Abstract
The Arabidopsis thaliana nuclear genome sequence revealed several open reading frames encoding proteins related to group II intron-encoded reverse transcriptase/maturases. Here, we show via sequence alignments that at least four such open reading frames are conserved in the nuclear genomes of A.thaliana and Oryza sativa (rice) and that they encode putative proteins belonging to two different classes (nMat-1 and nMat-2), neither of which is associated with a group II intron RNA structure. The two nMat-1 proteins have reverse transcriptase, maturase and DNA endonuclease domains characteristic of canonical group II intron-encoded proteins, while the two nMat-2 proteins have reverse transcriptase and maturase domains linked to a novel C-terminal domain. Although some nMat proteins have mutations expected to inactivate intron mobility functions, all could potentially retain the RNA splicing function. These nuclear maturase-like proteins may be imported into organelles to function in group II intron splicing and/or they may have assumed other cellular functions. Nuclear-encoded maturases could regulate organellar gene expression and may reflect a step in the evolution of mobile group II introns into spliceosomal introns.
INTRODUCTION
Mobile group II introns, which are present in bacterial, mitochondrial (mt) and chloroplast (cp) genomes, are widely believed to be the ancestors of nuclear pre-mRNA introns (1). These mobile introns encode reverse transcriptases (RTs) that function in intron mobility and also as maturases to promote RNA splicing by helping the intron RNA fold into the catalytically active structure (2,3). The mobile yeast mtDNA aI1 and aI2 and Lactococcus lactis Ll.LtrB group II introns, which have been studied as model systems, encode proteins with several conserved domains associated with different activities. These are: an N-terminal RT domain, with an upstream region Z and conserved sequence motifs (I–VII) characteristic of the fingers and palm domains of retroviral RTs; domain X, a putative RNA-binding domain associated with maturase activity; a C-terminal DNA-binding region and DNA endonuclease domain, which function in intron mobility (4–7). The mobility of these introns occurs by a novel target DNA-primed reverse transcription mechanism in which the intron RNA reverse splices directly into one strand of a DNA duplex, while the intron-encoded protein (IEP) uses the C-terminal DNA endonuclease domain to cleave the opposite strand and then uses the 3′ end of the cleaved strand as a primer for reverse transcription of the inserted intron RNA (8–10).
The reverse transcriptase/maturase proteins encoded by the yeast mtDNA and L.lactis Ll.LtrB introns are intron-specific splicing factors (11–13). However, there is suggestive evidence that the related matK proteins, which are encoded in tRNALys introns in higher plant chloroplasts and by free-standing open reading frames (ORFs) in the residual plastid genomes of non-photosynthetic plants, function in splicing multiple group II introns (14,15). Highly degenerate Euglena cp group II introns, referred to as group III introns, are also hypothesized to use a common splicing apparatus that includes a maturase encoded by one of the introns (16). Biochemical studies showed that the L.lactis Ll.LtrB intron protein (LtrA protein) interacts with both idiosyncratic and conserved regions of the group II intron catalytic core, suggesting how maturases in some organisms could evolve into general group II intron splicing factors (17,18). As noted previously, the evolution of an intron-specific maturase to function in splicing multiple group II introns could mirror a key step in the evolution of spliceosomal introns (2).
In this context, we noted with interest that recent genome sequencing projects have revealed putative proteins having similarity to group II intron maturases encoded by nuclear genes in Arabidopsis thaliana (19) and rice (Oryza sativa) (20,21). Here, we have analyzed four such ORFs and shown by sequence alignments that they can be divided into two classes, which are conserved in both A.thaliana and O.sativa. We speculate on the possible function and evolutionary significance of nuclear-encoded maturase-like proteins.
RESULTS AND DISCUSSION
Three ORFs in the A.thaliana genome had been annotated as encoding putative proteins having similarity to group II intron maturases (denoted here nMat-1b, nMat-2a and nMat-2b; accession nos BAB10231, NP_054444 and NP_177575, respectively). The fourth ORF (nMat-1a) had been annotated as an unknown protein (accession no. NM_102741) and we identified it as encoding a maturase-related protein by a BLAST search (22) of GenBank (release 131.0) with a conserved region of the LtrA protein encoded by the L.lactis Ll.LtrB group II intron (amino acids 303–489, encompassing conserved RT sequence motifs V–VII and domain X; accession no. Q57005). We also found four homologous ORFs in the draft rice genome sequence (public access at http://portal.tmri.org/rice/RiceAccess.html) (20); 67% identity, 80–82% similarity for nMat-1 proteins and 44–48% identity, 63–67% similarity for nMat-2 proteins. Based on BLAST searches of GenBank with the full-length ORFs, the closest relatives of the A.thaliana nMat-1 and nMat-2 proteins in GenBank (release 131.0) are the proteins encoded by Marchantia polymorpha mt group II introns coxII-I2 and cob-I3, respectively (25–31% identity based on the initial BLAST alignments). We also detected other weak matches to the M.polymorpha mt group II intron ORFs in the rice genome sequences, but the contigs were either incomplete or riddled with stops, so their significance could not be evaluated. All of the A.thaliana nMat ORFs and the rice nMat-1b ORF have putative mt import sequences and are classified as mt proteins by the TargetP computer program (v.1.0.1, available at http://www.cbs.dtu.dk/services/TargetP/) (23). The rice nMat-1a, nMat-2a and nMat-2b ORFs have sequence ambiguities at their N-termini, which prevented similar characterization.
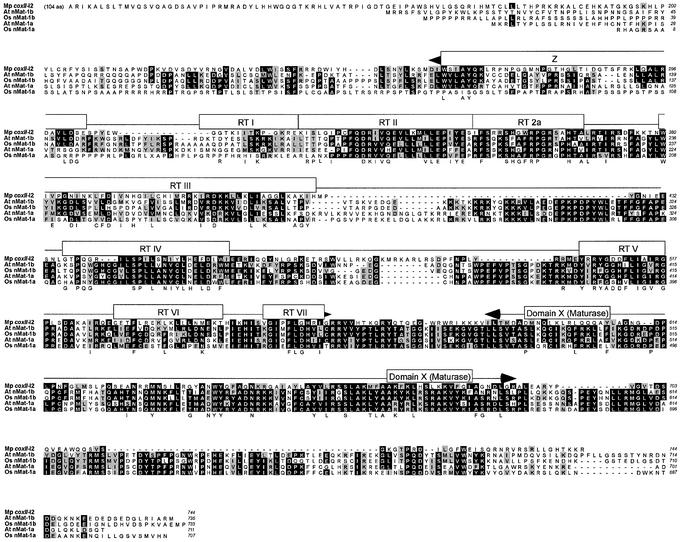
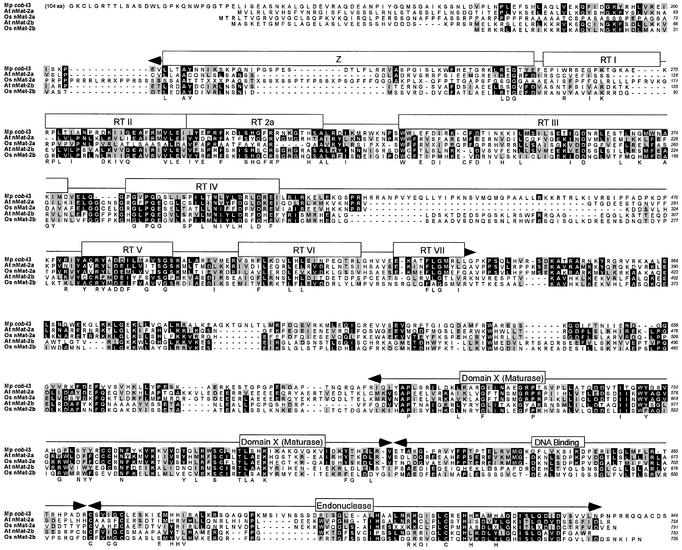
Figures 1 and 2 show refined sequence alignments for the two different ORF classes with the most closely related M.polymorpha group II intron-encoded protein. The nMat-1 proteins are homologous to the M.polymorpha coxII-I2 protein throughout the RT and X domains (26–30% identity, 37–42% similarity in the refined alignments), but their C-terminal domain is much larger and not homologous to the group II IEP DNA endonuclease domain or any other protein in the database. The nMat-2 proteins are homologous to the M.polymorpha cob-I3 protein throughout the RT, X and DNA-binding/DNA endonuclease regions (23–29% identity, 36–41% similarity). None of the nMat ORFs are associated with a recognizable group II intron RNA structure.
Figure 1.
Sequence alignment of nMat-1 putative proteins from A.thaliana (At) and rice (Os) with the mt coxII-I2 protein from M.polymorpha (Mp coxII-I2). Accession nos: M.polymorpha coxII-I2 protein NC_001660; At nMat-1a, NM_102741; At nMat-1b, BAB10231; Os nMat-1a, translated from CL009016.101; Os nMat-1b, AC087599. The rice nMat-1a protein was translated from the draft genome sequence, which contains sequence ambiguities (indicated by X in the alignments). The nMat-1b sequence of the Syngenta rice draft sequence is in clone CL027912.143, but has many sequence ambiguities and therefore is not shown in the alignment. Black shading with white letters indicates identical sequences in at least three of five proteins. Gray shading indicates similar amino acid residues according to the Henikoff matrix (32). Consensus sequences (75%) for RT motifs and domain X of mt group II encoded proteins are shown below the alignments (6).
Figure 2.
Sequence alignment of nMat-2 putative proteins from A.thaliana and rice with the cob-I3 protein from M.polymorpha. Accession nos: M.polymorpha cob-I3 maturase, NC_001660; At nMat-2a, NP_054444; At nMat-2b, NP_177575; Os nMat-2a, CLB8841.2; Os nMat-2b, CL000869.82.48. The rice proteins were translated from the draft genome sequence, which contains sequence ambiguities (indicated by X in the alignments). Highlighting and consensus sequences for the RT and X domains are as in Figure 1. Key amino acids in the DNA endonuclease domain based on analysis of the L.lactis LtrA protein are also shown below the alignment (7).
The nMat-1 proteins have deviations in the RT domain that are expected to inactivate RT activity (e.g. the conserved YGDD sequence of RT motif V is changed to YGGH, FGGH or FGSH), while the nMat-2 proteins have better matches to the RT consensus sequences, but have mutations in the DNA endonuclease domain that are expected to inactivate DNA endonuclease activity (e.g. in the ExHH motif involved in coordinating the catalytic metal ion) (7). Although the functional constraints on domain X sequences are not known, this region is conserved among the different nMat proteins and is a reasonable match to the consensus sequence for domain X of mt group II intron ORFs (6). Thus, some or all of the nMat proteins may lack mobility functions, but could retain the RNA splicing function. This situation is relatively common among maturase-related proteins. The cp matK proteins, for example, lack the DNA endonuclease domain and have a degenerate RT domain, but retain a well conserved domain X, which is presumably required for RNA splicing activity (5).
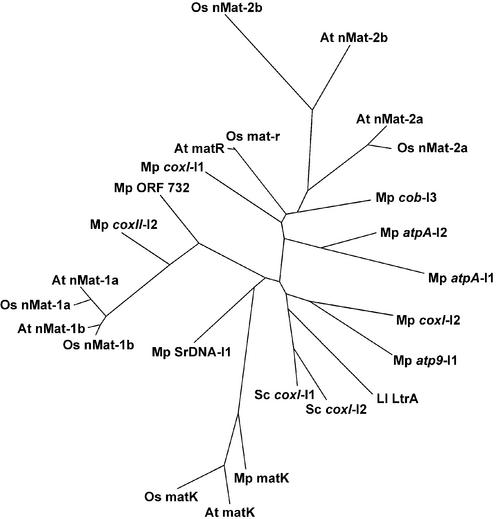
Figure 3 is an unrooted phylogenetic tree showing the relationship between the A.thaliana and O.sativa nMat-1 and nMat-2 proteins and various mt and cp group II IEPs, including the A.thaliana and O.sativa cp matK and mt matR/mat-r proteins; all the M.polymorpha mt group II IEPs and the free-standing ORF 732 maturase-related protein; and the well characterized yeast aI1 and aI2 and L.lactis Ll.LtrB IEPs. The tree was constructed based on alignment of RT regions V–VII and domain X, which are present in all of the proteins, using the PROML and DRAWTREE programs of the PHYLIP 3.6a3 package with default parameters (http://evolution.gs. washington.edu/phylip.html). Changing the amino acid similarity matrices (Jones–Taylor–Thornton model or Dayhoff PAM matrix model; 24,25) or other parameters gave substantially the same tree. The tree shows that the nMat-1 and nMat-2 proteins cluster with themselves and their closest relatives from M.polymorpha mitochondria. The branching order suggests that each nMat class was subjected to a sequence duplication after the divergence from the mt protein sequence. Further, each of the four nMat proteins in A.thaliana is more closely related to its homolog in rice than it is to the other A.thaliana nMat proteins, suggesting that all four proteins had already diverged in the last common ancestor of both plants (>100 000 000 years) (26). The branch lengths for the nMat-1a, nMat-1b and nMat-2a proteins appear generally similar to those of mt and cp group II IEPs, while the branch lengths for the nMat-2b proteins are somewhat longer, but still similar to the cp matK proteins, suggesting similar degrees of evolutionary constraint. The phylogenetic tree confirms the initial classification of the two related nMat classes based on BLAST similarity. The relatively close relationship between the nMat-2 proteins and the M.polymorpha cob-I3 IEP suggested by the tree is additionally supported by the presence of similar distinctively sized spacers between RT regions IV and V (∼50 amino acids) and RT region VII and domain X (∼180 amino acids) (6). In contrast, the neighboring A.thaliana and O.sativa mt matR/mat-r proteins lack RT regions Z and I and have a longer (∼150 amino acid) spacer between RT regions IV and V.
Figure 3.
Phylogenetic analysis. The regions encompassing RT motifs V–VII and domain X (∼100 amino acids each) were aligned using the CLUSTAL program (33) and the alignment was refined by hand. The boundaries of the aligned sequence blocks are as defined previously (5), with the variable spacer between the RT and X domains omitted. The tree was constructed using a Hidden Markov Model method, with the PROML and DRAWTREE programs of the PHYLIP 3.6a3 package with default parameters (http://evolution.gs.washington.edu/phylip.html). Accession nos: A.thaliana matK, AP000423; A.thaliana matR, X98300; L.lactis LtrA, Q57005; M.polymorpha matK, NC_001319; M.polymorpha mt proteins, NC_001660; O.sativa matK, NC_001320; O.sativa mat-r, AB076665; S.cerevisiae aI1 and aI2, NC_001224. nMat-1 and nMat-2 accession nos are indicated in the legends of Figures 1 and 2, respectively.
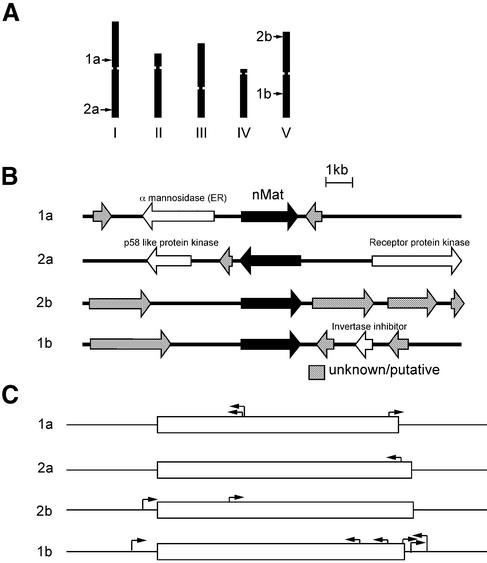
Figure 4 shows the chromosomal location of the four nMat ORFs in the A.thaliana genome. Two are on chromosome I and the other two are on chromosome V. All four ORFs have available T-element insertions, whose analysis may provide insight into function (Fig. 4C). Since maturase-encoding group II introns are present in plant mt and cp genomes, the most likely possibility is that the nMat ORFs were transferred from an organelle to the nucleus, as has been documented for other organellar genes (27). However, none of the ORFs are flanked by mt or cp protein genes, indicating either that they were not transferred with large segments of organelle DNA or that the surrounding regions have diverged. One particularly interesting possibility is that the ORFs were transferred by direct nuclear integration of mobile group II introns, whose conserved RNA structure subsequently degenerated. It is also possible that the RT/maturase proteins were associated with another type of mobile element that was ancestral to the group II intron ORFs (28,29). Dot plots comparing all eight A.thaliana and rice nMat genes and 2 kb flanking regions showed no extended DNA sequence similarity outside of the ORFs, and the neighboring genes differ in the two plant species.
Figure 4.
Chromosomal locations of and T-DNA insertions in or near the A.thaliana nMat ORFs. (A) Map of A.thaliana chromosomes I–V. The position of the nMat ORFs are indicated. (B) Genetic organization of a 10 kb region centered on the nMat ORFs. Genes are indicated by open arrows and identified according to GenBank annotation of the A.thaliana genome. Unknown proteins and potential proteins are shaded gray. (C) T-DNA insertions in and close to nMat ORFs. The T-DNA Express (http://signal. salk.edu/cgi-bin/tdnaexpress) and Garlic (http://www.tmri.org/pages/ collaborations/garlic_files/GarlicDescription.html) datbases were searched using nMat ORFs and flanking sequences and positive hits were mapped. ORFs are represented by open boxes with the ATG on the left. The orientations of the T-DNA insertions are indicated by arrows.
The high degree of conservation of the nMat proteins in A.thaliana and rice suggests that they have an essential function. Further, cDNA clones have been obtained for A.thaliana nMat-1b and nMat-2b, indicating that these genes are expressed (accession nos AF372929 and AY094457, respectively; http://signal.salk.edu/cgi-bin/sspsearch). Since the ORFs have mt localization signals and their maturase domains appear conserved, the most likely hypothesis is that the putative maturase proteins are transported into organelles to function in the splicing of group II introns. In addition, some or all of the proteins could have adapted to perform other cellular functions utilizing their putative RNA-binding activity (e.g. nuclear pre-mRNA splicing) and it remains possible that nMat2 proteins retain RT activity. In A.thaliana, only one of 22 mt group II introns (nad1-I4, which encodes matR, accession no. X98300) and only one of 26 cp group II introns (trnK-I1, which encodes matK) harbor a potential maturase (30,31). Thus, the nuclear-encoded maturases could potentially function as part of a common splicing apparatus for multiple organelle group II introns. The transfer of group II intron maturases to the nucleus has the potential advantage of facilitating the regulation of organelle gene expression by linking the splicing of one or more organellar introns to global signals that regulate gene expression in response to cellular energy state or environmental stimuli. In addition, transfer to the nucleus and the adaptation of maturases to function in splicing of multiple group II introns could reflect steps in the evolution of a common spliceosomal splicing apparatus for nuclear pre-mRNA introns.
Acknowledgments
ACKNOWLEDGEMENT
This work was supported by NIH grant GM37951.
REFERENCES
- 1.Michel F. and Ferat,J.L. (1995) Structure and activities of group II introns. Annu. Rev. Biochem., 64, 435–461. [DOI] [PubMed] [Google Scholar]
- 2.Lambowitz A.M., Caprara,M.G., Zimmerly,S. and Perlman,P.S. (1999) Group I and group II ribozymes as RNPs: clues to the past and guides to the future. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 451–485.
- 3.Belfort M., Derbyshire,V., Parker,M.M., Cousineau,B. and Lambowitz,A.M. (2002) Mobile introns: pathways and proteins. In Craig,N.L., Craige,R., Gellert,M. and Lambowitz,A.M. (eds), Mobile DNA, 2nd Edn. ASM Press, Washington, DC, pp. 761–783.
- 4.Michel F. and Lang,B.F. (1985) Mitochondrial class II introns encode proteins related to the reverse transcriptases of retroviruses. Nature, 316, 641–643. [DOI] [PubMed] [Google Scholar]
- 5.Mohr G., Perlman,P.S. and Lambowitz,A.M. (1993) Evolutionary relationships among group II intron-encoded proteins and identification of a conserved domain that may be related to maturase function. Nucleic Acids Res., 21, 4991–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zimmerly S., Hausner,G. and Wu,X.-c. (2001) Phylogenetic relationships among group II intron ORFs. Nucleic Acids Res., 29, 1238–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.San Filippo J. and Lambowitz,A.M. (2002) Characterization of the C-terminal DNA-binding/DNA endonuclease region of a group II intron-encoded protein. J. Mol. Biol., 324, 933–951. [DOI] [PubMed] [Google Scholar]
- 8.Zimmerly S., Guo,H., Eskes,R., Yang,J., Perlman,P.S. and Lambowitz,A.M. (1995) A group II intron RNA is a catalytic component of a DNA endonuclease involved in intron mobility. Cell, 83, 529–538. [DOI] [PubMed] [Google Scholar]
- 9.Zimmerly S., Guo,H., Perlman,P.S. and Lambowitz,A.M. (1995) Group II intron mobility occurs by target DNA-primed reverse transcription. Cell, 82, 545–554. [DOI] [PubMed] [Google Scholar]
- 10.Yang J., Zimmerly,S., Perlman,P.S. and Lambowitz,A.M. (1996) Efficient integration of an intron RNA into double-stranded DNA by reverse splicing. Nature, 381, 332–325. [DOI] [PubMed] [Google Scholar]
- 11.Carignani G., Groudinsky,O., Frezza,D., Schiavon,E., Bergantino,E. and Slonimski,P.P. (1983) An mRNA maturase is encoded by the first intron of the mitochondrial gene for the subunit I of cytochrome oxidase in S. cerevisiae. Cell, 35, 733–742. [DOI] [PubMed] [Google Scholar]
- 12.Moran J.V., Mecklenburg,K.L., Sass,P., Belcher,S.M., Mahnke,D., Lewin,A. and Perlman,P.S. (1994) Splicing defective mutants of the COXI gene of yeast mitochondrial DNA: initial definition of the maturase domain of the group II intron aI2. Nucleic Acids Res., 22, 2057–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saldanha R., Chen,B., Wank,H., Matsuura,M., Edwards,J. and Lambowitz,A.M. (1999) RNA and protein catalysis in group II intron splicing and mobility reactions using purified components. Biochemistry, 38, 9069–9083. [DOI] [PubMed] [Google Scholar]
- 14.Wolfe K.H., Morden,C.W. and Palmer,J.D. (1992) Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc. Natl Acad. Sci. USA, 89, 10648–10652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vogel J., Börner,T. and Hess,W.R. (1999) Comparative analysis of splicing of the complete set of chloroplast group II introns in three higher plant mutants. Nucleic Acids Res., 27, 3866–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Copertino D.W., Hall,E.T., Van Hook,F.W., Jenkins,K.P. and Hallick,R.B. (1994) A group III twintron encoding a maturase-like gene excises through lariat intermediates. Nucleic Acids Res., 22, 1029–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wank H., San Filippo,J., Singh,R.N., Matsuura,M. and Lambowitz,A.M. (1999) A reverse transcriptase/maturase promotes splicing by binding at its own coding segment in a group II intron RNA. Mol. Cell, 4, 239–250. [DOI] [PubMed] [Google Scholar]
- 18.Matsuura M., Noah,J.W. and Lambowitz,A.M. (2001) Mechanism of maturase-promoted group II intron splicing. EMBO J., 20, 7259–7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 408, 796–815. [DOI] [PubMed] [Google Scholar]
- 20.Goff S.A., Ricke,D., Lan,T.H., Presting,G., Wang,R., Dunn,M., Glazebrook,J., Sessions,A., Oeller,P., Varma,H. et al. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science, 296, 92–100. [DOI] [PubMed] [Google Scholar]
- 21.Yu J., Hu,S., Wang,J., Wong,G.K., Li,S., Liu,B., Deng,Y., Dai,L., Zhou,Y., Zhang,X. et al. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science, 296, 79–92. [DOI] [PubMed] [Google Scholar]
- 22.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nielsen H., Engelbrecht,J., Brunak,S. and von Heijne,G. (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng., 10, 1–6. [DOI] [PubMed] [Google Scholar]
- 24.Jones D.T., Taylor,W.R. and Thornton,J.M. (1992) The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci., 8, 275–282. [DOI] [PubMed] [Google Scholar]
- 25.Dayhoff M.O., Schwartz,R.M. and Orcutt,B.C. (1979) A model of evolutionary change in proteins. In Dayhoff,M.O. (ed.), Atlas of Protein Sequence and Structure, Vol. 5, Suppl. 3. National Biomedical Research Foundation, Silver Spring, MD, pp. 345–352.
- 26.Friis E.M., Pedersen,K.R. and Crane,P.R. (2001) Fossil evidence of water lilies (Nymphaeales) in the Early Cretaceous. Nature, 410, 357–360. [DOI] [PubMed] [Google Scholar]
- 27.Palmer J.D., Adams,K.L., Cho,Y., Parkinson,C.L., Qiu,Y.L. and Song,K. (2000) Dynamic evolution of plant mitochondrial genomes: mobile genes and introns and highly variable mutation rates. Proc. Natl Acad. Sci. USA, 97, 6960–6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kennell J.C., Moran,J.V., Perlman,P.S., Butow,R.A. and Lambowitz,A.M. (1993) Reverse transcriptase activity associated with maturase-encoding group II introns in yeast mitochondria. Cell, 73, 133–146. [DOI] [PubMed] [Google Scholar]
- 29.Curcio M.J. and Belfort,M. (1996) Retrohoming: cDNA-mediated mobility of group II introns requires a catalytic RNA. Cell, 84, 9–12. [DOI] [PubMed] [Google Scholar]
- 30.Marienfeld J., Unseld,M. and Brennicke,A. (1999) The mitochondrial genome of Arabidopsis is composed of both native and immigrant information. Trends Plant Sci., 4, 495–502. [DOI] [PubMed] [Google Scholar]
- 31.Sato S., Nakamura,Y., Kaneko,T., Asamizu,E. and Tabata,S. (1999) Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA Res., 6, 283–290. [DOI] [PubMed] [Google Scholar]
- 32.Henikoff S. and Henikoff,J.G. (1992) Amino acid substitution matrices from protein blocks. Proc. Natl Acad. Sci. USA, 89, 10915–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson J.D., Gibson,T.J., Plewniak,F., Jeanmougin,F. and Higgins,D.G. (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res., 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]