Abstract
The human DDB1 and DDB2 genes encode the 127 and 48 kDa subunits, respectively, of the damage-specific DNA-binding protein (DDB). Mutations in the DDB2 gene have been correlated with the hereditary disease xeroderma pigmentosum group E. We have investigated the proximal promoters of the DDB genes, both of which are G/C-rich and do not contain a TATA box. Transient expression analysis in HeLa cells using a luciferase reporter system indicated the presence of core promoters located within 292 bp (DDB1) and 220 bp (DDB2) upstream of the putative transcription initiation sites. Both core promoters contain multiple active Sp1 sites, with those of DDB1 at –123 to –115 and of DDB2 at –29 to –22 being critical determinants of promoter activity. In addition, an N-myc site at –56 to –51 for DDB1 is an essential transcription element, and mutations in a DDB1 NF-1 site at –104 to –92, a DDB2 NF-1 site at –68 to –56 and a DDB2 E2F site at +36 to +43 also reduce promoter activity. Taken together, these results suggest a regulation of basal transcription typical of cell cycle-regulated genes, and therefore support conjectures that the DDB heterodimer and/or its subunits have functions other than direct involvement in DNA repair.
INTRODUCTION
The rare recessive hereditary disease xeroderma pigmentosum (XP) presents clinically with a high incidence of skin cancer and hyperpigmentation in response to UV light exposure (1). Xeroderma pigementosum group E (XP-E) patients experience a mild form of the disease with generally a late onset of symptoms. The activity of the damaged DNA-binding protein (DDB) is absent from the cells of XP-E patients (2–6) due to mutations in the DDB2 gene (6–9). DDB is a heterodimer of p48 and p127, the products of DDB1 and DDB2, respectively (10,11). No naturally occurring mutations have been identified in DDB1, though knockout mutations of DDB1 homologs have been produced in Schizosaccharomyces pombe (12) and Drosophila melanogaster (13).
The high affinity of DDB for UV-induced DNA damage (14) and the absence of its activity from XP-E cells suggest a role for the protein in nucleotide excision repair (NER). However, evidence is conflicting in this regard. Micro injection of purified DDB corrected a mild deficiency in NER after UV damage in XP-E cells (15) and DDB has been implicated in global genomic repair (16,17). Recent reports show that DDB can stimulate the excision of cis-syn cyclobutane pyrimidine dimers but not (6–4) photoproducts in vitro through interaction with XPA and RPA proteins (18) and that DDB accumulated at DNA lesions after UV-irradiation and stimulated NER (19,20). However, in vitro reconstitution studies show that DDB is not essential for NER (21,22) and other studies find that XP-E cells that lack DDB activity have normal DNA NER characteristics (6).
Alternative roles for DDB have been proposed due to its interaction with other cellular proteins, specifically cell cycle regulating or regulated proteins. This is largely due to the fact that DDB2 is induced roughly 48 h after UV-irradiation, when DNA damage has been repaired (8). Hence, DDB1 is transported to the nucleus after repair is all but complete. Other observations show that DDB binds to the E2F1 transcription factor, which is involved in the transcriptional activation of the DNA replication genes, and the DDB–E2F1 complex stimulates E2F1-activated transcription (23). DDB is ubiquitinated by Cul-4A, with the consequent cell cycle regulation of DDB2, which peaks at the G1/S boundary (24,25). DDB1 also binds to several viral transcriptional activators, including hepatitis B virus X protein (26–28) and the V protein of paramyxovirus SV5, parainfluenza and mumps and measle viruses (29).
DDB heterodimer is an abundant protein with an estimated 100 000 copies per HeLa cell (10). DDB1 is mainly confined to the cytoplasm in undamaged cells, while DDB2 is found exclusively in the nucleus (8,30). After UV-irradiation, mRNA and protein levels of DDB2 are increased 3- to 4-fold (8), while DDB1 levels remain constant. The increase in DDB2 results in the translocation of DDB1 to the nucleus (30). Overall, current evidence suggests that DDB1 and DDB2 have multiple roles and that the subunits may function both independently and as a complex.
In this study we have analyzed the transcription regulatory region of the DDB1 and DDB2 genes to investigate whether there is a common basal control of each. Transient luciferase expression experiments identified core promoters that are normally thought to be associated with cell cycle-regulating/regulated genes: no TATA box, a G/C-rich region, a NF-1 element and multiple Sp1 elements. An N-myc consensus sequence was a critical determinant of DDB1 promoter activity, whereas an active E2F element was revealed in the DDB2 promoter. The latter is intriguing in light of the known interaction of DDB2 and E2F1 proteins (23). In general, these results would support a role(s) for DDB1 and DDB2 in functions other than DNA repair, since DNA repair is not thought to vary within the cell cycle.
MATERIALS AND METHODS
Cloning of DDB1 and DDB2 genomic DNAs
Fragments containing ∼5 kb of upstream genomic DNA were isolated from a human chromosome 11 genomic library (ATCC 57726). Briefly, the library was screened by a PCR-based method described by Israel (31) and the DDB1 and DDB2 fragments were subcloned into pBluescript SK (Stratagene) to produce DDB1/pBS and DDB2/pBS, respectively. Partial sequences were used to search the genomic nucleotide databases using the NCBI BLAST server and the BLASTN 2.2.3 program (32). All nucleotide numbering for human DDB1 and DDB2 are referenced to the putative transcription initiation sites (the 5′-end of the published cDNA sequences: DDB1, U18299; DDB2, U18300) as +1. Mouse DNA sequences were determined from a 129 SVJ genomic library (Stratagene) for DDB1 and from a C57 BL/6 mouse genomic library (a gift from Dr S. Aizawa) for DDB2.
Plasmid construction
Nucleotides 43–75 of the luciferase reporter plasmid pGL2-Basic (Promega) were deleted to produce the GLfuse vector, lacking a stop codon 5′ of the luciferase cDNA. The promoter regions of DDB1 which were studied included most of the first exon of DDB1 (coding for amino acids 1–19) and were inserted at the BglII site to produce an in-frame fusion of DDB1(amino acids 1–19) to luciferase. The same strategy was employed to produce a DDB2(amino acids 1–40)–luciferase fusion construct. The DDB/pBS plasmids were used as templates to engineer constructs containing varying lengths of upstream putative promoter region, employing standard PCR amplification and restriction digest protocols. Mutated promoter constructions were produced utilizing a two-step overlapping PCR method (33). All PCR amplified sequences were verified by DNA sequencing.
Cell culture, transfection and promoter activity measurements
HeLa S3 cells were grown in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (Hyclone) at 37°C, 5% CO2. Lipotransfection using log phase cells was performed with the Fugene6 reagent (Roche Applied Science). For DDB1 and DDB2 core promoter analysis, HeLa cells (4 × 105) were seeded onto 60 mm plates and incubated for 24 h before transfection with 1.2 µg (300 fmol) of test plasmid and 0.4 µg of GL2-Basic plasmid. For analysis of DDB1(–2298), 1.4 µg (270 fmol) of DDB1(–2298) test plasmid and 0.2 µg of GL2-Basic plasmid were transfected, while 1.1 µg (270 fmol) of DDB1(–370) plus 0.5 µg of GL2-Basic plasmid were used as a control transfection. To evaluate the DDB2(–5000) construct, 1.6 µg (225 fmol) of DDB2(–5000) test plasmid was transfected, while 0.9 µg (225 fmol) of the DDB2(–250) construct with 0.7 µg of GL2-Basic plasmid were used as a control transfection. An aliquot of 0.4 µg of CH110 plasmid (Pharmacia), which expresses β-galactosidase, was included in all transfections as an internal control to standardize transfection efficiency. For each transfection, 176 µl of DMEM without serum, 20 µl of plasmids in 2 mM Tris– HCl, pH 8.0, and 4 µl of Fugene6 were pre-incubated at room temperature. After 1 h the DNA/lipofectin mixture was added dropwise to the HeLa plates. The medium was not replaced and cells were harvested 48 h post-transfection into 500 µl of 1× reporter lysis buffer (Promega). Luciferase and β-galactosidase activities were measured using the reporter assay system as directed by the manufacturer (Promega). A LS3800 Beckman-Coulter liquid scintillation counter was used to measure luciferase activity. At least four extracts from two or more independent transfections were evaluated for each construct.
Identification of putative transcription factor-binding sites
Potential core promoters were identified by the NIH Proscan program (bimas.dcrt.nih.gov/molbio/proscan). This region was further analyzed by TRANSFAC 4.0 (transfac.gbf.de/TRANSFAC), using the MatInspector V2.2 (34) and PatSearch 1.1 (35) programs. Parameters for MatInspector were set for 1.0 core (a 4 nt highly conserved sequence) similarity and 0.85 matrix similarity, employing the vertebrates matrix group. Settings for PatSearch of the Consensi database were no core mismatches and 15% maximal flanking mismatches.
RESULTS
The core promoter of DDB1 is located within 292 bp upstream of the putative transcription initiation site
Clones containing 6 kb of DDB1 genomic DNA were isolated from an EcoRI-digested human chromosome 11 library and the DDB1 fragment was subcloned into a pBluescript vector. For all nucleotide numbering of DDB1 in this study, the 5′-end of the human DDB1 cDNA sequence (11) was designated as the putative transcription initiation site at +1. Restriction mapping, partial sequencing and subsequent database searches with the BLASTN 2.2.3 program determined that the insert contained DDB1 genomic sequence from approximately –5000 to +173 (the 3′-end of exon 1), plus 1123 nt of the first intron. Nucleotides 83403–88497 of the AC019071 contig sequence corresponded to DDB1 –3798 through 1123 nt of the first intron. Similarly, DNA was isolated from a mouse 129 SVJ genomic library. Mouse DDB1 –808 through the first 674 nt of the first intron matched nucleotides 909608–911263 of the mouse mgscv3 genomic sequence of WGS supercontig Mm19_WIFeb01_319.
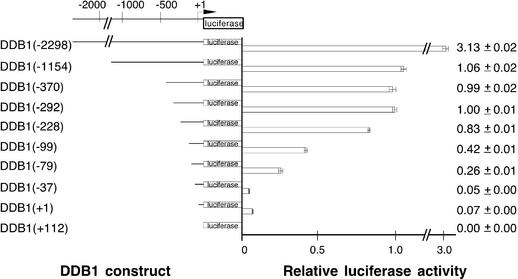
Evaluation of promoter activity utilized a chimeric promoter–reporter construct that included various DDB1 upstream genomic sequences through nucleotide +169 of the first exon. The DDB1 sequence was placed in-frame with the luciferase reporter cDNA, resulting in overexpression of a DDB1(amino acids 1–19)–luciferase fusion protein in the presence of an active promoter. Constructs containing DDB1 nucleotides –1154 to +169 and a series of 5′ unidirectional deletions were then created and the promoter activity of each construct was analyzed by luciferase assays of extracts from HeLa cells transfected with the construct. Activity was standardized by determination of β-galactosidase activity from co-transfected CH110 plasmid (Fig. 1).
Figure 1.
Transcription activities in log phase HeLa cells after deletions within the upstream region of the DDB1 gene. A series of 5′-unidirectional deletions were fused in-frame with the luciferase reporter gene and analyzed by transient transfection as described in Materials and Methods. Luciferase activities were normalized to β-galactosidase activity, which was also measured to control for transfection efficiency. Less than a 2-fold variation in transfection efficiency was observed. At least four extracts from two or more independent transfections were evaluated for each construct. The construct number in parentheses indicates the length of the tested promoter region upstream of the putative transcription initiation site (designated by the bent arrow at +1). Luciferase activity is shown relative to DDB1(–292). Means and standard deviations are indicated to the right.
Analysis of the sequence of DDB1 nucleotides –1154 to +169 by the NIH Proscan program identified a potential core promoter from –292 to –41, so the luciferase constructs of varying lengths of upstream sequence were compared to DDB1(–292) (Fig. 1). Promoter activity was stable unless deletions were made 3′ of the DDB1(–292) construct, confirming that the core proximal promoter was located within 292 bp upstream of the transcription initiation site. Results from separate experiments were very reproducible, as evidenced by moderate error bars. There was a progressive loss of activity upon deletions 3′ of nucleotide –292 and almost all activity was lost with the DDB1(–37) construct. The region from –292 to –41 is G/C-rich (74%) and does not contain a TATA box (Fig. 2).
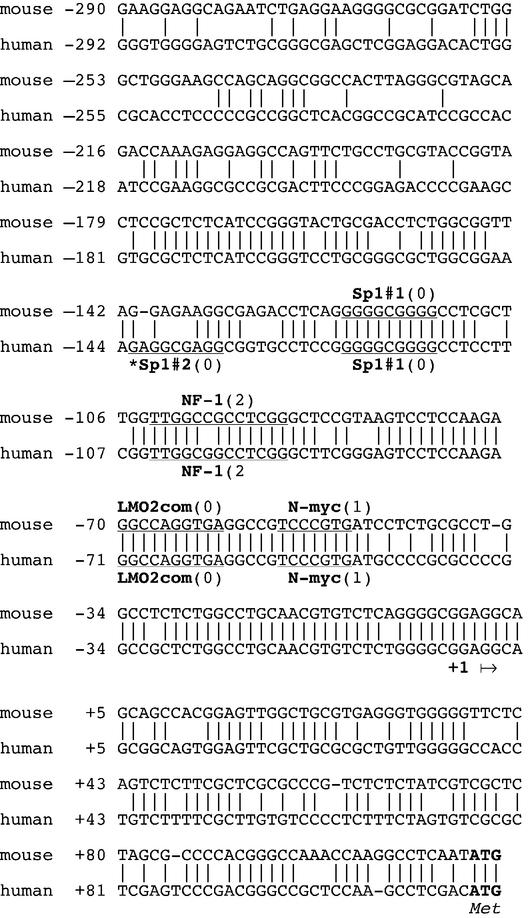
Figure 2.
Nucleotide sequence and potential regulatory elements of the promoter region of the DDB1 gene in mouse and human genomic DNAs. The consensus sequences of the potential transcription factor-binding sites are underlined. Numbers in parentheses are the number of mismatches as evaluated with the TRANSFAC database consensus sequence. Elements which were tested, although not found in both mouse and human DNAs, are marked by an asterisk. The putative transcription initiation site (+1) is indicated by an arrow, and ATG is the translation initiation site.
A longer upstream DDB1 sequence was also evaluated (Fig. 1). Interestingly, promoter activity of the DDB1(–2298) construct was 3-fold greater than that observed for the core promoter plasmid, DDB1(–292), suggesting that an important distal promoter is located within 2 kb of the putative transcription initiation site.
Mutations in N-myc, Sp1 and NF1 elements reduced DDB1 core promoter activity
The region from nucleotides –292 to +169 was further analyzed by the TRANSFAC 4.0 programs. The search produced a large number of potential transcription factor-binding sites. To limit the mutations to be tested, the human nucleotide sequence of DDB1 was compared to the mouse sequence. Although a high identity of 84% was found from nucleotide –178 to +1 (Fig. 2), the number of potential transcription factors to be tested was considerably reduced.
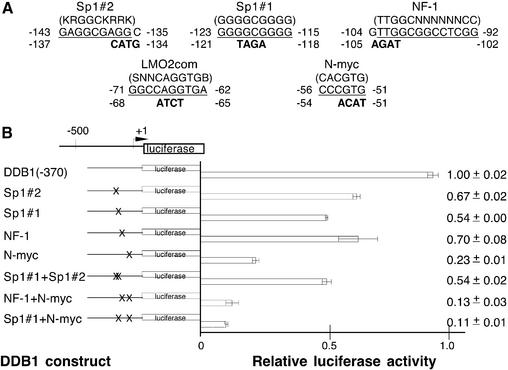
Based on the above information, mutant plasmids were constructed in which 3–4 nt were changed to abolish potential transcription factor-binding sites without creating new sites as identified by TRANSFAC analysis (Fig. 3A). The effects of these mutations on promoter activity are shown in Figure 3B. Mutation of the N-myc-binding site (–56 to –51) was most dramatic, with only 23% of the promoter activity remaining. Mutation of the Sp1#1 site (–123 to –115) was also significant, reducing promoter activity by 46%. A mutation of the potential Sp1#2 site at –143 to –135, which was not found in the mouse sequence, also reduced promoter activity by 23%, but a double mutation of both Sp1 sites did not result in a decrease greater than that found for the single Sp1#1 mutation. On the other hand, a double mutation of the N-myc and the Sp1#1 sites reduced promoter activity to 11%, accounting for almost all of the activity lost by the deletion of sequence upstream of nucleotide –37 with the construct DDB1(–37) (5%, Fig. 1). Mutation of a potential NF1 site (–104 to –92) produced a moderate decrease to 70%. A double mutation of the NF-1 and N-myc sites reduced promoter activity to 13%, which was approximately the same effect as seen with double mutation of the Sp1#1 and N-myc sites. Although the TRANSFAC search found a potential Lmo2 element in both mouse and human, mutation of this consensus sequence did not affect promoter activity (data not shown).
Figure 3.
Mutational analysis of putative proximal promoter transcription elements of the DDB1 gene. (A) The wild-type sequences that were analyzed by mutation are shown with their nucleotide position in the DDB1 gene. The consensus sequences are shown above in parentheses, the DDB1 wild-type element is underlined and mutations that were introduced are indicated below the wild-type sequence in bold. Mutations were introduced by overlapping PCR into the DDB1(–370) plasmid. (B) Potential transcription elements that were mutated are indicated to the left and correspond to those in (A). X on the construct schematic indicates the location of the mutation. The plasmids were transfected into log phase HeLa cells and luciferase activity was measured in cell extracts and standardized to β-galactosidase activity, which was also measured to control for transfection efficiency. Less than a 2-fold variation in transfection efficiency was observed. At least four extracts from two or more independent transfections were evaluated for each construct. Luciferase activity is shown relative to the control wild-type DDB1(–370). Means and standard deviations are indicated to the right.
The core promoter of DDB2 is located within 220 bp upstream of the putative transcription initiation site
Clones containing 6 kb of DDB2 genomic DNA were isolated from a human chromosome 11 library and the DDB2 fragment was subcloned into pBluescript plasmids. For all nucleotide numbering of DDB2 in this study, the 5′-end of the DDB2 cDNA sequence (11) was designated as the putative transcription initiation site at +1. Restriction mapping, partial sequencing and subsequent database searches (BLASTN 2.2.3) determined that the insert contained DDB2 genomic sequence from nucleotides approximately –5000 to +302, plus 1104 bp of the first intron. Nucleotides 75989–72478 in contig fragment 19 of the AC024045 sequence corresponded to DDB2 nucleotides –2105 to 1104 of the first inton. Similarly, DNA was isolated from a C57 BL/6 mouse genomic DNA library. Nucleotides 32389960–32389401 of the mouse mgscv3 genomic sequence of the WGS supercontig Mm2_ WIFeb01_27 matched the mouse DDB2 sequence of 488 nt upstream of the translation start site through 71 nt of the protein coding region.
Evaluation of promoter activity utilized a chimeric promoter–reporter plasmid that included various DDB2 upstream genomic sequences through nucleotide +295, with the DDB2 coding region in-frame with the luciferase sequence. A DDB2(amino acids 1–40)–luciferase fusion protein was overexpressed in the presence of an active promoter.
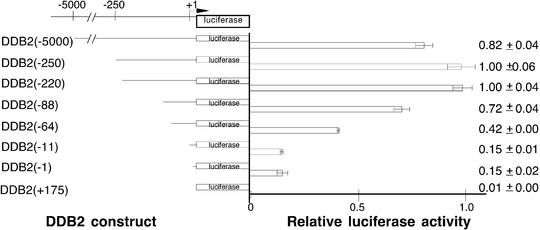
Analysis of the sequence of nucleotides –1055 to +296 by the NIH Proscan program predicted a core promoter from –220 to +32. DDB2(–250) and a series of 5′ unidirectional deletions using DDB2(–250) as a template were then created, and the promoter activity of each construct was analyzed by luciferase assays of extracts from HeLa cells transfected with the construct (Fig. 4). Unlike the case of DDB1, promoter activity was not completely lost by 5′ deletion to the putative transcription start site, suggesting that an important transcription element could be located downstream of nucleotide +1. Within the core promoter region, nucleotides –166 to –1 are G/C-rich (77%) and no TATA box was identified (Fig. 5).
Figure 4.
Transcription activities in log phase HeLa cells after deletions within the upstream region of the DDB2 gene. A series of 5′-unidirectional deletions were fused in-frame with the luciferase reporter gene and analyzed by transient transfection as described in Materials and Methods. Luciferase activity was standardized to β-galactosidase activity to control for variations in transfection activity. Less than a 2-fold variation in transfection efficiency was observed. At least four extracts from two or more independent transfections were evaluated for each construct. The construct number in parentheses indicates the length of the tested promoter region upstream of the putative transcription initiation site (designated by the bent arrow at +1). Luciferase activity is shown relative to DDB1(–220). Means and standard deviations are indicated to the right.
Figure 5.
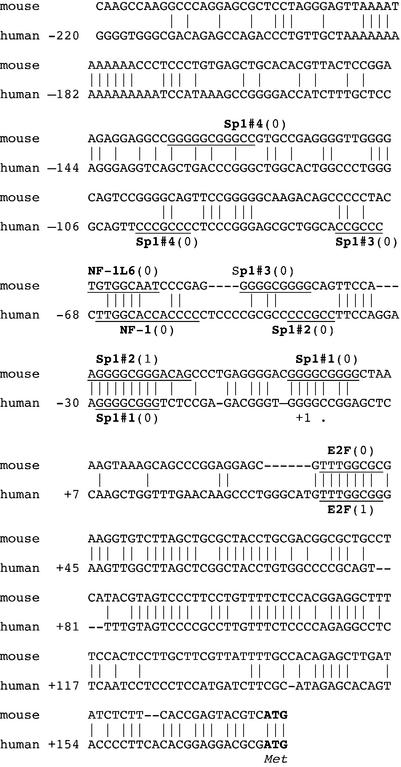
Nucleotide sequence and potential regulatory elements of the promoter region of the DDB2 gene in mouse and human genomic DNAs. The consensus sequences of the potential transcription factor-binding sites are underlined. Numbers in parentheses are the number of mismatches as evaluated with the TRANSFAC database consensus sequence. Elements which were tested, although not found in both mouse and human DNAs, are marked by an asterisk. The putative transcription initiation site (+1) is indicated by an arrow, and ATG is the translation initiation site.
A longer upstream DDB2 sequence was also tested, but no increase over core promoter activity was seen with the DDB2(–5000) construct (Fig. 4).
Mutations of Sp1, NF-1 and E2F elements reduced DDB2 core promoter activity
The region from nucleotides –220 to +176 was further analyzed by the TRANSFAC 4.0 programs. As seen with DDB1, many potential transcription factor elements were predicted. Whereas the DDB2 human and mouse nucleotide sequences upstream of the putative transcription initiation site have only isolated regions of homology, several potential promoter elements were common to both (Fig. 5). However, homology was too poor to designate a putative mouse transcription initiation site by comparison to the human sequence.
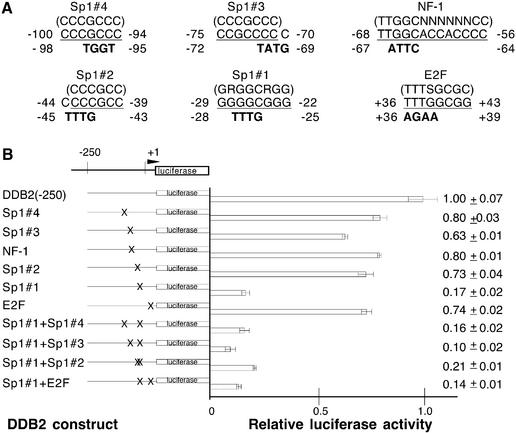
Based upon the above information, mutant plasmids were constructed in which 3–4 nt were changed to abolish these potential transcription factor-binding sites without creating new sites (Fig. 6A). The effects of these mutations on promoter activity are shown in Figure 6B. Mutation of the Sp1#1 site (–29 to –22) most drastically decreased the promoter activity level to 17%, accounting for all of the activity lost by the deletion of sequence upstream of nucleotide –11 with the DDB2(–11) construct (15%, Fig. 4). A smaller loss was observed for each of the three other Sp1 sites, but double mutations of these with Sp1#1 did not significantly decrease activity below that seen with mutation of Sp1#1 alone. A mutation of the E2F site (+36 to +43) decreased promoter activity to 74%, confirming the presence of an important element downstream of the putative transcription initiation site. A double mutation of Sp1#1 and E2F resulted in no significant further reduction. Mutation of the NF-1 site (–68 to –56) decreased promoter activity to 80%.
Figure 6.
Mutational analysis of putative proximal promoter transcription elements of the DDB2 gene. (A) The wild-type sequences that were analyzed by mutation are shown with their nucleotide position in the DDB2 gene. The consensus sequences are shown above in parentheses, the DDB2 wild-type element is underlined and mutations that were introduced are indicated below the wild-type sequence in bold. Mutations were introduced by overlapping PCR into the DDB2(–250) plasmid. (B) Potential transcription elements that were mutated are indicated to the left and correspond to those in (A). X on the construct schematic indicates the location of the mutation. The plasmids were transfected into log phase HeLa cells and luciferase activity was measured in cell extracts and standardized to β-galactosidase activity, which was also measured to control for transfection efficiency. Less than a 2-fold variation in transfection efficiency was observed. At least four extracts from two or more independent transfections were evaluated for each construct. Luciferase activity is shown relative to the control wild-type DDB2(–250). Means and standard deviations are indicated to the right.
DISCUSSION
DDB is highly abundant in HeLa cells and binds to multiple forms of DNA damage with high affinity in vitro, but the question remains as to what purpose this strong DNA damage recognition serves. Moreover, are there independent roles for the individual subunits? In this report we have investigated the basal control of the two DDB genes, and striking similarities and differences in their transcription regulation have been observed. The regions directly upstream of the DDB1 and DDB2 transcription initiation sites are TATA-less and G/C-rich and they each contain important Sp1 and NF-1 elements. These are common characteristics of promoters of housekeeping genes and cell cycle-regulated human promoters (36–38).
Sp1 is a well-investigated factor that regulates transcription through specific sequences in G/C-rich promoter regions and is often critical for transcription initiation of TATA-less promoters (39). In mammalian cells, promoters lacking TATA boxes generally have several Sp1 elements (40). Sp1 is a regulator of the cell cycle in G1 phase (41). Four Sp1 sequences were identified in the promoter of DDB2 and mutation of Sp1#1 (–29 to –22), which is closest to the transcription initiation site, most drastically reduced transcription activity to 17%. Mutations of the other three Sp1 elements resulted in more modest decreases of activity to 63–80%. Double Sp1 mutations that included Sp1#1 did not decrease activity below that observed with Sp1#1 alone. These findings, combined with the fact that 5′ deletion of the core promoter to –11 produced 15% promoter activity, indicated that Sp1#1 is a critical basal transcription factor element in the DDB2 gene.
In the DDB1 promoter, Sp1 elements were also found to be essential for maximal activity, but less important compared to their role in DDB2 transcriptional activation. Individual mutations of the Sp1 consensus sequences, Sp1#2 (–143 to –135) and Sp1#1 (–123 to –115), reduced promoter activity to 67 and 54%, respectively. A double Sp1 mutation did not reduce transcription below that found for Sp1#1 alone. Similar to the Sp1 elements in DDB2, mutation of the Sp1 site in closest proximity to the initiation site resulted in a greater effect on promoter activity.
The myc family of proto-oncogenes, which includes mainly c-myc, N-myc and L-myc, are central mediators of cell proliferation (42,43). In DDB1 the N-myc site (–56 to –51) was identified as the critical promoter element. Mutation of this site drastically reduced activity to 23%. A double mutation with Sp1#1 resulted in a further reduction to 11%, not quite accounting for the loss of activity to 5% with 5′ deletion of the core promoter to –37.
An E2F element was also identified downstream of the putative transcription initiation site in DDB2. E2F elements are often only functional when they are proximal to the transcription initiation site (44). Mutation of this E2F consensus sequence decreased promoter activity to 74%. A double mutation of Sp1#1 and E2F did not result in any significant further reduction over that seen with Sp1#1 alone. The E2F1–E2F6 family are transcription factors that form a heterodimer complex with DP1–DP3. Overexpression of E2F1–E2F3 activates many proteins which are regulated by cell cycle progression (45). Sp1 and E2F1 have been observed to form a functional complex in cell cycle-regulated transcription (46,47). It also has been demonstrated that DDB2 protein alone can interact with E2F1, though both subunits of DDB are required to stimulate E2F1-activated transcription (23). Since E2F-controlled transcription appears to be a tightly regulated system, it is feasible that the activation of DDB2 would be regulated by E2F, thereby determining the number of DDB–E2F1 complexes present in the cell.
The mutation (+36 to +39) of the E2F consensus site also deleted a putative p53 element, REhDDB2 (+8 to +38), reported by Tan and Chu (48). While the search engines that we used did not recognize this p53 site, our laboratory has identified a second putative p53 site. However, no effect of a p53 element would have been detected in the system that we employed to evaluate DDB2 promoter activity because, while HeLa cells do produce p53 mRNA, p53 protein levels are undetectable. HPV18 present in HeLa cells produces active E6 that forms a complex with the ubiquitin ligase, E6AP, and together they target p53 for degradation by proteosome mechanisms (49). With regard to the putative p53-binding site in the DDB2 promoter which is reported to be active in human cells but not mouse cells, we do not believe that its presence or absence from human cells would affect the results of this study that concerns the basal levels of DDB2 under promoter control. Contrary to misconceptions present in the literature, mouse cells contain the same basal level of DDB activity as do human cells (50).
The original intent of this undertaking was to explore the regulation of basal transcription of the DDB genes. Essentially, both the DDB1 and DDB2 promoters are typical of cell cycle-regulated and housekeeping genes: TATA-less and G/C-rich, with active Sp1 and NF-1 elements. In addition, DDB1 contains a critical proto-oncogene N-myc element, again implying that DDB1 is regulated during progression through the cell cycle. Possibly most interesting was the finding that DDB2 has an important E2F-binding site, suggesting a tightly controlled regulation of the number of DDB–E2F1 complexes present in the cell. It has been proposed that in undamaged cells the association of DDB with E2F1 stimulates E2F1 transcription activation of DNA replication genes and progression from G1 to S phase (23). However, after UV-irradiation DDB2 could be bound to damaged DNA, decreasing the number of DDB complexes formed, resulting in cell cycle arrest at the G1/S checkpoint. In any event, the apparent regulation of both DDB1 and DDB2 in a cell cycle-dependent manner lends credence to a role for these proteins aside from NER.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr S. Aizawa (Center for Developmental Biology, RIKEN, Kobe, Japan) for the kind gift of the C57 BL/6 mouse genomic DNA library and Ms Ann Fischer for helping with cell culture. This work was supported by NIH grant RO1-GM59424 and P30-ES08196 (to S.L.) and a grant from the Nakatomi Foundation (to T.I.).
REFERENCES
- 1.Cleaver J.E. and Kraemer,K.H. (1995) Xeroderma pigmentosum and Cockayne Syndrome. In Scriver,C.R., Beaudet,A.L., Sly,W.S. and Valle,D. (eds), The Metabolic and Molecular Bases of Inherited Disease, Vol. III. McGraw-Hill, New York, NY, pp. 4393–4419.
- 2.Chu G. and Chag,E. (1988) Xeroderma pigmentosum group E cells lack a nuclear factor that binds to damaged DNA. Science, 242, 564–567. [DOI] [PubMed] [Google Scholar]
- 3.Kataoka H. and Fujiwara,Y. (1991) UV-damage-specific DNA-binding protein in xeroderma pigmentosum complementation group E. Biochem. Biophys. Res. Commun., 175, 1139–1143. [DOI] [PubMed] [Google Scholar]
- 4.Keeney S., Wein,H. and Linn,S. (1992) Biochemical heterogeneity in xeroderma pigmentosum complementation group E. Mutat. Res., 273, 49–56. [DOI] [PubMed] [Google Scholar]
- 5.Otrin V.R., Kuraoka,I., Nardo,T., McLenigan,M., Eker,A.P.M., Stefanini,M., Levine,A.S. and Wood,R.D. (1998) Relationship of the xeroderma pigmentosum group E defect to chromatin and DNA binding proteins UV-DDB and replication protein A. Mol. Cell. Biol., 18, 3182–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Itoh T., Mori,T., Ohkubo,H. and Yamaizumi,M. (1999) A newly identified patient with clinical xeroderma pigmentosum phenotype has a non-sense mutation in the DDB2 gene and incomplete repair in (6–4) photoproducts. J. Invest. Dermatol., 113, 251–257. [DOI] [PubMed] [Google Scholar]
- 7.Nichols A.F., Ong,P. and Linn,S. (1996) Mutations specific to the xeroderma pigmentosum group E Ddb– phenotype. J. Biol. Chem. 271, 24317–24320. [DOI] [PubMed] [Google Scholar]
- 8.Nichols A.F., Itoh,T., Graham,J.A., Liu,W., Yamaizumi,M. and Linn,S. (2000) Human damage-specific DNA-binding protein p48. Characterization of XP-E mutations and regulation following UV irradiation. J. Biol. Chem., 275, 21422–21428. [DOI] [PubMed] [Google Scholar]
- 9.Itoh T., Linn,S., Ono,T. and Yamaizumi,M. (2000) Reinvestigation of the classification of five cell strains of xeroderma pigmentosum group E with reclassification of three of them. J. Invest. Dermatol., 114, 1022–1029. [DOI] [PubMed] [Google Scholar]
- 10.Keeney S., Chang,G.J. and Linn,S. (1993) Characterization of a human DNA dmage binding protein implicated in xeroderma pigmentosum. J. Biol. Chem., 268, 1293–1300. [PubMed] [Google Scholar]
- 11.Dualan R., Brody,T., Keeney,S., Nichols,A.F., Admon,A. and Linn,S. (1995) Chromosomal localization and cDNA cloning of the genes (DDB1 and DDB2) for the p127 and p48 subunits of a human damage-specific DNA binding protein. Genomics, 29, 62–69. [DOI] [PubMed] [Google Scholar]
- 12.Zolezzi F., Fuss,J., Uzawa,S. and Linn,S.M. (2002) Characterization of a Schizosaccharomyces pombe strain deleted for a sequence homologue of the human damaged DNA binding 1 (DDB1) gene. J. Biol. Chem., 277, 41183–41191. [DOI] [PubMed] [Google Scholar]
- 13.Takata K., Ishikawa,G., Hirose,F. and Sakaguchi,K. (2002) Drosophila damage-specific DNA-binding protein 1 (D-DDB1) is controlled by the DRE/DREF system. Nucleic Acids Res., 30, 3795–3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reardon J.T., Nichols,A.F., Keeney,S., Smith,C.A., Taylor,J.S., Linn,S. and Sancar,A. (1993) Comparative analysis of binding of human damaged DNA-binding protein (XP-E) and Escherichia coli damage recognition protein (UvrA) to the major ultraviolet photoproducts: T[c,s]T, T[t,s]T, T[6-4]T, and T[Dewar]T. J. Biol. Chem., 268, 21301–21308. [PubMed] [Google Scholar]
- 15.Keeney S., Eker,A.P., Brody,T., Vermeulen,W., Bootsma,D., Hoeijmakers,J.H. and Linn,S. (1994) Correction of the DNA repair defect in xeroderma pigmentosum group E by injection of a DNA damage-binding protein. Proc. Natl Acad. Sci. USA, 91, 4053–4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hwang B.J., Ford,J.M., Hanawalt,P.C. and Chu,G. (1999) Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc. Natl Acad. Sci. USA, 96, 424–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang J.Y., Hwang,B.J., Ford,J.M., Hanawalt,P.,C. and Chu,G (2000) Xeroderma pigmentosum p48 gene enhances global genomic repair and suppresses UV-induced mutagenesis. Mol. Cell, 5, 737–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wakasugi M., Shimizu,M., Morioka,H., Linn,S., Nikaido,O. and Matsunaga,T. (2001) Damaged DNA-binding protein DDB stimulates the excision of cyclobutane pyrimidine dimers in vitro in concert with XPA and replication protein A. J. Biol. Chem., 276, 15434–15440. [DOI] [PubMed] [Google Scholar]
- 19.Rapic-Otrin V., McLenigan,M.P., Bisi,D.C., Gonzalez,M. and Levine,A.S. (2002) Sequential binding of UV DNA damage binding factor and degradation of the p48 subunit as early events after UV irradiation. Nucleic Acids Res., 30, 2588–2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wakasugi M., Kawashima,A., Morioka,H., Linn,S., Sancar,A., Mori,T., Nikaido,O. and Matsunaga,T. (2002) DDB accumulates at DNA damage sites immediately after UV irradiation and directly stimulates nucleotide excision repair. J. Biol. Chem., 277, 1637–1640. [DOI] [PubMed] [Google Scholar]
- 21.Mu D., Park,C.-H., Matsunaga,T., Hsu,D.S., Reardon,J.T. and Sancar,A. (1995) Reconstitution of human DNA repair excision nuclease in a highly defined system. J. Biol. Chem., 270, 2415–2418. [DOI] [PubMed] [Google Scholar]
- 22.Kazantsev. A., Mu,D., Nichols,A.F., Zhao,X., Linn,S. and Sancar,A. (1996) Functional complementation of xeroderma pigmentosum complementation group E by replication protein A in an in vitro system. Proc. Natl Acad. Sci. USA, 93, 5014–5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayes S., Shiyanov,P., Chen,X. and Raychaudhuri,P. (1998) DDB, a putative DNA repair protein, can function as a transcriptional partner of E2F1. Mol. Cell. Biol., 18, 240–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shiyanov P., Nag,A. and Raychaudhuri,P. (1999) Cullin 4A associates with the UV-damaged DNA-binding protein DDB. J. Biol. Chem., 274, 35309–35312. [DOI] [PubMed] [Google Scholar]
- 25.Nag A., Bondar,T., Shiv,S. and Raychaudhuri,P. (2001) The xeroderma pigmentosum group E gene product DDB2 is a specific target of cullin 4A in mammalian cells. Mol. Cell. Biol., 21, 6738–6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee T.H., Elledge,S.J. and Butel,J. (1995) Hepatitis B virus X protein interacts with a probable cellular DNA repair protein. J. Virol., 69, 1107–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sitterlin D., Lee,T.H., Prigent,S., Tiollais,P., Butel,J.S. and Transy,C. (1997) Interaction of the UV-damaged DNA-binding protein with hepatitis B virus X protein is conserved among mammalian hepadnaviruses and restricted to transactivation-proficient X-insertion mutants. J. Virol., 71, 6194–6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nag A., Datta,A., Yoo,K., Bhattacharyya,D., Chakrabortty,A., Wang,X., Slagle,B.L., Costa,R.H. and Raychaudhuri,P. (2001) DDB2 induces nuclear accumulation of the hepatitis B virus X protein independently of binding to DDB1. J. Virol., 75, 10383–10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin G.Y., Paterson,R.G., Richardson,C.D. and Lamb,R.A. (1998) The V protein of the paramyxovirus SV5 interacts with damage-specific DNA binding protein. Virology, 249, 189–200. [DOI] [PubMed] [Google Scholar]
- 30.Liu W., Nichols,A., Graham,J., Dualan,R., Abbas,A. and Linn,S. (2000) Nuclear transport of human DDB protein induced by ultraviolet light. J. Biol. Chem., 275, 21429–21434. [DOI] [PubMed] [Google Scholar]
- 31.Israel D.I. (1993) A PCR-based method for high stringency screening of DNA libraries. Nucleic Acids Res., 21, 2627–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ho S.N., Hunt,H.D., Horton,R.M., Pullen,J.K. and Pease,L.R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene, 77, 51–59. [DOI] [PubMed] [Google Scholar]
- 34.Quandt K., Frech,K., Karas,H., Wingender,E. and Werner,T. (1995) MatInd and MatInspector—new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res., 23, 4878–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heinemeyer T., Wingender,E., Reuter,I., Hermjakob,H., Kel,A.E., Kel,O.V., Ignatieva,E.V., Ananko,E.A., Podkolodnaya,O.A., Kolpakov,F.A. et al. (1998) Databases on transcriptional regulation: TRANSFAC, TRRD, and COMPEL. Nucleic Acids Res., 26, 362–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pearson B.E., Nasheuer,H.P. and Wang,T.S. (1991) Human DNA polymerase alpha gene: sequences controlling expression in cycling and serum-stimulated cells. Mol. Cell. Biol., 11, 2081–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao L. and Chang,L.S. (1997) The human POLD1 gene. Identification of an upstream activator sequence, activation by Sp1 and Sp3, and cell cycle regulation. J. Biol. Chem., 272, 4869–4882. [PubMed] [Google Scholar]
- 38.Huang D., Knuuti,R., Palosaari,H., Pospiech,H. and Syvaoja,J.E. (1999) cDNA and structural organization of the gene Pole1 for the mouse DNA polymerase epsilon catalytic subunit. Biochim. Biophys. Acta, 1445, 363–371. [DOI] [PubMed] [Google Scholar]
- 39.Fry C.J. and Farnham,P.J. (1999) Context-dependent transcriptional regulation. J. Biol. Chem., 274, 29583–29586. [DOI] [PubMed] [Google Scholar]
- 40.Emami K.H., Burke,T.W. and Smale,S.T. (1998) Sp1 activation of a TATA-less promoter requires a species-specific interaction involving transcription factor IID. Nucleic Acids Res., 26, 839–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grinstein E., Jundt,F., Weinert,I., Wernet,P. and Royer,H.-D. (2002) Sp1 as G1 cell cycle phase specific transcription factor in epithelial cells. Oncogene, 21, 1485–1492. [DOI] [PubMed] [Google Scholar]
- 42.Grandori C., Cowley,S.M., James,L.P. and Eisenman,R.N. (2000) The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol., 16, 653–699. [DOI] [PubMed] [Google Scholar]
- 43.Luscher B. (2001) Function and regulation of the transcription factors of the Myc/Max/Mad network. Gene, 277, 1–14. [DOI] [PubMed] [Google Scholar]
- 44.Fry C.J., Slansky,J.E. and Farnham,P.J. (1997) Position-dependent transcriptional regulation of the murine dihydrofolate reductase promoter by the E2F transactivation domain. Mol. Cell. Biol., 17, 1966–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu L., Timmers,C., Maiti,B., Saavedra,H.I., Sang,L., Chong,G.T., Nuckolls,F., Giangrande,P., Wright.,F.A., Field,S.J. et al. (2001) The E2F1-3 transcription factors are essential for cellular proliferation. Nature, 414, 457–462. [DOI] [PubMed] [Google Scholar]
- 46.Karlseder J., Rotheneder,H. and Wintersberger,E. (1996) Interaction of Sp1 with the growth- and cell cycle-regulated transcription factor E2F. Mol. Cell. Biol., 16, 1659–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang D., Jokela,M., Tuusa,J., Skog,S., Poikonen,K. and Syvaoja,J.E. (2001) E2F mediates induction of the Sp1-controlled promoter of the human DNA polymerase ε B-subunit gene POLE2. Nucleic Acids Res., 29, 2810–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan T. and Chu,G. (2002) p53 binds and activates the xeroderma pigmentosum DDB2 gene in humans but not mice. Mol. Cell. Biol., 22, 3247–3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scheffner M., Werness,B.A., Huibregtse,J.M., Levine,A.J. and Howley,P.M. (1990) The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell, 63, 1129–1136. [DOI] [PubMed] [Google Scholar]
- 50.Zolezzi F. and Linn,S. (2000) Studies of the murine DDB1 and DDB2 genes. Gene, 245, 151–159. [DOI] [PubMed] [Google Scholar]