, a series of deletion mutants flanking the Y start codons were generated in the AUG81 background (Δ8 until Δ16; Fig. 2A). The constructs Δ1 to Δ7, which are not listed in this series and extended to the C protein start codon at position 114, did not ablate Y expression (data not shown). The deletion upstream of the Y1 start codon spanning nucleotides 159–179 (referred to as Δ8AUG81), did not negatively affect Y1/Y2 expression; on the contrary, it produced a marked increase in the levels of the Y1 protein relative to C′ (Fig. 2B, lanes 4 and 5). In all these experiments the C′ protein serves as an internal control since it is made from the same mRNA and has a similar t1/2 as the Y proteins (data not shown). However, deletion of nucleotides 201–227 (Δ10AUG81) ablated Y1/Y2 protein expression (Fig. 2B, lanes 6 and 7). This deletion effectively replaces the Y2 AUG with an ACA codon (Fig. 2A). At this point it should be remembered that Y initiation is largely codon independent (28) (also see Fig. 3C, lane 10) suggesting that the Δ10 phenotype did not arise simply due to the loss of the Y2 AUG (see below). Indeed, simply reinserting the Y2 AUG codon into the Δ10 clone did not rescue the phenotype (data not shown). Further deletions downstream of the Y2 start codon (Δ11 until Δ16; the latter deletion just precedes the next AUG in the C ORF, namely AUG408) did not alter the translation phenotype (Fig. 2). Curiously, although the Δ13 and Δ14 deletions were of similar size (i.e. nine amino acids), the truncated C′ proteins migrated very differently on SDS gels (Fig. 2D). This aberrant mobility of C mutants has been observed previously (34,37), and is probably related to the very charged nature of the protein (pI ∼ 10). We also examined deletions between the Y1 and Y2 start codons. For this, two clones were constructed: Δ9AUG81 deleted nucleotides 189–197 (three of the five codons between Y1 and Y2), whereas Δ9NsiAUG81 also inverted the CG nucleotide pair just upstream of the Y2 AUG thereby generating an NsiI restriction site. This inversion also created an AUG codon in the P ORF between the Y start sites (Fig. 2A). Both clones produced a marked increase in the level of Y protein expression (Fig. 2C). Indeed in some experiments the Y proteins were as abundant as C′. Pulse-chase demonstrated no change in the half-life of these truncated proteins compared with the wild-type, suggesting that this effect was a result of increased initiation rates (data not shown). Curiously, Δ9AUG81 expressed a single band that co-migrated with the Y2 protein (lanes 4 and 5), whereas Δ9NsiAUG81 produced both Y proteins at roughly equivalent amounts (lanes 3 and 6). It should be noted that in both these clones the ‘Kozak’ consensus sequences flanking both start sites are poor suggesting that start site selection is not dictated by these cis-acting elements. In addition, the AUG introduced into the P ORF was silent (data not shown) confirming earlier observations indicating that the shunted ribosome initiates only at the Y start sites (28). To exclude the possibility that sequence elements in the 3′ half of the C ORF play a role, two further large deletions were also constructed (referred to as Δ17 and Δ18; Fig. 2A). The Δ17 deletion removed all sequences 3′ of the AUG408, whereas Δ18 removed the 3′ terminal 127 nt. Expression of Y proteins was readily detected in both mutants (Fig. 2E, lanes 7–10); however, their levels in the Δ17 construct appeared to be somewhat lower. We therefore re-examined this effect by introducing these mutations into a Δ9AUG81 background, i.e. a background that significantly enhances shunt-mediated Y protein expression (Fig. 2C, lanes 4 and 5, and Fig. 3B, lane 3). To ensure the unambiguous identification of the deleted Y protein products, we also removed all the sequences upstream of the NarI site at position 155 (Fig. 3A) in both the Δ9Δ17 and Δ9Δ18 clones thereby generating a series of constructs referred to by the prefix Δ5′. The new capped 5′ end was now ∼30 nt upstream of the Y1 initiation codon, and based on earlier work we reasoned that recruitment of initiation factors and the 40S ribosome so close to the Y start sites would compromise shunting but not linear scanning (see below) (28). The products of this scanning dependent initiation provided positional markers for the Δ9Δ17Y and Δ9Δ18Y proteins (Fig. 3B, lanes 4 and 5, and 8 and 9, respectively). Expression of the clones Δ9Δ17AUG81 and Δ9Δ18AUG81 produced two proteins in equimolar amounts, a phenotype reminiscent of the parent Δ9AUG81 (Fig. 3B, lanes 6 and 7, 10 and 11, and 3, respectively). The lower band co-migrated with the corresponding truncated Y protein confirming that shunting was occurring efficiently in both the Δ17 and Δ18 clones. The upper band is the truncated C′ protein.
The Δ10 region is required for shunt-mediated initiation
Deletion analysis maps a single region between nucleotides 201–227 (referred to as Δ10) that is required for Y protein expression (Table 1). However, is the phenotype of the Δ10 clone a feature of shunt-mediated initiation? One could envisage that this mutation alters the stability of RNA structural elements that in turn modulate initiation at Y starts whose contexts are already very poor. A role for structural elements in modulating initiation at weak AUG codons has already been described (38). To answer this question we examined Y protein expression when the start sites were encountered by a linear scanning ribosome. For this we once again exploited the Δ5′ deletion approach outlined earlier (namely, deletion of all sequences upstream of a unique NarI site at position 155 in the cDNA; see Fig. 3A). The Δ5′ deletion was introduced into the clones AUG81, Δ9AUG81, Δ9Y1UGUY2UGUAUG81 (a Δ9 clone in which the Y1/Y2 start codons have been changed to UGU) and Δ10AUG81. Expression profiles from these constructs are depicted in Figure 3C (the MVC clone in this figure will be described later). That most of the ribosomes are accessing the Y sites by scanning is confirmed by a comparison of lane 10 with lanes 11 and 12. Shunted ribosomes continued to express Y when the start codon was changed to UGU, although the level of Y relative to C′ was nearly 2-fold lower that in the Δ9 parent clone (compare lanes 7 and 10). However, no Y expression was detected when this change was examined in the Δ5′ background (lanes 11 and 12), although a faster migrating doublet band was observed. We reasoned that this doublet probably arose due to scanning-dependent initiation at downstream AUG codons, namely, AUG408 that would give rise to a Y3 protein, and AUG482 that would give rise to a Y4 protein (Fig. 3A). We suspected that both sites might be utilised because the Kozak context of the former is poor (…CACAUGC…) and the latter much better (…GCG AUGG…). To examine this point we also expressed a clone in which all the sequences upstream of the AUG408 were deleted (ΔY3). The two proteins expressed by this construct (Fig. 3C, lanes 2 and 3) co-migrated with those of the Δ9Y1UGUY2UGUAUG81 clone, and when the AUG408 codon was changed to GGG (ΔY3GGG) the upper band of the doublet was lost demonstrating that it was indeed a Y3 protein (Fig. 3D). Interestingly, Y3 and Y4 proteins have been detected in SeV-infected cells and frequently appear in our transfection assays (data not shown). The Δ5′AUG81 (lanes 5 and 6), Δ5′Δ9 (lanes 8 and 9) and Δ5′Δ10 (lanes 14 and 15) all continued to express Y proteins (Fig. 3C). In addition, the presence of a Δ10Y protein produced by scanning confirms that the absence of such a product via shunting is not because the protein is highly unstable. The results therefore indicate that the Δ10 deletion has touched a sequence element required exclusively for shunt-mediated initiation at the Y1/Y2 starts (Fig. 3C, compare lane 13 with lanes 14 and 15).
Table 1. A résumé of the results of the deletion analysis in Figure 2.
| Clones | Nucleotides deleted | Y protein expression |
|---|---|---|
| AUG81 | WT | + |
| Δ8 | 159–179 | +++ |
| Δ9 | 189–197 | ++++ |
| Δ10 | 201–227 | – |
| Δ11 | 227–253 | + |
| Δ12 | 254–280 | + |
| Δ13 | 281–307 | + |
| Δ14 | 308–334 | + |
| Δ15 | 341–373 | + |
| Δ16 | 341–406 | + |
| Δ17 | 408–725 | + |
| Δ18 | 599–725 | + |
A single + indicates that Y proteins were detected at levels comparable to those observed in the AUG81 clone. (Note: both 3HA tagged and untagged versions of these clones were analysed and gave identical results.)
Since the Δ10 mutation also removed the Y2 AUG start codon we examined to what extent loss of this initiation codon contributed to the phenotype. For this, we introduced the Δ10 mutation into the Δ9NsiAUG81 background, producing Δ9NsiΔ10AUG81 (Fig. 4A). As observed previously, Δ9NsiAUG81 significantly enhanced the levels of Y1/Y2 relative to C′ (Fig. 4B, compare lanes 2 and 3), whereas Δ10AUG81 effectively eliminated both Y1 and Y2 (lane 4). The double mutant produced a dramatic reduction in the levels of Y1/Y2 (compare lanes 3 and 5). Starting from Δ9NsiΔ10AUG81 we then re-introduced the Y2 AUG (Δ9NsiΔ10Y2AUGAUG81). This produced a marginal increase in the levels of Y1/Y2 relative to Δ9Δ10NsiAUG81 (lane 6), which nonetheless remained at very low levels relative to the parent Δ9NsiAUG81. Therefore, the major determinant of the Δ10 phenotype appears to be contained within the 24 nt downstream of the Y2 AUG.
To further characterise the Δ10 region we examined the effect of point mutations. To select the sites of mutation we aligned regions downstream of the Y1/Y2 start codons from SeV and hPIV1 (a virus closely related to SeV which also expresses a set of Y proteins) (39,40), as well as the region downstream of the SeV X protein (a protein whose initiation is also shunt mediated) (31). What is immediately striking is the presence of two AUG codons with identical spacing (Fig. 5A). However, the Δ9 mutation series suggest that this spacing is not a critical element of the Y1/Y2 shunt. Conserved nucleotides in the three-way alignment were then changed by transversion within a P/Cwt background (i.e. a construct that will express C′, P, C, Y1 and Y2). These changes severely reduced Y protein expression, although trace amounts of what appears to be a Y2 protein were sometimes observed (Fig. 5B). This result supports the notion that the region immediately downstream of the Y2 codon is playing a role in modulating shunt-mediated initiation, but is it the acceptor site?
Figure 5.
Point mutations within the Δ10 region severely perturb shunt-mediated initiation. (A) An alignment of the sequences downstream of the Y start sites in SeV and hPIV1, and the SeV X protein. Sites chosen for mutagenesis and the changes made to generate the P/C mutant clone are indicated by the arrows. (B) Vaccinia-T7 infected A549 cells were transfected with the plasmids indicated above the panel. Transfections were metabolically labelled at 18–20 h.p.i. with 35S-translabel. Proteins were recovered by immunoselection with polyclonal anti-C antiserum, resolved on a 17.5% SDS–polyacrylamide gel and visualised by fluorography.
The Δ10 region cannot itself promote shunting
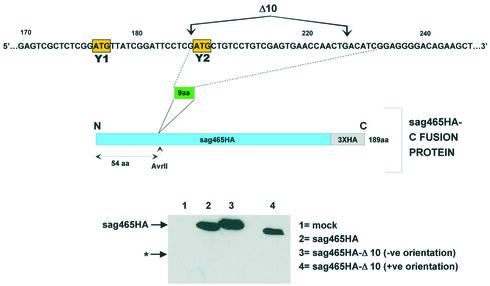
The Δ10 region is clearly required for Y1/Y2 expression, but is it sufficient to promote ribosomal shunting, i.e. is it an acceptor site? To resolve this point we decided to ask if insertion of this region into a heterologous mRNA would promote the expression of a truncated protein due to shunting. For this we choose a human endogenous retrovirus (HERV) super antigen (sag), which carries a triple HA tag at its C-terminus (sag465HA) (36). A double-stranded oligonucleotide fragment carrying the Δ10 (equivalent to nine amino acids) region and flanked by AvrII restriction sites was inserted 54 codons (at a unique AvrII site) from the sag465HA AUG initiation codon (which is in an excellent context, …ACCAUGG…) generating the clone sag465HA-Δ10 (Fig. 6). The ORF in this fragment is open in both orientations, which permitted us to use clones carrying the fragment in the reverse orientation as a negative control. Insertion of the Δ10 region in the negative orientation produced a protein that migrated slightly more slowly than sag465HA consistent with the insertion of nine amino acids (compare lanes 2 and 3). However, the Δ10 region in the positive orientation produced a protein that migrated slightly faster than the parent sag465HA although the insertion was confirmed by sequencing of the clone. Presumably, these differences arise due to the nature of the primary sequence inserted. Shunting in this latter construct should have permitted the expression of a protein nearly 50 amino acids shorter than the sag465HA protein (its estimated mobility on the SDS gel based upon protein size markers is indicated by a star in the figure). However, immunoblots performed with the anti-HA antibody in cell extracts expressing these constructs did not reveal the presence of any shorter protein forms, even after extensive over-exposure (Fig. 6, lower panel). These results indicate that the Δ10 region is necessary but not sufficient to promote ribosomal translocation, and suggest that it may not act as a discrete functional unit in a manner analogous to an IRES element.
Figure 6.
The Δ10 region fails to promote ribosomal shunting on a heterologous mRNA. As illustrated in the upper panel, the Δ10 region (nucleotides 201–227) was inserted at a unique AvrII site within the construct sag465HA generating the fusion protein sag463HA-Δ10 (with the Δ10 region inserted in both orientations, indicated as +ve and –ve). These plasmid clones plus the sag465HA parent were transfected into vaccinia-T7 infected A549 cells. Infected but non-transfected cells provided a mock control. Cytoplasmic extracts were prepared at 20 h.p.i and resolved on a 17.5% SDS–polyacrylamide gel. HA-tagged proteins were visualised by immunoblotting with an anti-HA monoclonal antibody using the CDP-star light detection system (lower panels). The star (*) represents the position at which one should observe a protein product initiated by a shunting event at the inserted Y2 AUG (based upon estimated molecular mass).
A virus strain that expresses low levels of the Y protein: phenotype maps to the Δ10 region
The C/Y cDNA used throughout this study was derived from the Harris strain of SeV (SeVH) (35). It was noted that another SeV strain, the strain MVC (SeVMVC) (41), expressed very low levels of the Y proteins. We investigated this further by first infecting HeLa cells with both SeVH and SeVMVC, and metabolically labelling these infections at 18–20 h post-infection (h.p.i.). Translation products of the C ORF were analysed by immunoprecipitation with the anti-C antiserum. The SeVH strain produced the characteristic pattern of C′, C, Y1 and Y2 proteins expressed from a P/Cwt mRNA (27) (Fig. 7B, lane 2). However, the profile observed with the strain SeVMVC was markedly different (lane 3). As already reported, the C′ and C proteins migrated slower on the SDS– polyacrylamide gel (42), a mobility shift that is consistent with a number of primary sequence changes in the C ORF, most of which are located in the N-terminal 85 amino acids of C′ (Fig. 7A). In addition, products of the C ORF smaller than the C protein (i.e. putative Y proteins indicated by stars in Fig. 7B, lane 3) were considerably less abundant than in the SeVH strain suggesting that shunting was impaired. Alignment of the nucleotide sequences around the Y1/Y2 start codons showed a cluster of four changes in the Δ10 region (Fig. 7A). It also revealed that although SeVMVC had conserved the Y1 AUG codon, it had an ACG threonine codon at the position of Y2 (Fig. 7A). Using our transient expression assay we investigated to what extent these changes contributed to the reduction in shunt-mediated Y protein expression. Initially, we introduced the AUG81 mutation (i.e. changing the normal ACG start codon for C′ to AUG) into an MVC P/C cDNA clone (AUG81MVC). Since the MVC C′/C proteins, and presumably the Y proteins, migrate differently from those of the Harris strain, we generated a marker for the MVC Y proteins using the Δ5′ approach outlined earlier (i.e. expression directed by a scanning ribosome). Expression of Δ5′MVC produced a single protein band, presumably the Y1 protein (Fig. 3C, lanes 17 and 18). Y2 is probably not expressed in this construct since it is an ACG codon. Expression from the AUG81MVC clone produced essentially undetectable levels of Y protein (Fig. 3C, lane 16, and Fig. 7C, lanes 5 and 6), a phenotype reminiscent of that observed with the Δ10AUG81 clone from the Harris strain (Fig. 3C, lane 13, and Fig. 7C, lane 4). Although shunt-mediated Y protein expression is codon independent, we initially evaluated to what extent the presence of a Y2 ACG codon would perturb shunting in the AUG81 background of the Harris strain (referred to as AUG81HY2ACG in Fig. 7C). Not surprisingly, introduction of this ACG change had itself little effect on Y protein expression (lanes 9 and 10). We next examined to what extent the Δ10 region of MVC (Fig. 7A) was responsible for the reduced Y protein expression observed in this strain by introducing all four changes into the AUG81H and Δ9AUG81H clones (these mutants are referred to as AUG81H* and Δ9AUG81H*, respectively, in Fig. 7C). Expression from the AUG81H* clone produced a C′ protein which co-migrated with the MVC protein (compare lanes 5 and 6 with 7 and 8) indicating that the altered mobility of the C proteins between these two strains resided in the primary sequence changes within the Δ10 region. This clone also showed impaired shunt-mediated initiation since Y proteins were essentially undetectable; a similar result was observed with AUG81MVC (Fig. 7C). Likewise, although the introduction of the four changes into Δ9AUG81H did not totally ablate Y expression (Fig. 7C, lanes 11 and 12), Y levels relative to C′ were 2-fold lower than in the parent Δ9AUG81 (lane 3) indicating that shunt-mediated initiation was impaired. Therefore, it would appear that changes within the Δ10 region might have been exploited by SeV strains to modulate the level of Y protein expression.
DISCUSSION
In this work we have extended the characterisation of the SeV ribosomal shunt, a shunt that is used to direct the expression of a subset of the viral C proteins, namely Y1 and Y2. The shunt functions in mammalian cells outside the context of a SeV infection indicating that no viral functions are required in trans. Since only cellular trans-acting factors are implicated, this shunt may represent a prototype for a novel mechanism of initiation in eukaryotes. It is characterised by the apparent absence of a donor site (S.de Breyne, T.Pelet, V.Simonet and J.Curran, manuscript in preparation) and the degeneracy in the initiation codon (28).
Cis-acting elements: the nature of the acceptor site
An extensive deletion analysis spanning the entire C ORF has identified a 24 nt region just downstream of the Y2 AUG codon (referred to as Δ10) that is crucial for shunt-mediated Y protein expression. This deletion did not perturb Y expression when the start codons were viewed by a linear scanning ribosome. In addition, mutations in this element were associated with impaired Y protein expression in the MVC strain of SeV. These results clearly identify this region as an important element of the acceptor site. It may function to recruit the ribosome by directly interacting with a component of the pre-initiation complex, or it may bind a cellular protein that in turn serves to bridge the interaction between acceptor site and ribosome. Ribosome re-positioning (which is effectively a form of shunting) has been observed on the HCV IRES. This event correlates with the binding of a 25 kDa cellular protein that has been characterised both as the S5 (43) and S9 (11) ribosomal protein. Alternatively, cellular protein(s) may be required to induce the correct RNA fold for recruitment (i.e. they serve as RNA chaperones) (44); a role analogous to that proposed for the RNA binding proteins that interact with IRES elements (45). However, the failure of this region to promote shunting in a heterologous mRNA and in a bicistronic construct (S.de Breyne, T.Pelet, V.Simonet and J.Curran, manuscript in preparation), suggests that this short linear sequence, although necessary, is not sufficient to promote ribosome translocation. Deletion analysis has failed to identify any other element within the C ORF or the 5′ UTR that is required for shunting. On the contrary, a series of deletions flanking the Y start codons actually enhanced significantly shunt-mediated initiation. These ‘up-mutations’ also functioned when the Y start codons were changed to UGU confirming that this phenotype was shunt-specific (non-AUG Y initiation codons were not recognised by a linear scanning ribosome). We suspect that the missing element in this equation is RNA structure, and that correct mRNA folding functions to juxtaposition the site of initial ribosomal recruitment (namely the 5′ cap) and the acceptor site in a way analogous to the stem–loop structure present in the CaMV shunt (18). Failure to place these two elements in close proximity would explain the failure to translocate on the heterologous and bicistronic mRNAs. RNA structure in solution is dynamic, and those deletions that enhance shunting may do so by stabilising a tertiary fold that facilitates recruitment, because it spatially co-localises the 5′ end and the acceptor region. The relieving of negative acting structural constraints was observed with the SeV X protein shunt in which an RNaseH induced cleavage of the mRNA upstream of the X start codon (the two fragments of RNA remaining associated non-covalently) significantly enhanced X protein expression (31). However, computer generated RNA folds have not permitted the identification of structural elements around the Y1/Y2 start sites whose alterations could be correlated with the shunt phenotypes observed with the deletion mutants. One may therefore envisage this shunt functioning in three steps: first, ribosomal loading (via a 5′ cap); secondly, recruitment of the ribosome by the acceptor; and thirdly, start site selection.
Why shunting?
When answering this question it is perhaps informative to regard the literature on IRES function. Because IRESs use a limited set of initiation factors they tend to promote translation under conditions in which linear scanning is compromised. This arises either because the ribosome is unable to load at the 5′ cap, and/or linear scanning is blocked, and occurs during viral infection (13) or conditions of cellular stress (e.g. heat shock, apoptosis) (5,8). In a similar vein, it is now clear that both cellular and viral IRESs can be modulated during the cell cycle presumably due to regulation of RNA binding proteins that enhance IRES function (44,46–48). Extrapolat ing from these points, one can speculate that shunts in which the ribosome enters via the 5′ cap will respond negatively to alterations that affect loading of the 4F component, but may actually be selected for under conditions that limit subsequent linear scanning. This could arise due to modulation of factors such as eIF4B, which is cleaved during apoptosis (49), or eIF1 and eIF1A, factors involved in scanning and initiation codon selection (50). Indeed, a similar type of regulation was proposed for the CaMV shunt (21). Alternatively, the shunt may be controlled via RNA binding protein(s) that are physiologically regulated.
Finally, shunts may not exist simply because they confer regulated flexibility in protein expression. On the contrary, a shunt may function only to expand the genetic complexity of a given mRNA by permitting the ribosome access to internal ORFs that may not even start with an AUG codon. Such shunts would be regulated only at the site of ribosomal loading, i.e. the 5′ cap.
With specific regard to the SeV shunt, some insights into its regulation may be gleaned by examination of Y protein function in the viral life cycle. The Ys are part of a nested set of C proteins, which play a central role in blocking the interferon (IFN)-mediated establishment of the cellular antiviral state, and have also been linked to apoptosis (34,37,51,52). Both IFN signalling and apoptosis impinge on translation (8,13), and it is not improbable that shunting has been selected to ensure translation of this subset of C proteins under these particular physiological conditions. However, attempts to directly demonstrate an effect of IFN on Y1/Y2 protein expression have yielded inconclusive results (data not shown). The SeVMVC strain that showed impaired Y expression was itself generated by the repeated passage in cell culture of a virulent isolate (SeVM), and was later shown to be attenuated in animal studies (41,42). However, this attenuation is unlikely to be related to the shunt phenotype since the highly virulent SeVM also expresses low levels of the Y proteins and the sequence of the Δ10 region is identical to that found in SeVMVC (data not shown). Shunt regulation and its consequence for the viral life cycle in its host remains therefore an open question.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Dr Bernadino Conrad for reading the manuscript. The work was supported by a grant from the Swiss National Science Foundation (No. 31-57434.99) and the Roche Research Foundation.
REFERENCES
- 1.Chakrabarti A. and Maitra,U. (1991) Function of eukaryotic initiation factor 5 in the formation of an 80 S ribosomal polypeptide chain initiation complex. J. Biol. Chem., 266, 14039–14045. [PubMed] [Google Scholar]
- 2.Huang H.K., Yoon,H., Hannig,E.M. and Donahue,T.F. (1997) GTP hydrolysis controls stringent selection of the AUG start codon during translation initiation in Saccharomyces cerevisiae. Genes Dev., 11, 2396–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pestova T.V., Lomakin,I.B., Lee,J.H., Choi,S.K., Dever,T.E. and Hellen,C.U. (2000) The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature, 403, 332–335. [DOI] [PubMed] [Google Scholar]
- 4.Dever T.E. (1999) Translation initiation: adept at adapting. Trends Biochem. Sci., 24, 398–403. [DOI] [PubMed] [Google Scholar]
- 5.Gingras A.C., Raught,B. and Sonenberg,N. (1999) eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem., 68, 913–963. [DOI] [PubMed] [Google Scholar]
- 6.Sonenberg N. and Gingras,A.C. (1998) The mRNA 5′ cap-binding protein eIF4E and control of cell growth. Curr. Opin. Cell Biol., 10, 268–275. [DOI] [PubMed] [Google Scholar]
- 7.Jackson R.J. (1996) A comparative view of initiation site selection mechanisms. In Hershey,J.W.B., Mathews,M.B and Sonenberg,N. (eds), Translational Control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 71–112.
- 8.Holcik M., Sonenberg,N. and Korneluk,R.G. (2000) Internal ribosome initiation of translation and the control of cell death. Trends Genet., 16, 469–473. [DOI] [PubMed] [Google Scholar]
- 9.Duncan R., Milburn,S.C. and Hershey,J.W. (1987) Regulated phosphorylation and low abundance of HeLa cell initiation factor eIF-4F suggest a role in translational control. Heat shock effects on eIF-4F. J. Biol. Chem., 262, 380–388. [PubMed] [Google Scholar]
- 10.Hiremath L.S., Webb,N.R. and Rhoads,R.E. (1985) Immunological detection of the messenger RNA cap-binding protein. J. Biol. Chem., 260, 7843–7849. [PubMed] [Google Scholar]
- 11.Pestova T.V., Shatsky,I.N., Fletcher,S.P., Jackson,R.J. and Hellen,C.U. (1998) A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev., 12, 67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hohn T., Dominguez,D.I., Scharer-Hernandez,N., Pooggin,M., Schmidt-Puchta,W., Hemmings-Mieszczak,M. and Futterer,J. (1998) Ribosome shunting in eukaryotes: what the viruses tell me. In Bailey-Serres,J. and Gallie,D.R. (eds), A Look Beyond Transcription: Mechanisms Determining mRNA Stability and Translation in Plants. American Society of Plant Physiologists, New York, NY, pp. 84–95.
- 13.Gale M. Jr, Tan,S.L. and Katze,M.G. (2000) Translational control of viral gene expression in eukaryotes. Microbiol. Mol. Biol. Rev., 64, 239–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Futterer J., Gordon,K., Sanfacon,H., Bonneville,J.M. and Hohn,T. (1990) Positive and negative control of translation by the leader sequence of cauliflower mosaic virus pregenomic 35S RNA. EMBO J., 9, 1697–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Futterer J., Kiss-Laszlo,Z. and Hohn,T. (1993) Nonlinear ribosome migration on cauliflower mosaic virus 35S RNA. Cell, 73, 789–802. [DOI] [PubMed] [Google Scholar]
- 16.Yueh A. and Schneider,R.J. (1996) Selective translation initiation by ribosome jumping in adenovirus-infected and heat-shocked cells. Genes Dev., 10, 1557–1567. [DOI] [PubMed] [Google Scholar]
- 17.Yueh A. and Schneider,R.J. (2000) Translation by ribosome shunting on adenovirus and hsp70 mRNAs facilitated by complementarity to 18S rRNA. Genes Dev., 14, 414–421. [PMC free article] [PubMed] [Google Scholar]
- 18.Dominguez D.I., Ryabova,L.A., Pooggin,M.M., Schmidt-Puchta,W., Futterer,J. and Hohn,T. (1998) Ribosome shunting in cauliflower mosaic virus. Identification of an essential and sufficient structural element. J. Biol. Chem., 273, 3669–3678. [DOI] [PubMed] [Google Scholar]
- 19.Pooggin M.M., Hohn,T. and Futterer,J. (2000) Role of a short open reading frame in ribosome shunt on the cauliflower mosaic virus RNA leader. J. Biol. Chem., 275, 17288–17296. [DOI] [PubMed] [Google Scholar]
- 20.Ryabova L.A. and Hohn,T. (2000) Ribosome shunting in the cauliflower mosaic virus 35S RNA leader is a special case of reinitiation of translation functioning in plant and animal systems. Genes Dev., 14, 817–829. [PMC free article] [PubMed] [Google Scholar]
- 21.Ryabova L.A., Pooggin,M.M., Dominguez,D.I. and Hohn,T. (2000) Continuous and discontinuous ribosome scanning on the cauliflower mosaic virus 35 S RNA leader is controlled by short open reading frames. J. Biol. Chem., 275, 37278–37284. [DOI] [PubMed] [Google Scholar]
- 22.Park H.S., Himmelbach,A., Browning,K.S., Hohn,T. and Ryabova,L.A. (2001) A plant viral “reinitiation” factor interacts with the host translational machinery. Cell, 106, 723–733. [DOI] [PubMed] [Google Scholar]
- 23.Pooggin M.M., Futterer,J., Skryabin,K.G. and Hohn,T. (2001) Ribosome shunt is essential for infectivity of cauliflower mosaic virus. Proc. Natl Acad. Sci. USA, 98, 886–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cuesta R., Xi,Q. and Schneider,R.J. (2000) Adenovirus-specific translation by displacement of kinase Mnk1 from cap- initiation complex eIF4F. EMBO J., 19, 3465–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Curran J., Latorre,P. and Kolakofsky,D. (1998) Translational gymnastics on the Sendai virus P/C mRNA. Semin. Virol., 8, 351–357. [Google Scholar]
- 26.Curran J. (1998) A role for the Sendai virus P protein trimer in RNA synthesis. J. Virol., 72, 4274–4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Curran J. and Kolakofsky,D. (1989) Scanning independent ribosomal initiation of the Sendai virus Y proteins in vitro and in vivo. EMBO J., 8, 521–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Latorre P., Kolakofsky,D. and Curran,J. (1998) Sendai virus Y proteins are initiated by a ribosomal shunt. Mol. Cell. Biol., 18, 5021–5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Futterer J., Potrykus,I., Bao,Y., Li,L., Burns,T.M., Hull,R. and Hohn,T. (1996) Position-dependent ATT initiation during plant pararetrovirus rice tungro bacilliform virus translation. J. Virol., 70, 2999–3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reynolds J.E., Kaminski,A., Kettinen,H.J., Grace,K., Clarke,B.E., Carroll,A.R., Rowlands,D.J. and Jackson,R.J. (1995) Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J., 14, 6010–6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Curran J. and Kolakofsky,D. (1988) Scanning independent ribosomal initiation of the Sendai virus X protein. EMBO J., 7, 2869–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fuerst T.R., Earl,P.L. and Moss,B. (1987) Use of a hybrid vaccinia virus-T7 RNA polymerase system for expression of target genes. Mol. Cell. Biol., 7, 2538–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rose J.K., Buonocore,L. and Whitt,M.A. (1991) A new cationic liposome reagent mediating nearly quantitative transfection of animal cells. Biotechniques, 10, 520–525. [PubMed] [Google Scholar]
- 34.Garcin D., Latorre,P. and Kolakofsky,D. (1999) Sendai virus C proteins counteract the interferon-mediated induction of an antiviral state. J. Virol., 73, 6559–6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giorgi C., Blumberg,B.M. and Kolakofsky,D. (1983) Sendai virus contains overlapping genes expressed from a single mRNA. Cell, 35, 829–836. [DOI] [PubMed] [Google Scholar]
- 36.Stauffer Y., Marguerat,S., Meylan,F., Ucla,C., Sutkowski,N., Huber,B., Pelet,T. and Conrad,B. (2001) Interferon-alpha-induced endogenous superantigen. A model linking environment and autoimmunity. Immunity, 15, 591–601. [DOI] [PubMed] [Google Scholar]
- 37.Garcin D., Curran,J. and Kolakofsky,D. (2000) Sendai virus C proteins must interact directly with cellular components to interfere with interferon action. J. Virol., 74, 8823–8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kozak M. (1989) Context effects and inefficient initiation at non-AUG codons in eucaryotic cell-free translation systems. Mol. Cell. Biol., 9, 5073–5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsuoka Y., Curran,J., Pelet,T., Kolakofsky,D., Ray,R. and Compans,R.W. (1991) The P gene of human parainfluenza virus type 1 encodes P and C proteins but not a cysteine-rich V protein. J. Virol., 65, 3406–3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Power U.F., Ryan,K.W. and Portner,A. (1992) The P genes of human parainfluenza virus type 1 clinical isolates are polycistronic and microheterogeneous. Virology, 189, 340–343. [DOI] [PubMed] [Google Scholar]
- 41.Itoh M., Isegawa,Y., Hotta,H. and Homma,M. (1997) Isolation of an avirulent mutant of Sendai virus with two amino acid mutations from a highly virulent field strain through adaptation to LLC-MK2 cells. J. Gen. Virol., 78, 3207–3215. [DOI] [PubMed] [Google Scholar]
- 42.Garcin D., Itoh,M. and Kolakofsky,D. (1997) A point mutation in the Sendai virus accessory C proteins attenuates virulence for mice, but not virus growth in cell culture. Virology, 238, 424–431. [DOI] [PubMed] [Google Scholar]
- 43.Fukushi S., Okada,M., Stahl,J., Kageyama,T., Hoshino,F.B. and Katayama,K. (2001) Ribosomal protein S5 interacts with the internal ribosomal entry site of hepatitis C virus. J. Biol. Chem., 276, 20824–20826. [DOI] [PubMed] [Google Scholar]
- 44.Pilipenko E.V., Pestova,T.V., Kolupaeva,V.G., Khitrina,E.V., Poperechnaya,A.N., Agol,V.I. and Hellen,C.U. (2000) A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev., 14, 2028–2045. [PMC free article] [PubMed] [Google Scholar]
- 45.Kaminski A. and Jackson,R.J. (1998) The polypyrimidine tract binding protein (PTB) requirement for internal initiation of translation of cardiovirus RNAs is conditional rather than absolute. RNA, 4, 626–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Honda M., Kaneko,S., Matsushita,E., Kobayashi,K., Abell,G.A. and Lemon,S.M. (2000) Cell cycle regulation of hepatitis C virus internal ribosomal entry site-directed translation. Gastroenterology, 118, 152–162. [DOI] [PubMed] [Google Scholar]
- 47.Pyronnet S., Pradayrol,L. and Sonenberg,N. (2000) A cell cycle-dependent internal ribosome entry site. Mol. Cell, 5, 607–616. [DOI] [PubMed] [Google Scholar]
- 48.Sachs A.B. (2000) Cell cycle-dependent translation initiation: IRES elements prevail. Cell, 101, 243–245. [DOI] [PubMed] [Google Scholar]
- 49.Clemens M.J., Bushell,M., Jeffrey,I.W., Pain,V.M. and Morley,S.J. (2000) Translation initiation factor modifications and the regulation of protein synthesis in apoptotic cells. Cell Death Differ., 7, 603–615. [DOI] [PubMed] [Google Scholar]
- 50.Pestova T.V., Borukhov,S.I. and Hellen,C.U. (1998) Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature, 394, 854–859. [DOI] [PubMed] [Google Scholar]
- 51.Itoh M., Hotta,H. and Homma,M. (1998) Increased induction of apoptosis by a Sendai virus mutant is associated with attenuation of mouse pathogenicity. J. Virol., 72, 2927–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kato A., Ohnishi,Y., Kohase,M., Saito,S., Tashiro,M. and Nagai,Y. (2001) Y2, the smallest of the Sendai virus C proteins, is fully capable of both counteracting the antiviral action of interferons and inhibiting viral RNA synthesis. J. Virol., 75, 3802–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]