Abstract
Short interfering RNA (siRNA), the active agent of RNA interference, shows promise of becoming a valuable tool in both basic and clinical research. We explore the tolerance to mutations and chemical modifications in various parts of the two 21-nt strands of a siRNA targeting the blood clotting initiator Tissue Factor. The mutations were G/C transversions. The chemical modifications were 2′-O-methylation, 2′-O-allylation and phosphorothioates. We found that siRNA generally tolerated mutations in the 5′ end, while the 3′ end exhibited low tolerance. This observation may facilitate the design of siRNA for specific targeting of transcripts containing single nucleotide polymorphisms. We further demonstrate that in our system the single antisense strand of the wild-type siRNA is almost as effective as the siRNA duplex, while the corresponding methylated M2+4 version of the antisense had reduced activity. Most of the chemically modified versions tested had near-wild-type initial activity, while the long-term activity was increased for certain siRNA species. Our results may improve the design of siRNAs for in vivo experiments.
INTRODUCTION
RNA interference (RNAi), the process of depleting RNA targets by the use of double-stranded RNA (dsRNA), was first reported in 1998 (1). Since the demonstration of the efficacy of short interfering RNA (siRNA) in mammalian cells (2–4), this new tool has been used to successfully target various infectious agents like HIV, poliovirus and RSV (5–9). In some cases, newly constructed vectors were used to produce short hairpin RNA (shRNA) that is processed to siRNA intracellularly (10–14).
The mechanism of RNAi is not well understood (15–22). The triggering long dsRNA or aberrant RNA is assumed to be processed by the RNase III-like enzyme Dicer to approximately 21–23 nt siRNA, which is then incorporated into the RNA-induced silencing complex (RISC). This step can be bypassed by transfection of chemically synthesised siRNA. Dicer has also been implicated in processing of the siRNA-related short temporal RNA let-7, shown in Caenorhabditis elegans to be involved in control of larval development, and widely conserved in other species, among them humans (23–25).
The physiological function of RNAi is assumed to be defence against viral infections. This has been convincingly demonstrated in plants, where some viruses have evolved counter-measures against RNAi (26,27). An insect virus has recently been shown to both activate RNA silencing and express a suppressor protein in Drosophila cells (28). In C.elegans, mutations in RNAi genes have resulted in the activation of transposons (29,30), arguing for their involvement in the defence against these genomic parasites.
The tolerance of the effect of siRNA for mutations is still unclear. Boutla et al. (31) reported that a mutated siRNA with a single centrally located mismatch relative to the mRNA target sequence retained substantial activity in Drosophila. In contrast, Elbashir et al. (32) found that a single mismatch was deleterious to activity in an in vitro Drosophila embryo lysate assay. In previous work (33) we tried to reconcile these conflicting results by depicting the RNAi process in vivo as a dynamic one where several factors influenced the final outcome, among them siRNA target position, siRNA concentration, mRNA concentration, mRNA synthesis rate and siRNA’s inherent cleavage activity, an activity that can be reduced gradually by mismatch mutations.
In the present work we explore how various mutations and chemical modifications alter the efficacy and duration of our most effective siRNA (hTF167i) targeting the human Tissue Factor (hTF) mRNA. The objectives were, firstly, to find regions less tolerant in their siRNA effect for single mutations, thus possibly facilitating the design of siRNA for specific targeting of transcripts containing single nucleotide polymorphisms. Secondly, we wished to improve the long-term activity of our siRNA by stabilising the RNA strands against nucleases through introducing phosphorothioates and modifications of the 2′-OH.
We find that hTF167i has a general tolerance to mutations, with less tolerance for mutations at 3′ end of the siRNA. Furthermore, with the exception of certain allyl-modifications, the backbone modifications did not reduce the activity of the siRNA to a significant degree. Extensive use of phosphorothioate modifications resulted in cytotoxicity. The 2′-O-methylation modifications, on the other hand, showed promise as they resulted in increased persistence of activity with no toxicity to cells.
MATERIALS AND METHODS
SiRNA preparation
RNAs (21 nt) were chemically synthesised using phosphoramidites (Glen Research and PerSeptive Biosystems). Deprotected and desilylated synthetic oligoribonucleotides were purified by reverse-phase HPLC. Ribonucleotides were annealed at 10 µM in 200 µl 10 mM Tris–HCl pH 7.5 by boiling and gradual cooling in a water bath. Successful annealing was confirmed by non-denaturing (15%) polyacrylamide gel electrophoresis.
Cell culture and transfections
The human keratinocyte cell line HaCaT was cultured in serum-free keratinocyte medium (Gibco BRL) supplemented with 2.5 ng/ml epidermal growth factor and 25 µg/ml bovine pituitary extract. The cell line was regularly passaged at sub-confluence and plated 1 or 2 days before transfection. HaCaT cells in 6-well plates were transfected at low confluency (<40%) with 1.0 ml 100 nM siRNA in serum-free medium, using Lipofectamine 2000. For complexation, 10 µM stock solution of siRNA was diluted with 10× vol. of serum-free medium and mixed with an equal volume of medium-diluted Lipofectamine 2000, at a v/w ratio of liposome to siRNA of 5:2. Batch dilutions of liposomes were performed for each 6-well plate and allowed to pre-incubate at room temperature for 5–7 min before addition to the medium-diluted siRNA. Complexes were replaced with full medium 5 h after initiation of transfection. For standard assays of activity, cells were harvested the day after transfection. For longer incubations and time-course experiments, medium was replaced every second day after transfection.
Northern analysis
Polyadenylated mRNA was isolated using Dynabeads oligo(dT)25 (Dynal). Isolated mRNA was fractionated by electrophoresis for 16–18 h on 1.3% agarose/formalde hyde (0.8 M) gels and blotted on to nylon membranes (MagnaCharge, Micron Separations). Membranes were hybridised with random-primed Tissue Factor (TF) (position 61–1217 in cDNA) and GAPDH (1.2 kb) cDNA probes in PerfectHyb hybridisation buffer (Sigma).
RESULTS
Mutational scanning of siRNA targeting hTF167
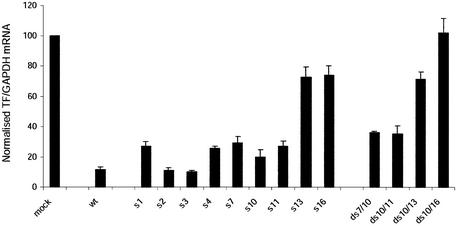
We have previously demonstrated that one and two central mutations in siRNA targeting position 167 in hTF did not abolish its ability to deplete endogenous TF mRNA levels (33). We decided to map this siRNA more systematically in order to determine whether mutations were tolerated equally well within the whole siRNA. To simplify analysis, only GC pairs were mutated by inversion of the pairs. A total of eight different single-mutant siRNAs were designed and named according to the position (starting from the 5′ of the sense strand) of the mutation (s1, s2, s3, s4, s7, s11, s13, s16) (Fig. 1). The previously described centrally located single-mutant M1 (33), was included in this analysis and renamed s10.
Figure 1.
Mutated and wild-type (wt) versions of the siRNA hTF167i. The sequence of the sense strand of wild-type siRNA corresponds to position 167–187 in hTF (Acc.No. M16553). Single (s1, s2, s3, s4, s7, s10, s11, s13, s16) and double (ds7/10, ds10/11, ds10/13, ds10/16) mutants are all named according to the position of the mutation, counted from the 5′ end of the sense strand. All mutations (in bold) are GC inversions relative to the wild-type.
These mutant siRNAs were analysed for their ability to deplete endogenous TF mRNA in HaCaT cells (Fig. 2). The wild-type siRNA, and the mutant s10, included as positive controls, depleted TF mRNA to 10 and 20% residual levels, as reported previously (33). The activities of the other mutants could be categorised into three groups depending on their position along the siRNA. Mutations in the extreme 5′ end of the siRNA were well tolerated, exhibiting essentially the same activity as the wild-type. Mutations localised towards the 3′ end, and up to the approximate midpoint of the siRNA (s4, s7, s10, s11), were slightly impaired in their activity, resulting in depletion of mRNA to 20–30% residual levels. Both of the mutations in the 3′ half of the siRNA (s13, s16), however, exhibited severely impaired activity, suggesting a bias in the tolerance for mutations in the reaction(s) involving siRNA. The activities of four double mutants, in which the central position (s10) was mutated in conjunction with one additional position (s7, s11, s13, s16), were also analysed. The bias in mutation tolerance was also evident for these double mutants, as the rank order of their activity mirrored that of the activity of the single mutants of the variant position. This observation strengthens the proposition that the differential activity of mutants is due to an intrinsic bias in the tolerance for target mismatches along the sequence of the siRNA.
Figure 2.
Activity of mutant siRNAs against endogenous hTF mRNA. HaCaT cells were harvested for mRNA isolation 24 h post-transfection. TF expression was normalised to that of GAPDH. Normalised expression in mock-transfected cells was set as 100%. Data are averages + s.d. of at least three independent experiments.
Chemical modification of siRNAs for increased persistence of silencing
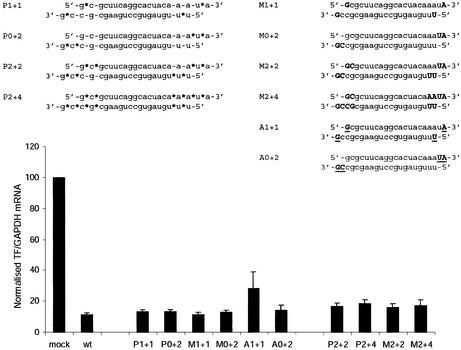
The effect of siRNA in human cells is transient and typically decreases during a few days in cell culture (18,33). The ability to extend the period of effective silencing would be of importance for the possible use of siRNA in therapy. To increase the intracellular stability of siRNA, we introduced a gradual increase of various chemical modifications to both ends of the siRNA. We have previously used phosphorothioate linkages and 2′-O-modifications in the form of methylation and allylation to stabilise hammerhead ribozymes (34), and decided to try a similar strategy for siRNAs. In all modified siRNA species, the same modifications were introduced in both strands. It has been reported that siRNA with a general 2′-O-methylation in either strand have no activity (32). We therefore started with less extensive modification. Initially, siRNAs with one modification in each of the extreme 5′ and 3′ ends of their strands (P1+1, M1+1, A1+1, Fig. 3) were synthesised. The 5′ end of the siRNAs might be more sensitive to modification since it must be phosphorylated in order to be active in vivo (22). We therefore included siRNAs with modifications only in the non-basepairing 3′ overhangs (P0+2, M0+2 and A0+2, Fig. 3), which were known to tolerate various types of modifications (2,3,32,33).
Figure 3.
Activity of chemically modified versions of the siRNA hTF167i. Non-modified ribonucleotides are in lower case. Phosphorothioate linkages are indicated by asterisks (*), while 2′-O-methylated and 2′-O-allylated ribonucleotides are in normal and underlined bold upper case, respectively. Expression of TF and GAPDH mRNA was determined 24 h post-transfection of HaCaT cells. Experiments were performed and analysed as in Figure 2. Data are averages + s.d. of at least three independent experiments.
Northern analysis of transfected HaCaT cells demonstrated essentially undiminished activity of all the chemically modified siRNAs, with the exception of the siRNA with allylation at both ends (Fig. 3). Allyl-modification in the 3′ end only had no effect on activity. The presence of a bulky substituent in the 2′-hydroxyl of the 5′ terminal nucleotide might interfere with the in vivo phosphorylation of the siRNA necessary for its activity (22). Further modification of the siRNA by methylation resulted in gradual attenuation of activity. One day after transfection with siRNAs carrying 4+4, 4+6, 6+6 or 6+8 methyl groups in their ends, the cells exhibited residual reporter gene expression levels of 28, 42, 68 and 75%, respectively.
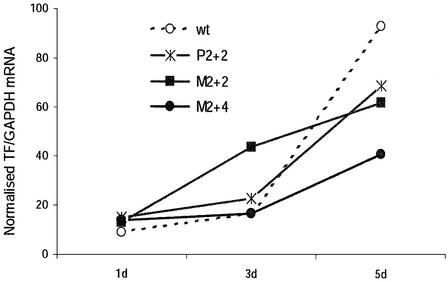
We next wanted to know if any of these modifications increased the persistence of siRNA-mediated silencing. The level of endogenous TF mRNA recovers gradually 3–5 days after transfection with wild-type siRNA targeting hTF167 (33). Transfecting HaCaT cells with unmodified and chemically modified siRNAs in parallel did not result in any significant differences in their silencing activities at 3 and 5 days post-transfection until a rather high degree of modification was used. Allylated siRNAs were not tested in this experiment, since they showed reduced effectivity even with only one substituent in the 5′ end (Fig. 3). SiRNAs methylated in the 2′-OH in up to 2+4 positions showed well conserved activity at 24 h [16–18% residual levels compared with 11% in cells transfected with the wild-type (Fig. 3)]. The most extensively phosphorothioated siRNA proved to be cytotoxic, resulting in ∼70% cell death compared with mock-transfected cells (measured as the expression level of the control mRNA GAPDH). This siRNA species was therefore not included in the further analysis. The remaining siRNA species were evaluated for increased persistence of silencing by analysing TF mRNA expression 5 days after a single transfection of 100 nM siRNA. At this point, TF expression in cells transfected with wild-type siRNA had recovered almost completely (80% residual expression compared with mock-transfected cells) (data not shown). In cells transfected with the most extensively 2′-O-methylated siRNA (M2+4), however, strong silencing was still evident (32% residual expression). The less extensively modified siRNA species (P2+2, M2+2), although less effective than M2+4, consistently resulted in lower TF expression 5 days post-transfection (55–60%) than the wild-type. Fewer modifications (P1+1, P0+2, M1+1, M0+2, A0+2) did not improve the silencing effect 5 days post-transfection. Time-course experiments demonstrated that the wild-type siRNA was still the most effective 3 days post-transfection, when silencing was relatively unimpaired. However, silencing drops off at a much higher rate thereafter for the unmodified siRNA (Fig. 4), possibly due to a faster depletion of the intracellular siRNA pool.
Figure 4.
Persistence of TF silencing by chemically modified siRNAs in HaCaT cells harvested 1, 3 and 5 days after a single transfection of 100 nM siRNA. Medium was replaced every second day. Data are from one representative out of three independent experiments.
Comparison of the effect of duplex and single-stranded antisense siRNA in whole cells
The antisense strand of siRNA is incorporated into RISC in HeLa cell extracts and supports RISC-specific target RNA cleavage, although at lower efficiency than the siRNA duplex (35,36). The antisense RNA was shown to silence transgene expression in a dose-dependent manner (36). We investigated whether antisense siRNA could silence endogenous gene expression with an efficiency comparable to duplex siRNA at moderate concentrations of the antisense oligo. HaCaT cells were transfected in parallel with 100 nM siRNA or 200 nM of the antisense strand, with or without pre-phosphorylation of the oligo (Fig. 5). In our quantitative assay the antisense RNA was as efficient as the siRNA duplex in depleting endogenous TF mRNA. A clear cleavage product was seen for both agents. The observation that antisense RNA exhibits almost full activity most likely reflects the high efficiency and excess capacity of this particular siRNA species. For the slightly less efficient chemically modified siRNA M2+4, on the other hand, there is a clear difference in depletion rate between the duplex siRNA and antisense siRNA (Fig. 5), suggesting that higher concentrations are required for maximum effect of this antisense RNA. Phosphorylation prior to transfection did not improve the efficacy of either antisense strand, consistent with recent observations that single-stranded RNA can be phosphorylated in vivo (36).
Figure 5.
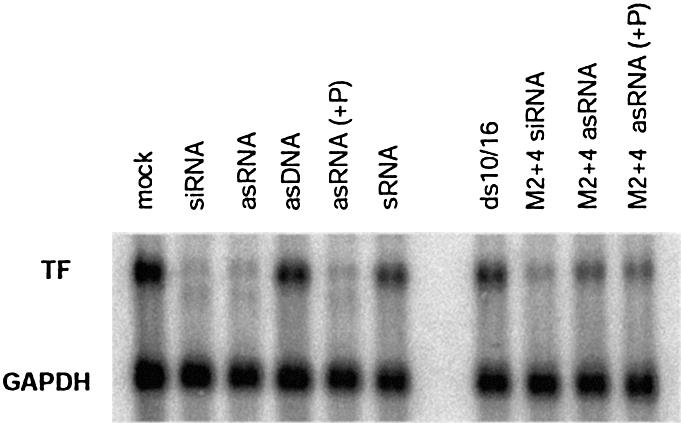
Levels of TF and control mRNA (GAPDH) in cells 24 h after transfection with various siRNA derivatives (see Figs 1 and 3). Cells were transfected with 100 nM siRNA duplex or 200 nM single-stranded RNA or DNA. RNA samples phosphorylated prior to transfection are indicated with ‘(+P)’. Phosphorylation was by polynucleotide kinase (New England Biolabs), followed by phenol/chloroform extraction and desalting on G25 Sephadex Quick-Spin columns (Roche).
DISCUSSION
This work has demonstrated tolerance of a highly effective siRNA, hTF167i, for a wide range of mutational and chemical modifications, as well as for removal of the sense strand from the duplex. Although allyl-modification of both terminal nucleotides resulted in some loss of activity, a total of six modifications in the form of either methylation of the sugar moiety or thiolation of the backbone were well tolerated, causing only a marginal reduction in the maximal silencing observed with the unmodified siRNA. Toxicity was, however, observed with longer stretches of phosphorothioates, but not with the same level of methyl-modification. SiRNA with 2+4 2′-O-methylated terminal nucleotides (M2+4) demonstrated both conserved initial activity and its increased duration in time-course experiments. These active end-methylated siRNAs are in possible contrast to the observations by Elbashir et al. (32) that fully 2′-O-methylated siRNAs are inactive. While full substitution of either strand with 2′-deoxynucleotides destroys siRNA activity (31–33), up to four deoxynucleotide modifications in the 3′ ends were found to support good activity in an in vitro cleavage assay (32). Taken together, the above data suggest the existence of different degrees of tolerance for chemical modification of siRNAs.
We find that single-stranded antisense siRNA can be highly effective in depleting endogenous gene expression at relatively moderate concentrations. Although antisense siRNA is still less effective than the corresponding siRNA duplex, this observation should help reduce the cost of selecting the best siRNA site, as the initial screening can be accomplished through the synthesis of the antisense strand only.
We have demonstrated a lower tolerance to mutations in the 3′ end of our most active siRNA. This bias cannot be due to differences in siRNA duplex stability as all mutations were G-C inversions and thus to a first approximation energetically equal. The stability of the duplex between a mutated siRNA antisense strand and the mRNA should be more severely affected by a central than a peripheral target mismatch. Consistent with this, we observe that 5′ end mutations have a negligible effect on activity compared with more centrally located mutations. The observed bias might be linked to the proposed existence of a ‘ruler’ region in the siRNA which is primarily used by the RISC complex to ‘measure’ the target mRNA for cleavage (32). It was demonstrated that the 5′ end of the antisense strand of siRNA sets the ruler for target RNA cleavage. This is likely to occur by a physical interaction of RISC with this region of the siRNA, which should therefore be more sensitive to mismatches with the target RNA. The universality of this observation is currently being investigated, using siRNAs targeting other sites in TF. The identification of a region of generally increased sensitivity to the effect of mismatches with the target mRNA, would be of great importance for the potential use of siRNAs to specifically target transcripts of disease-associated alleles in various dominant-negative disorders.
Highly diverging effects of mutations on siRNA activity have been reported. Jacque et al. (6) found that a single mismatch in siRNA targeting the HIV LTR resulted in partial loss of activity, while another siRNA targeting the HIV VIF exhibited almost full activity. Four mutations, however, abolished activity completely. Complete abolishment of activity has been reported by Gitlin et al. (8), Klahre et al. (37) and Garrus et al. (38), for siRNAs with 5, 6 and 7 mutations, respectively. Kisielow et al. (39) reported that a siRNA resulting in essentially complete knockdown of the expression of its target gene, was unable to inhibit the expression of a transgene with two non-contiguous mutations in the recognition sequence of the siRNA. The positions of these mutations correspond to our ds10/13 double mutant, which also exhibited low activity in our assay. A central double mutation, reported by Boutla et al. (31) and ourselves (33) to support partial activity, led to severe loss of activity for Yu et al. (40) and Wilda et al. (41), the latter using a siRNA with only 17 base pairs. On the other hand, abolishment of in vivo activity by a single mutation has been reported. Brummelkamp et al. (10), using a shRNA assumed to be processed to siRNA by Dicer (13), achieved inactivation by single mutations in either the second or ninth position from the putative 5′ end of the shRNA. Gitlin et al. (8), argued the case for single mutation inactivation more strongly by isolating siRNA-resistant poliovirus strains containing single mutations in the target site on the genomic RNA, corresponding to the sixth or ninth nucleotide of the siRNA, counted from the 5′ end of the sense strand. On balance, different siRNAs seem to be inactivated by mutations to different degrees, the outcome being at least in part target-sequence dependent.
In previous work (33) we established that the siRNA effect depends at least in some instances on the siRNA target position, mirroring earlier observations for ribozymes and antisense oligos (34,42,43). Further support for siRNA position effects has now emerged from several different sources. Several examples of inactive siRNAs in mammalian cells have recently been described (11,44,45), while a weak positional effect in Drosophila lysates can be inferred from published data (32). In C.elegans, Simmer et al. (46) managed to activate dozens of previously inactive dsRNA stretches, using the RNAi-sensitive rrf-3 negative strain, and in some cases created RNAi knockout phenotypes from genes that had hitherto not been responsive to dsRNA. Finally, Yang et al. (47) recently reported some inactive chemically synthesised and in vitro transcribed siRNA, as well as inactive shRNA. The authors demonstrated that the new technique of esiRNA, in which an overlapping set of siRNAs are produced in vitro by partial digestion with Escherichia coli RNase III, can be superior even to dsRNA. Processing of dsRNA by Dicer starts from a fixed end and proceeds in a sequential manner (3,48,49), producing a largely non-overlapping set of siRNAs. Cleavage of a dsRNA by E.coli RNase III will thus create a larger and more complete set of siRNAs than the non-overlapping set produced by Dicer from dsRNA. The higher activity of the esiRNA supports the argument that different siRNAs have different activities. Otherwise any set of siRNA from a dsRNA would have the same activity.
The inactivation of siRNA by mismatches has implications for the proposed function of RNAi as a defence system against retro-transposons and viruses. It is unclear why a viral defence mechanism should allow escape of a virus by a single mismatch. A differentiated population of siRNA with widely differing activities seems more likely. Some siRNAs with an intermediate activity can thus be more vulnerable to siRNA:target mismatch, while intrinsically stronger siRNAs have excess capacity and tolerate a single mismatch, as clearly exemplified by the tolerance exhibited to chemical and mutational modifications of hTF167i in the present study. This tolerance in highly active siRNAs should make viral escape more difficult, and our model is therefore consistent with both the published data and the proposed biological role of RNAi as a viral defence.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by grants from the Norwegian Cancer Society, Health and Rehabilitation, and the Research Council of Norway (RCN) to H.P. T.H. is a fellow of RCN.
REFERENCES
- 1.Fire A., Xu,S., Montgomery,M.K., Kostas,S.A., Driver,S.E. and Mello,C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- 2.Elbashir S.M., Harborth,J., Lendeckel,W., Yalcin,A., Weber,K. and Tuschl,T. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature, 411, 494–498. [DOI] [PubMed] [Google Scholar]
- 3.Elbashir S.M., Lendeckel,W. and Tuschl,T. (2001) RNA interference is mediated by 21 and 22 nt RNAs. Genes Dev., 15, 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caplen N.J., Parrish,S., Imani,F., Fire,A. and Morgan,R.A. (2001) Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl Acad. Sci. USA, 98, 9742–9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee N.S., Dohjima,T., Bauer,G., Li,H., Li,M.J., Ehsani,A., Salvaterra,P. and Rossi,J. (2002) Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol., 19, 500–505. [DOI] [PubMed] [Google Scholar]
- 6.Jacque J.M., Triques,K. and Stevenson,M. (2002) Modulation of HIV-1 replication by RNA interference. Nature, 418, 435–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Novina C.D., Murray,M.F., Dykxhoorn,D.M., Beresford,P.J., Riess,J., Lee,S.K., Collman,R.G., Lieberman,J., Shankar,P. and Sharp,P.A. (2002) siRNA-directed inhibition of HIV-1 infection. Nat. Med., 8, 681–686. [DOI] [PubMed] [Google Scholar]
- 8.Gitlin L., Karelsky,S. and Andino,R. (2002) Short interfering RNA confers intracellular antiviral immunity in human cells. Nature, 418, 430–434. [DOI] [PubMed] [Google Scholar]
- 9.Hu W., Myers,C., Kilzer,J., Pfaff,S. and Bushman,F. (2002) Inhibition of retroviral pathogenesis by RNA interference. Curr. Biol., 12, 1301–1311. [DOI] [PubMed] [Google Scholar]
- 10.Brummelkamp T.R., Bernards,R. and Agami,R. (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science, 296, 550–553. [DOI] [PubMed] [Google Scholar]
- 11.Miyagishi M. and Taira,K. (2002) U6 promoter driven siRNAs with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat. Biotechnol., 20, 497–500. [DOI] [PubMed] [Google Scholar]
- 12.Paul C.P., Good,P.D., Winer,I. and Engelke,D.R. (2002) Effective expression of small interfering RNA in human cells. Nat. Biotechnol., 20, 505–508. [DOI] [PubMed] [Google Scholar]
- 13.Paddison P.J., Caudy,A.A., Bernstein,E., Hannon,G.J. and Conklin,D.S. (2002) Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev., 16, 948–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sui G., Soohoo,C., Affar,B., Gay,F., Shi,Y., Forrester,W.C. and Shi,Y. (2002). A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl Acad. Sci. USA, 99, 5515–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zamore P.D. (2002). Ancient pathways programmed by small RNAs. Science, 296, 1265–1269. [DOI] [PubMed] [Google Scholar]
- 16.Plasterk R.H.A. (2002). RNA silencing: the genome’s immune system. Science, 296, 1263–1265. [DOI] [PubMed] [Google Scholar]
- 17.Ahlquist A. (2002). RNA-dependent RNA polymerases, viruses and RNA silencing. Science, 296, 1270–1273. [DOI] [PubMed] [Google Scholar]
- 18.Tuschl T. and Borkhardt,A. (2002). Small interfering RNAs: a revolutionary tool for the analysis of gene function and gene therapy. Mol. Interv., 2, 158–167. [DOI] [PubMed] [Google Scholar]
- 19.Bernstein E., Caudy,A.A., Hammond,S.M. and Hannon,G.J. (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature, 409, 363–366. [DOI] [PubMed] [Google Scholar]
- 20.Hammond S.M., Bernstein,E., Beach,D. and Hannon,G.J. (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature, 404, 293–296. [DOI] [PubMed] [Google Scholar]
- 21.Hammond S.M., Boettcher,S., Caudy,A.A., Kobayashi,R. and Hannon,G.J. (2001) Argonaute2, a link between genetic and biochemical analyses of RNAi. Science, 293, 1146–1150. [DOI] [PubMed] [Google Scholar]
- 22.Nykänen A., Haley,B. and Zamore,P.D. (2001) ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell, 107, 309–321. [DOI] [PubMed] [Google Scholar]
- 23.Hutvagner G., McLachlan,J., Pasquinelli,A.E., Bálint,É., Tuschl,T. and Zamore,P.D. (2001) A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science, 293, 834–838. [DOI] [PubMed] [Google Scholar]
- 24.Lee R.C., Feinbaum,R.L. and Ambros,V. (1993) The C.elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell, 75, 843–854. [DOI] [PubMed] [Google Scholar]
- 25.Reinhart B.J., Slack,F.J., Basson,M., Pasquinelli,A.E., Bettinger,J.C., Rougvie,A.E., Horvitz,H.R. and Ruvkun,G. (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature, 403, 901–906. [DOI] [PubMed] [Google Scholar]
- 26.Vance V. and Vaucheret,H. (2001) RNA silencing in plants – defense and counterdefense. Science, 292, 2277–2280. [DOI] [PubMed] [Google Scholar]
- 27.Waterhouse P.M., Wang,M.B. and Lough,T. (2001) Gene silencing as an adaptive defence against viruses. Nature, 411, 834–842. [DOI] [PubMed] [Google Scholar]
- 28.Li H., Li,W.X. and Ding,S.W. (2002) Induction and suppression of RNA silencing by an animal virus. Science, 296, 1319–1321. [DOI] [PubMed] [Google Scholar]
- 29.Ketting R.F., Haverkamp,T.H., van Luenen,H.G. and Plasterk,R.H. (1999) Mut-7 of C.elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell, 99, 133–141. [DOI] [PubMed] [Google Scholar]
- 30.Ketting R.F. and Plasterk,R.H. (2000). A genetic link between co-suppression and RNA interference in C.elegans. Nature, 404, 296–298. [DOI] [PubMed] [Google Scholar]
- 31.Boutla A., Delidakis,C., Livadaras,I., Tsagris,M. and Tabler,M. (2001) Short 5′-phosphorylated double-stranded RNAs induce RNA interference in Drosophila. Curr. Biol., 11, 1776–1780. [DOI] [PubMed] [Google Scholar]
- 32.Elbashir S.M., Martinez,J., Patkaniowska,A., Lendeckel,W. and Tuschl,T. (2001) Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J., 20, 6877–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holen T., Amarzguioui,M., Wiiger,M.T., Babaie,E. and Prydz,H. (2002). Positional effects of short interfering RNAs targeting the human coagulation trigger Tissue Factor. Nucleic Acids Res., 30, 1757–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amarzguioui M., Brede,G., Babaie,E., Grotli,M., Sproat,B. and Prydz,H. (2000) Secondary structure prediction and in vitro accessibility of mRNA as tools in the selection of target sites for ribozymes. Nucleic Acids Res., 28, 4113–4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martinez J., Patkaniowska,A., Urlaub,H., Lührmann,R. and Tuschl,T. (2002) Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell, 110, 563–574. [DOI] [PubMed] [Google Scholar]
- 36.Schwarz D.S., Hutvágner,G., Haley,B. and Zamore,P.D. (2002) Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol. Cell, 10, 537–548. [DOI] [PubMed] [Google Scholar]
- 37.Klahre U., Crete,P., Leuenberger,S.A., Iglesias,V.A. and Meins,F.,Jr (2002) High molecular weight RNAs and small interfering RNAs induce systemic post-transcriptional gene silencing in plants. Proc. Natl Acad. Sci. USA, 99, 11981–11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garrus J.E., von Schwedler,U.K., Pornillos,O.W., Morham,S.G., Zavitz,K.H., Wang,H.E., Wettstein,D.A., Stray,K.M., Cote,M., Rich,R.L. et al. (2001). Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell, 107, 55–65. [DOI] [PubMed] [Google Scholar]
- 39.Kisielow M., Kleiner,S., Nagasawa,M., Faisal,A. and Nagamine,Y. (2002). Isoform-specific knockdown and expression of adaptor protein ShcA using small interfering RNA. Biochem J., 363, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu J.Y., DeRuiter,S.L. and Turner,D.L. (2002). RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc. Natl Acad. Sci. USA, 99, 6047–6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilda M., Fuchs,U., Wossmann,W. and Borkhardt,A. (2002). Killing of leukemic cells with a BCR/ABL fusion gene by RNA interference (RNAi). Oncogene, 21, 5716–5724. [DOI] [PubMed] [Google Scholar]
- 42.Amarzguioui M. and Prydz,H. (1998). Hammerhead ribozyme design and application. Cell. Mol. Life Sci., 54, 1175–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mir A.A., Lockett,T.J. and Hendry,P. (2001). Identifying ribozyme-accessible sites using NUH triplet-targeting gapmers. Nucleic Acids Res., 29, 1906–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harborth J., Elbashir,S.M., Bechert,K., Tuschl,T. and Weber,K. (2001) Identification of essential genes in cultured mammalian cells using small interfering RNAs. J. Cell Sci., 114, 4557–4565. [DOI] [PubMed] [Google Scholar]
- 45.Krichevsky A.M. and Kosik,K.S. (2002) RNAi functions in cultured mammalian neurons. Proc. Natl Acad. Sci. USA, 99, 11926–11929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simmer F., Tijsterman,M., Parrish,S., Koushika,S., Nonet,M., Fire,A., Ahringer,J. and Plasterk,R. (2002) Loss of the putative RNA-directed RNA polymerase RRF-3 makes C.elegans hypersensitive to RNAi. Curr. Biol., 12, 1317–1319. [DOI] [PubMed] [Google Scholar]
- 47.Yang D., Buchholz,F., Huang,Z., Goga,A., Chen,C.-Y., Brodsky,F.M. and Bishop,J.M. (2002) Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. Proc. Natl Acad. Sci. USA, 99, 9942–9947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ketting R.F., Fischer,S.E., Bernstein,E., Sijen,T., Hannon,G.J. and Plasterk,R.H. (2001) Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C.elegans. Genes Dev., 15, 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang H., Kolb,F.A., Brondani,V., Billy,E. and Filipowicz,W. (2002). Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J., 21, 5875–5885. [DOI] [PMC free article] [PubMed] [Google Scholar]