Abstract
The 5′-untranslated region of Bag-1 mRNA contains an internal ribosome entry segment (IRES) and the translation of Bag-1 protein can be initiated by both cap-dependent and cap-independent mechanisms. In general, cellular IRESs require non-canonical trans-acting factors for their activity, however, very few of the proteins that act on cellular IRESs have been identified. Proteins that interact with viral IRESs have also been shown to stimulate the activity of cellular IRESs and therefore the ability of a range of known viral trans-acting factors to stimulate the Bag-1 IRES was tested. Two proteins, poly r(C) binding protein 1 (PCBP1) and polypyrimidine tract binding protein (PTB), were found to increase the activity of the Bag-1 IRES in vitro and in vivo. The regions of the Bag-1 IRES RNA to which they bind have been determined, and it was shown that PCBP1 binds to a short 66 nt section of RNA, whilst PTB interacts with a number of sites over a larger area. The minimum section of the RNA that still retained activity was determined and both PCBP1 and PTB interacted with this region suggesting that these proteins are essential for Bag-1 IRES function.
INTRODUCTION
The human Bag-1 gene (Bcl-2 associated athanogene) encodes three major isoforms generated from a single transcript, p50, p46 and p36 (BAG-1L, BAG-1M and BAG-1S respectively) that differ at their N-termini. BAG-1L, which initiates from a non-canonical CUG codon, contains an SV40-like nuclear localisation signal (NLS) at its N-terminus, and this is thought to be responsible for the mainly nuclear distribution of this isoform (1). The AUG-initiated isoforms, BAG-1M and BAG-1S, lack this NLS and as such are localised predominantly cytoplasmically (1,2). The variation in the N-termini of the Bag-1 proteins also leads to different protein binding specificities and the isoforms have diverse cellular roles. Bag-1 was originally identified as RAP46 (receptor associated protein) through its interaction with the glucocorticoid receptor (3) and has been found to associate with numerous other members of the steroid hormone receptor superfamily. It was also documented as HAP46 (Hsc70/Hsp70 associated protein) through its interaction with the 70 kDa heat-shock proteins (4,5) where it has a role as a co-chaperone in the protein folding response (6–8). The murine homologue, Bag-1, was identified through its interaction with Bcl-2, an anti-apoptotic gene (9). Bag-1 has been shown to interact with Bcl-2 and to promote the anti-apoptotic properties of this protein, blocking a step in the apoptotic pathway (10).
It was originally suggested that the three main isoforms of Bag-1, in addition to a minor isoform, p29, are translated by leaky scanning (1), however, expression of Bag-1 in vivo does not support this hypothesis (1,2,11). Recently we have demonstrated that the p36 isoform of Bag-1 can be translated by internal ribosome entry through use of an internal ribosome entry segment (IRES) in addition to the cap-dependent scanning mechanism (11). IRES elements are used to initiate translation under conditions where cap-dependent scanning is compromised (12) and our data suggest that the IRES of Bag-1 is required to maintain translation of the p36 isoform following heat shock (11).
The internal ribosome entry mechanism of translation was first identified in picornaviruses and these have been widely studied in terms of structure, mechanisms and trans-acting factor requirements (13). Viral IRESs vary widely in their dependence on trans-acting factors. For example, some viral IRESs such as the encephalomyocarditis virus are able to function well in vitro (14,15), in contrast, other viral IRESs such as polio virus or the human rhino virus (HRV) require the addition of extracts derived from HeLa cells to in vitro systems before they are active (16–18). In the case of HRV, two proteins that are required have been identified, upstream of N-ras (unr) (19), and polypyrimidine tract binding protein (PTB) (20). A number of additional viral IRES binding/activating proteins have been identified, including the La autoantigen which is used by polio virus IRES (21), and poly r(C) binding protein 2 (PCBP2) which binds to polio virus IRES (22) and has been shown to activate entero/rhino virus IRESs in vitro (23). These proteins are thought to act as RNA chaperones to either maintain or aid the RNA to form a structure that is competent for ribosome recruitment.
Recently, a large number of cellular IRESs have been identified but the mechanisms by which they initiate translation are poorly understood (12). The protein factor requirements for cellular IRESs are much less well defined although data produced thus far would suggest that each IRES has a requirement for a specific set of trans-acting factors. First, cellular IRESs show considerable cell tropism and this would suggest that levels of endogenous trans-acting factors vary between cell lines (24–26). Secondly, the proteins that interact with two cellular IRESs studied so far are different, thus the Apaf-1 IRES requires PTB and unr for function in vitro and in vivo (27), whilst the XIAP IRES has a requirement for La (28).
In this study the protein factor requirements for efficient Bag-1 IRES activity has been investigated. We demonstrate a direct and specific interaction of PTB and poly r(C) binding protein 1 (PCBP1) with the Bag-1 IRES, which stimulates IRES function both in vitro and in vivo.
MATERIALS AND METHODS
Plasmid constructs
The plasmids pRF and pRBF harbouring deletion segments of Bag-1 are described in Figures 1A, 2A and 4A. The plasmid pSKL is based upon the vector pSK+bluescript (Stratagene); the Bag-1 5′-untranslated region (5′-UTR) or deletions were cloned into this vector in frame with the firefly luciferase gene (Figs 3A and 5A). Deletion fragments were generated by PCR using specific primers to the regions required. For expression of proteins used, the cDNAs were present in PET28a vectors, enabling expression of protein in Escherichia coli and subsequent purification of the protein. For expression in tissue culture cells, the cDNAs were subcloned into pCDNA3.1 and for expression in insect cells (for purification of protein) subcloned into pBlueBac4 (Invitrogen).
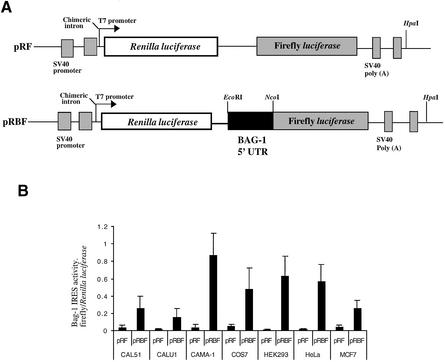
Figure 1.
A comparison of the efficiency of Bag-1 IRES-mediated translation in cell lines of different origin. (A) Schematic representation of the dicistronic reporter constructs pRF and pRBF where pRBF contains the Bag-1 5′-UTR inserted into the vector pRF and fused in-frame with the firefly luciferase gene. (B) The plasmids pRF and pRBF were transfected into the cell lines indicated. CAL51, CAMA-1 and MCF7s are of human breast carcinoma origin; CALU1, human lung cancer; COS7, a monkey epithelial cell line (CV-1) immortalised with SV40 DNA; HEK293, a human embryonic kidney cell line immortalised with adenovirus; and HeLa S3, of human cervical epitheloid carcinoma origin. IRES activity was expressed as the ratio of downstream cistron expression to upstream cistron expression (firefly/Renilla luciferase), with any differences in transfection efficiencies corrected for using the β-galactosidase transfection control (PJ7lacZ). Error bars indicate standard deviations as determined from at least three independent experiments performed in triplicate.
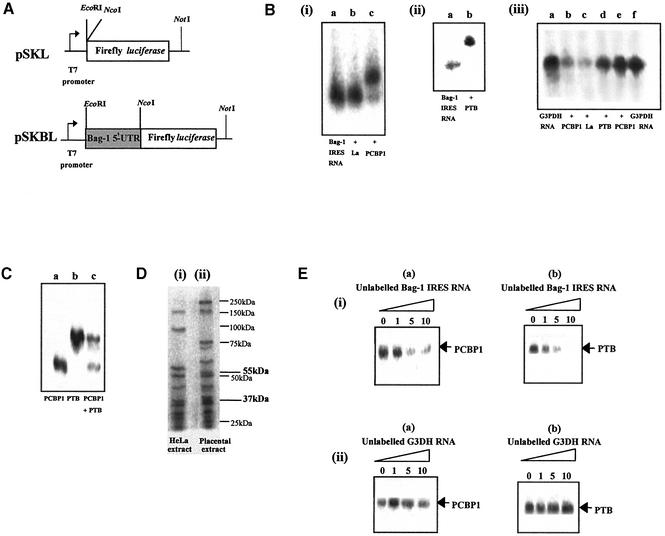
Figure 2.
PTB and PCBP1 bind directly to Bag-1 IRES RNA. (A) Schematic diagram of the monocistronic plasmids pSKL and pSKBL where pSKBL contains the Bag-1 5′-UTR fused in-frame with the luciferase gene. (B) (i and ii) EMSAs of radiolabelled Bag-1 IRES RNA alone (i and ii, lane a) and in combination with 0.2 µg of protein (i, lane b), La (i, lane c PCBP1). A gel retardation is observed with PTB and PCBP1, suggesting complex formation, but not with La. (iii) Control EMSAs were carried out with non-specific radiolabelled RNA of the same size from G3PDH (lanes a and f, G3PDH RNA alone). No gel retardation is observed with 0.2 µg of any protein tested (lanes b–e). (C) UV-crosslinking assay of radiolabelled Bag-1 IRES RNA in combination with 0.2 µg of PTB and/or PCBP1, again showing both proteins binding to Bag-1 RNA. (D) UV-crosslinking assay of radiolabelled Bag-1 IRES RNA with cell extracts. (Lane i) HeLa extract and (lane ii) placental extract. Bands corresponding in size to PTB (55 kDa) and PCBP1 (37 kDa) can be observed in both lanes, marked in bold type. Sizes and position of markers are indicated in plain type. There is also a marker at 37 kDa. (E) (i) UV-crosslinking assay of radiolabelled Bag-1 IRES RNA in combination with (a) 0.2 µg of PCBP1, where binding is competed by the addition of a 5× molar excess of unlabelled Bag-1 IRES RNA and (b) 0.2 µg of PTB, where binding is competed by the addition of an equimolar quantity of unlabelled Bag-1 IRES RNA. (ii) UV-crosslinking assay of radiolabelled Bag-1 RNA in combination with (a) 0.2 µg of PCBP1 or (b) 0.2 µg of PTB, where there is no competition observed with a 10× molar excess of unlabelled G3DH RNA.
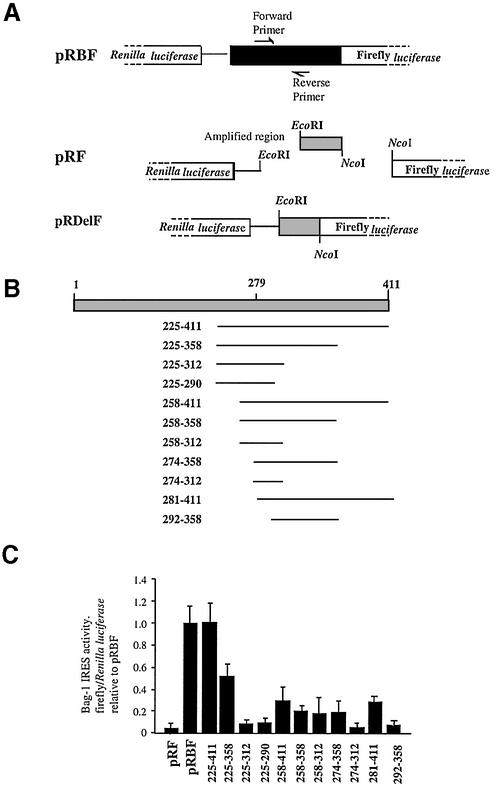
Figure 4.
PCBP1 and PTB bind to the minimal active element of the Bag-1 IRES. (A) Schematic diagram showing construction of deletion constructs from the discistronic plasmid pRBF (Fig. 1A) by PCR where delF indicates the forward primer, which introduces an EcoRI site and delR the reverse primer, which introduces an NcoI site, primer sequences are described in Materials and Methods. (B) Representation of the sections of the Bag-1 5′-UTR amplified in comparison with the full-length 5′-UTR. (C) Relative IRES activity of the deletion constructs in HeLa cells taken as a ratio of firefly/Renilla luciferase, normalised to a β-galactosidase transfection control and expressed relative to pRBF, which is assigned a value of 1.
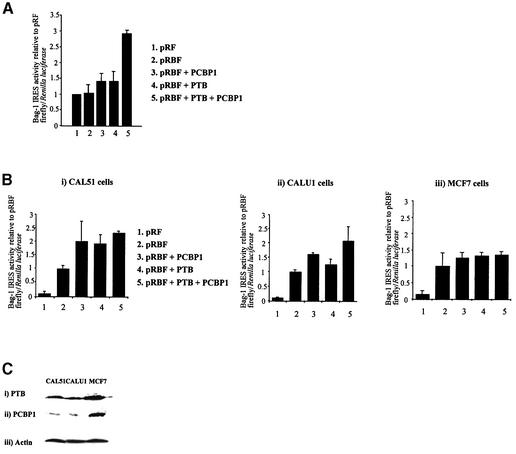
Figure 3.
PTB and PCBP1 stimulate Bag-1 IRES activity in vitro and in vivo. (A) 200 ng of PTB and/or PCBP1 can stimulate IRES activity from the dicistronic plasmid pRBF (Fig. 1A). IRES activity is expressed as a ratio of the downstream cistron to the upstream cistron (firefly/Renilla luciferase), in rabbit reticulocyte lysates primed with 100 ng of capped pRBF RNA. (B) The cell lines (i) CAL51 (human breast carcinoma), (ii) CALU1 (human lung cancer) and (iii) MCF7 (human breast carcinoma) were co-transfected with plasmids containing the pRBF (Fig. 1A) and/or PCBP1/PTB. Activity of the Bag-1 IRES is expressed using the ratio of downstream cistron expression to upstream cistron expression (firefly/Renilla luciferase) with any differences in transfection efficiencies corrected for using the β-galactosidase transfection control and expressed relative to pRBF alone to show the increase in IRES activity produced by each trans-acting factor. Error bars indicate standard deviations as determined from at least three independent experiments performed in triplicate. (C) Western blots of cell lysates for endogenous protein levels with (i) anti-PTB antibody, (ii) anti-PCBP1 antibody and (iii) anti-actin antibody as a loading control show a correlation with the level of activation shown by transfection of each protein in each cell line.
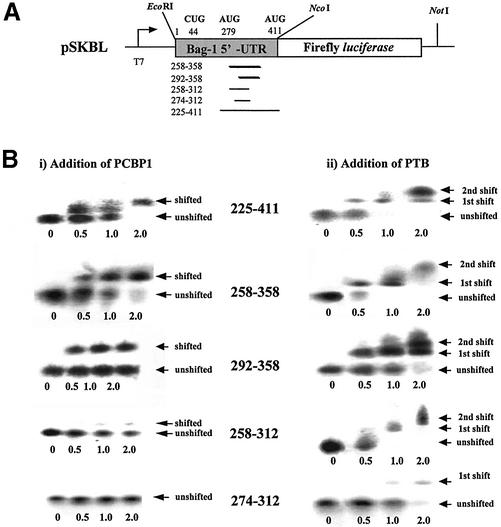
Figure 5.
PCBP1 and PTB bind to specific fragments of the Bag-1 5′-UTR. (A) Schematic diagram of the monocistronic constructs pSKL and pSKBL showing sites for run-off transcription. (B) EMSAs of radiolabelled segments of the Bag-1 5′-UTR with the addition of 2.5, 5 or 10 µg/µl (0.5, 1 or 2 µl total per reaction, respectively) of (i) PCBP1 or (ii) PTB. Arrows indicate positions of protein–RNA complexes (shifted) or RNA alone (unshifted).
Protein expression
Proteins were overexpressed in E.coli from the pET28a vector by the addition of isopropyl-β-d-thiogalactopyranoside to the growth medium. The proteins that contained a His tag were purified using a nickel affinity column (Qiagen). Alternatively, unr was purified from cultures of Sf9 cells that had been infected with a recombinant baculovirus expressing unr-His (Invitrogen). Cells were harvested and lysed in phosphate-buffered saline containing 0.1% Triton X-100, and the tagged protein purified on a nickel affinity column.
Cell culture and transient transfections
Cells were typically grown in Dulbecco’s modified Eagle’s medium (Gibco-BRL) containing 10% fetal calf serum, under humidified atmosphere containing 5% CO2. The cell lines CALU1, CAL51 and CAMA1 were a kind gift from Dr G. Packham (CRUK Unit, Southampton, UK). MCF7 and HeLa were originally purchased from ATCC. Cells were transfected using FuGene 6 (Roche) as specified by the manufacturer. Alternatively, calcium phosphate-mediated transfections were performed as described, with minor modifications (29). Lysates were prepared from transfected cells using 1× passive lysis buffer. Firefly and Renilla luciferase activities were measured using the ‘Stop and glo’ dual-luciferase reporter assay system (Promega) according to the manufacturer’s instructions with the exception that only 25 µl of each reagent was used. Light emission was measured over 10 s using an OPTOCOMP I luminometer. Activity of the β-galactosidase transfection control was measured using a Galactolight Plus assay system (Tropix). All transfections were carried out in triplicate on at least three independent occasions.
In vitro transcription reactions
Vector DNA was linearised by restriction digestion using a site downstream of the region of interest (HpaI for dicistronic, NcoI for monocistronic); transcripts were synthesised in a reaction mixture containing 1× transcription buffer [40 mM HEPES–KOH (pH 7.9), 6 mM MgCl2, 2 mM spermidine, 10 mM dithiothreitol (DTT), 10 mM NaCl], 40 U RNAguard or RNasin, 1 mM ATP, 1 mM UTP, 1 mM CTP, 0.5 mM GTP, 1 µM m7G(5′)ppp(5′)G, 1 µg of DNA template and 20 U T7 or T3 RNA polymerase to a final volume of 50 µl. For radiolabelled RNAs, CTP was replaced with 50 µCi [α-32P]CTP. The reaction mixture was incubated at 37°C for 1.5 h and the RNA purified.
In vitro translation reactions
The Promega rabbit reticulocyte flexi-lysate in vitro translation system was primed with 5 ng/µl RNA and used according to the manufacturer’s instructions. The reaction was performed in a final volume of 12.5 µl and 0.1 µg of each protein was added where appropriate. Luciferase activities were assayed as described above, and the firefly and Renilla values expressed relative to the control plasmid pRF, which was assigned a value of 1. All experiments were performed in triplicate on at least three independent occasions.
UV-crosslinking analysis
Radiolabelled transcript was generated from pSKBL linearised with NcoI. Approximately 2.5 pmol per reaction was incubated with 0.25 µg of protein in 1× UV-crosslinking buffer [10 mM HEPES (pH 7.4), 3 mM MgCl2, 100 mM KCl, 5 mM creatine phosphate, 1 mM DTT, 1 mM ATP, 6% glycerol, 0.1 µg/µl tRNA] for 15 min at room temperature. For competition assays, unlabelled competitor RNAs were added with labelled RNA. The reaction mixtures were UV irradiated using a 305 nm UV light source for 30 min on ice. RNase A and RNase V1 (0.2 mg/ml) were added to the mixture to degrade any unprotected RNA by incubation at 37°C for 30 min. Sample buffer was added and the samples separated on a 10% polyacrylamide gel by SDS–PAGE. Gels were dried at 80°C under vacuum for 2 h and analysed on a Molecular Dynamics Phosphorimager.
Electrophoretic mobility shift assays (EMSAs)
Approximately 20 pmol of RNA was incubated with protein as appropriate in a buffer mix containing 40 U RNAguard, 2 µl of 5× transcription buffer [200 mM Tris–-HCl (pH 8.0), 40 mM MgCl2, 10 mM spermidine, 250 mM NaCl, 50 mM DTT, 15 µg tRNA], and 2 µl of 10 mM ATP in a reaction volume of 15 µl for 10 min at room temperature. DNA loading dye was added (50% sucrose, 0.1 M EDTA, 0.2% bromophenol blue) and samples loaded onto 0.7× Tris-borate-EDTA (TBE) agarose gels. Samples were electrophoresed at 100 V for ∼3 h in 1× TBE loading buffer. All buffers and loading dyes were filter-sterilised. The gels were dried under vacuum at 60°C for 2 h and exposed on a phosphoimager.
RESULTS
Comparison of Bag-1 IRES-mediated internal initiation between cell types
The majority of cellular IRESs studied thus far show considerable cell tropism in that they do not work efficiently in all cell types (24–26). This is presumably because the expression of specific IRES trans-acting factors varies between cell lines (24–26). The ability of the Bag-1 IRES to function in a variety of cell lines was tested by transfecting HeLa, COS7, HEK293, MCF7, CAL51, CALU1 and CAMA1 cell lines with the dicistronic construct pRBF or the control construct pRF. The expression from both Renilla and firefly luciferase cistrons was assayed and normalised to the transfection control β-galactosidase. The efficiency of the IRES is represented as a ratio of firefly luciferase to Renilla luciferase expression from pRBF (Fig. 1A). As expected, the Bag-1 IRES showed a wide range of activities in different cell types and firefly luciferase activity was found to vary considerably according to cell line (Fig. 1B). The Bag-1 IRES was highly active in CAMA-1 and active in HeLa cells, COS7 and HEK293 cells, yet relatively inactive in MCF7, CAL51 and CALU1 cells (Fig. 1B). The expression of the Bag-1 IRES in these cell lines differed from that observed with the c-myc IRES (25) and the Apaf-1 IRES (30), which would again suggest that cellular IRESs have different requirements for trans-acting factors.
PCBP1 and PTB bind to the Bag-1 IRES
EMSAs were performed to identify putative Bag-1 IRES trans-acting factors by using a range of known IRES interacting proteins including PCBP1 and PCBP2 (22), PTB (20), DAP5 (31,32), La (28) and unrip (19). Radiolabelled Bag-1 IRES RNA was generated from in vitro transcription reactions primed with DNA derived from the monocistronic constructs (Fig. 2A); the resulting RNA was incubated with protein and the products separated on 0.5% TBE agarose gels. Only when BAG-1 IRES RNA was incubated with PTB or PCBP1 was a decrease in the mobility of the RNA observed (Fig. 2B and B.M.Pickering and A.E.Willis, unpublished data), suggesting both of these proteins bind the IRES directly. No difference in mobility of Bag-1 IRES RNA was observed with the other proteins tested, for example La (Fig. 2B, i, lane b). To test the specificity of this interaction, the proteins were also incubated with a non-specific RNA segment from glyceraldehyde-3-phosphate dehydrogenase (G3PDH) of approximately the same size. No alterations in mobility of this RNA were observed with any of the proteins tested (Fig. 2B, iii). To confirm this interaction, UV-crosslinking analysis was performed. Thus, radiolabelled Bag-1 IRES RNA was incubated with PCBP1 and/or PTB, samples were exposed to UV light, any RNA not bound to protein digested with RNases and the products separated by PAGE. Both PTB and PCBP1 either singly or in combination interacted with Bag-1 IRES RNA (Fig. 2C). Crosslinking analysis was also undertaken with cell extracts made from HeLa or placenta. Thus, radiolabelled Bag-1 IRES RNA was incubated with HeLa or placental extracts, exposed to UV light and unbound RNA was digested with RNases. The products were separated by PAGE. A number of proteins that interact with the Bag-1 IRES were identified in both extracts with sizes of approximately 150, 55, 45, 37, 30 and 29 kDa in addition to some extract-specific proteins (Fig. 2D). The 55 and 37 kDa proteins are the same size as PTB and PCBP1, respectively. The other proteins remain to be identified.
To determine the specificity of the interaction between the Bag-1 IRES and these proteins, UV-crosslinking experiments were performed in the presence of excess unlabelled Bag-1 IRES RNA or G3PDH mRNA (Fig. 2E). Both proteins bind specifically to the Bag-1 IRES. Hence, there was a reduction in the binding of protein to the radiolabelled transcripts with a 1-fold molar excess of unlabelled BAG-1 IRES RNA, but not with a 10-fold molar excess of G3PDH RNA (Fig. 2E, i and ii). Crosslinking analysis was also performed with La, unrip, DAP5 and PCBP2 but no protected RNA was detected (data not shown).
PCBP1 and PTB stimulate the Bag-1 IRES in vitro and in vivo
In general, cellular IRESs work very inefficiently (if at all) in vitro, but we have shown that it is possible to stimulate certain cellular IRESs by the addition of known viral trans-acting factors (27). The activity of the Bag-1 IRES in a dicistronic RNA (generated from pRBF and pRF; Fig. 1A) was tested in the rabbit reticulocyte lysate system with the addition of PCBP1 and PTB. The Bag-1 IRES functioned very inefficiently in vitro and no appreciable luciferase activity was detected over that produced from RNA derived from the vector pRF which does not contain an IRES (Fig. 1A). However, activation was observed with PTB and PCBP1, when added singly, each stimulating IRES activity to 1.4-fold (Fig. 3A). Moreover, addition of PCBP1 and PTB had an additive effect in combination producing 3-fold stimulation of the IRES (Fig. 3A).
To test whether these proteins could stimulate the function of the Bag-1 IRES in vivo, co-transfections were carried out using pRBF with plasmids expressing PCBP1 or PTB either singly or in combination into the cell lines which showed low Bag-1 IRES activity (Fig. 1). Thus, transfection of CAL51 and CALU1 with either PTB or PCBP1 alone had a stimulatory effect, which was additive when the plasmids containing DNA encoding these proteins were transfected in combination (Fig. 3C, i and ii). In contrast, transfection of MCF7s with PTB and/or PCBP1 did not significantly stimulate Bag-1 IRES activity (Fig. 3C, iii). To test whether there was a correlation between the expression of PTB/PCBP1 and IRES function western analysis was performed and cell lysates were immunoblotted for PTB, PCBP1 or actin as a loading control (Fig. 3D). There is a good correlation between endogenous protein levels and activation of the IRES by these proteins. For example, MCF7s which are not stimulated by co-transfection with PTB and/or PCBP1 have comparatively high levels of these proteins. In contrast, CALU1 or CAL51 cell lines which are stimulated by these proteins have lower expression of PTB and PCBP1 than MCF7 cells (Fig. 2D).
A comprehensive deletion analysis was then performed to identify the minimal element that harboured IRES activity to determine whether PTB or PCBP1 were essential for Bag-1 IRES function.
The minimum active fragment of the Bag-1 IRES is 186 nt
Regions of the Bag-1 5′-UTR DNA were obtained by PCR and subcloned into the dicistronic reporter vector pRF (Fig. 4A and B). Deletions were transfected into HeLa cells and cell lysates assayed for luciferase activity (Fig. 4C). A region of 218 nt from 225 to 411 was identified as the minimal element since this region of RNA retained 100% IRES activity. The 5′ end of this section of RNA must be critical for function of the IRES since deleting a further 25 nt at the 5′ end reduced the activity of the 258–441 fragment to 25% (Fig. 4C). It is possible to delete an additional 107 nt from the 3′ end and retain 50% of the IRES function with the 225–358 fragment, but the removal of a further 46 nt to generate the 225–312 fragment resulted in an inactive IRES (Fig. 4C). These data would suggest that the first 225 nt are not required for an active IRES, but this section of RNA may bind other, perhaps regulatory, proteins.
PCBP1 and PTB bind to the minimal active fragment
EMSAs were then performed to identify the regions of Bag-1 IRES RNA that could be bound by PCBP1 and PTB. Fragments of the DNA encoding Bag-1 IRES RNA were obtained by PCR and inserted into the vector pSKL (Fig. 5A). Radiolabelled RNAs were generated from these plasmids by in vitro transcription reactions and these were incubated with PTB or PCBP1 (Fig. 5B, i). The minimum active fragment 225–411 binds both PTB and PCBP1 as efficiently as the full-length RNA (Figs 5B and 2B). To refine the binding sites further, RNA was generated from additional deletion constructs (Fig. 5A) and these were used in EMSAs. Thus, incubating the 258–358 segment of radiolabelled Bag-1 IRES RNA with increasing amounts of PCBP1 showed that PCBP1 bound directly to this region of the IRES. A decrease in mobility of the radiolabelled 292–358 fragment was also observed and this would imply that PCBP1 binds to a 66 nt fragment. The 3′ end of this fragment is important for binding since no protein–RNA complexes were observed with the RNA generated from either 258–312 or 274–312 segments of the 5′-UTR (Fig. 5B, i).
Incubating increasing amounts of PTB with the 258–358 and 292–358 segments of RNA shows that this protein binds to these RNA segments (Fig. 5B, ii). Interestingly, addition of a 2-fold molar excess of PTB to RNA gave rise to a shifted band of a position that would correspond to a dimer of PTB. PTB also interacts with the 258–312 fragment and the 274–312 fragment, but this only occurs at a 2-fold molar excess of the protein and may be non-specific. Thus, this analysis refined the PCBP1 binding site to the 292–312 region of the 5′-UTR, whilst PTB appeared to bind at multiple sites along the 258–358 region.
Thus, both proteins interact strongly with the fragment that retains 50% of the activity (225–358) suggesting that these are indeed trans-acting factors for the Bag-1 IRES.
DISCUSSION
To obtain a better understanding about the function and mechanisms of action of cellular IRESs it is essential that the trans-acting factors that mediate internal ribosome entry are elucidated. This is a complex task since the data suggest that each cellular IRES has a distinct set of trans-acting factor requirements (Fig. 1) (24–28). However, it would appear that some of the proteins that mediate internal ribosome entry on viral IRESs are also used by cellular IRESs. Thus, both the Apaf-1 IRES and HRV IRESs require unr and PTB for function (27) and the XIAP and polio virus IRESs require La for activity (28). In order to identify some of the trans-acting factors that are used by the Bag-1 IRES, known viral IRES trans-acting factors were tested to determine which could interact with the Bag-1 IRES RNA. Of the seven proteins that were tested, only two, PTB and PCBP1, could bind in EMSAs (Fig. 2B). UV-crosslinking studies showed that both PCBP1 and PTB interact directly and specifically with Bag-1 IRES RNA (Fig. 2C and D). Interestingly, these proteins would appear to work in concert since a combination of these two proteins stimulated the Bag-1 IRES 3-fold in vitro (Fig. 3B). Moreover, it was possible to increase the activity of the Bag-1 IRES in the cell lines that had low Bag-1 IRES activity by co-transfection of pRBF with plasmids containing cDNAs encoding these proteins (Fig. 3C, i and ii).
PCBP1, which is a member of the KH domain family of single-stranded nucleic acid binding proteins (33), was found to bind strongly to a 66 nt fragment (Fig. 5). The proteins in the KH domain family generally bind to cytidine-rich sequences (33), however, there is not a cytidine-rich stretch in the linear sequence to which PCBP1 binds. It is likely, therefore, that this protein is recognising secondary or tertiary structural motifs in the Bag-1 IRES. Computer predictions have been carried out using the mfold program, although, in the absence of experimentally derived data it is difficult to predict the structure of RNA that is being recognised. It is of interest to note that PCBP2, which has 90% amino acid similarity to PCBP1, does not bind to Bag-1 IRES RNA (data not shown). Therefore, these proteins must recognise distinct structural motifs. In this regard, it has been shown that PCBP2, but not PCBP1, is able to restore the activity of entero/rhinoviruses in PCBP-depleted extracts (23). In addition, PCBP1 and PCBP2 have been found to bind to different structural motifs of the polio virus IRES and mediate stability of the mRNA (34).
PTB contains four RNA recognition motifs and has been shown to interact with both cellular (35) and viral IRESs (13). The PTB binding site on the Bag-1 IRES was less well defined than the PCBP1 site (Fig. 5), although it was found to bind strongly to segments of the minimal Bag-1 IRES element (Figs 4 and 5). The EMSA data would suggest that PTB is able to bind to the RNA as a dimer (Fig. 5B) and it is thought that PTB exists in solution as a dimer (36). The binding of PTB to viral IRESs is consistent with the notion that this protein interacts with multiple structures and brings them into the correct conformation for ribosome binding (13) and it is likely that the same is true for the Bag-1 IRES.
Both the binding sites for PCBP1 and PTB are located within the minimum element of the IRES that has full activity (from 225 to 411) and indeed both bind within the region which has 50% activity of the full-length IRES (Fig. 5). Since the activity of the Bag-1 IRES in vitro is enhanced by the addition of both PTB and PCBP1, and given that PTB seems to have multiple contact points on the RNA, we suggest that these proteins act as RNA chaperones and allow the Bag-1 IRES to attain the correct structure that is competent for 40S ribosomal subunit entry. To test this theory, work is being carried out to obtain a secondary structural model for the Bag-1 IRES in the presence of these proteins.
Acknowledgments
ACKNOWLEDGEMENTS
This work was funded by grants from the BBSRC (fellowship to A.E.W.) and the Wellcome Trust (S.A.M.). B.M.P. and J.R.E. hold MRC studentships.
REFERENCES
- 1.Yang X., Chernenko,G., Hao,Y., Ding,Z., Pater,M.M., Pater,A. and Tang,S.-C. (1998) Human BAG-1/RAP46 protein is generated as four isoforms by alternative translation initiation and overexpressed in cancer cells. Oncogene, 17, 981–989. [DOI] [PubMed] [Google Scholar]
- 2.Packham G., Brimmell,M. and Cleveland,J.L. (1997) Mammalian cells express two differently localised Bag-1 isoforms generated by alternative translation initiation. Biochem. J., 328, 807–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeiner M. and Gehring,U. (1995) A protein that interacts with members of the nuclear hormone receptor family: identification and cDNA cloning Proc. Natl Acad. Sci. USA, 92, 11465–11469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeiner M., Niyaz,Y. and Gehring,U. (1999) The hsp70-associating protein Hap46 binds to DNA and stimulates transcription. Proc. Natl Acad. Sci. USA, 96, 10194–10199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stuart J.K., Myszka,D.G., Joss,L., Mitchell,R.S., McDonald,S.M., Xie,Z., Takayama,S., Reed,J.C. and Ely,K.R. (1998) Characterization of interactions between the anti-apoptotic protein BAG-1 and Hsc70 molecular chaperones. J. Biol. Chem., 273, 22506–22514. [DOI] [PubMed] [Google Scholar]
- 6.Nollen E.A.A., Kabakov,A.E., Brunstung,J.F., Kanon,B., Hohfeld,J. and Kampinga,H.H. (2001) Modulation of in vivo HSP70 chaperone activity by Hip and Bag-1. J. Biol. Chem., 276, 4677–4682. [DOI] [PubMed] [Google Scholar]
- 7.Gassler C.S., Wiederkehr,T., Brehmer,D., Bukau,B. and Mayer,M.P. (2001) Bag-1M accelerates nucleotide release for human Hsc70 and Hsp70 and can act concentration-dependent as positive and negative cofactor. J. Biol. Chem., 276, 32538–32544. [DOI] [PubMed] [Google Scholar]
- 8.King F.W., Wawrzynow,A., Hohfeld,J. and Zylicz,M. (2001) Co-chaperones Bag-1, Hop and Hsp40 regulate Hsc70 and Hsp90 interactions with wild-type or mutant p53. EMBO J., 20, 6297–6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takayama S., Sato,T., Krajewski,S., Kochel,K., Irie,S., Millan,J.A. and Reed,J.C. (1995) Expression and location of Hsp70/Hsc-binding anti-apoptotic protein BAG-1 and its variants in normal tissues and tumor cell lines. Cell, 80, 279–284.7834747 [Google Scholar]
- 10.Takayama S., Krajewski,S., Krajewska,M., Kitada,S., Zapata,J.M., Kochel,K., Knee,D., Scudiero,D., Tudor,G., Miller,G.J. et al. (1998) Cloning and functional analysis of BAG-1: a novel Bcl-2-binding protein with anti-cell death activity. Cancer Res., 58, 3116–3131.9679980 [Google Scholar]
- 11.Coldwell M.J., deSchoolmeester,M.L., Fraser,C.A., Pickering,B.M., Packham,G. and Willis,A.E. (2001) The p36 isoform of BAG-1 is translated by internal ribosome entry following heat shock. Oncogene, 20, 4095–4100. [DOI] [PubMed] [Google Scholar]
- 12.Hellen C.U.T. and Sarnow,P. (2001) Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev., 15, 1593–1612. [DOI] [PubMed] [Google Scholar]
- 13.Belsham G.J. and Jackson,R.J. (2000) Translation initiation on picornavirus RNA. In Sonenberg,N., Hershey,J.W.B. and Mathews,M.B. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 869–900.
- 14.Pestova T.V., Shatsky,I.N. and Hellen,C.U.T. (1996) Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol. Cell. Biol., 16, 6870–6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pestova T.V., Hellen,C.U.T. and Shatsky,I.N. (1996) Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol., 16, 6859–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown B. and Ehrenfeld,E. (1979) Translation of poliovirus RNA in vitro: changes in cleavage pattern and initiation sites by ribosomal salt wash. Virology, 97, 396–405. [DOI] [PubMed] [Google Scholar]
- 17.Borman A., Howell,M.T., Patton,J.G. and Jackson,R.J. (1993) The involvement of a spliceosome component in internal initiation of human rhinovirus RNA translation. J. Gen. Virol., 74, 1775–1788. [DOI] [PubMed] [Google Scholar]
- 18.Borman A.M., Bailly,J.-L., Girard,M. and Kean,K.M. (1995) Picornavirus internal ribosome entry segments: comparison of translation efficiency and the requirements for optimal internal initiation of translation in vitro. Nucleic Acids Res., 23, 3656–3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunt S.L., Hsuan,J.J., Totty,N. and Jackson,R.J. (1999) unr, a cellular cytoplasmic RNA-binding protein with five cold-shock domains, is required for internal initiation of human rhinovirus RNA. Genes Dev., 13, 437–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunt S.L. and Jackson,R.J. (1999) Polypyrimidine tract binding protein (PTB) is necessary, but not sufficient, for efficient internal initiation of translation of human rhinovirus-2 RNA. RNA, 5, 344–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Svitkin Y.V., Meerovitch,K., Lee,H.S., Dholakia,J.N., Kenan,D.J., Agol,V.I. and Sonenberg,N. (1994) Internal translation initiation on poliovirus RNA; further characterisation of La function in poliovirus translation in vitro. J. Virol., 68, 1544–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gamarnik A.V. and Adino,R. (1997) Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA, 3, 882–892. [PMC free article] [PubMed] [Google Scholar]
- 23.Walter B.L., Nguyen,J.H.C., Ehrenfeld,E. and Semler,B.L. (1999) Differential ultilization of poly (rC) binding protein 2 directed by picornavirus IRES elements. RNA, 5, 1570–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jopling C.L. and Willis,A.E. (2001) N-myc translation is initiated via an internal ribosome entry segment that displays enhanced activity in neuronal cells. Oncogene, 20, 2664–2670. [DOI] [PubMed] [Google Scholar]
- 25.Stoneley M., Subkhankulova,T., Le Quesne,J.P.C., Coldwell,M.J., Jopling,C.L., Belsham,G.J. and Willis,A.E. (2000) Analysis of the c-myc IRES; a potential role for cell-type specific trans-acting factors and the nuclear compartment. Nucleic Acids Res., 28, 687–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Creancier L., Mercier,P., Prats,A.-C. and Morello,D. (2001) c-myc internal ribosome entry site activity is developmentally controlled and subjected to a strong translational repression in adult transgenic mice. Mol. Cell. Biol., 21, 1833–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitchell S.A., Brown,E.C., Coldwell,M.J., Jackson,R.J. and Willis,A.E. (2001) Protein factor requirements of the Apaf-1 internal ribosome entry segment: roles of polypyrimidine tract binding protein and upstream of N-ras. Mol. Cell. Biol., 21, 3364–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holcik M. and Korneluk,R.G. (2000) Functional characterization of the X-linked inhibitor of apoptosis (XIAP) internal ribosome entry site element: role of La autoantigen in XIAP translation. Mol. Cell. Biol., 20, 4648–4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stoneley M. (1998) Functional analysis of the 5′ untranslated region of the c-myc proto-oncogene. PhD Thesis, University of Leicester, UK.
- 30.Coldwell M.J., Mitchell,S.A., Stoneley,M., MacFarlane,M. and Willis,A.E. (2000) Initiation of Apaf-1 translation by internal ribosome entry. Oncogene, 19, 899–905. [DOI] [PubMed] [Google Scholar]
- 31.Henis-Korenblit S., Levy Strumpf,N., Goldstaub,D. and Kimchi,A. (2000) A novel form of DAP5 protein accumulates in apoptotic cells as a result of caspase cleavage and internal ribosome entry site-mediated translation. Mol. Cell. Biol., 20, 496–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henis-Korenblit S., Shani,G., Sines,T., Marash,L., Shohat,G. and Kimchi,A. (2002) The caspase-cleaved DAP5 protein supports internal ribosome entry site-mediated translation of death proteins. Proc. Natl Acad. Sci. USA, 99, 5400–5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Makeyev A.V. and Liebhaber,S.A. (2002) The poly (C)-binding proteins: a multiplicity of functions and a search for mechanisms. RNA, 8, 265–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murray K.E., Roberts A.W. and Barton,D.J. (2001) Poly r(C) binding proteins mediate polio virus mRNA stability. RNA, 7, 1126–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim Y.K., Hahm,B. and Jang,S.K. (2000) Polypyrimidine tract-binding protein inhibits translation of Bip mRNA. J. Mol. Biol., 304, 119–133. [DOI] [PubMed] [Google Scholar]
- 36.Perez I., McAfee,J.G. and Patton,J.G. (1997) Multiple RRMs contribute to RNA binding specificity and affinity for polypyrimidine tract binding protein. Biochemistry, 36, 11881–11890. [DOI] [PubMed] [Google Scholar]