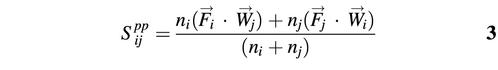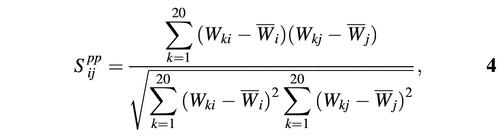Abstract
To improve the recognition of weak similarities between proteins a method of aligning two sequence profiles is proposed. It is shown that exploring the sequence space in the vicinity of the sequence with unknown properties significantly improves the performance of sequence alignment methods. Consistent with the previous observations the recognition sensitivity and alignment accuracy obtained by a profile–profile alignment method can be as much as 30% higher compared to the sequence–profile alignment method. It is demonstrated that the choice of score function and the diversity of the test profile are very important factors for achieving the maximum performance of the method, whereas the optimum range of these parameters depends on the level of similarity to be recognized.
INTRODUCTION
The observation that structures and functions of proteins can be inferred by protein sequence comparisons led to the fast development of sequence alignment methods. It has been shown that at short evolutionary distances structures and sequences of proteins are very similar and easily aligned by pairwise sequence alignment methods (1,2). For more distantly related proteins, where only certain sequence features or structural motifs are conserved, the similarity between two proteins cannot easily be recognized by pairwise alignment methods.
There are different ways to infer the connections between protein sequences in the sequence space; one group of methods tries to relate the two sequences being matched through the third sequence (3,4), while others detect increasingly divergent members of a given family by constructing multiple sequence alignments (5–10). Multiple alignments of related sequences properly translated into position-specific score matrices (PSSMs), profiles or hidden Markov models (HMMs) indeed contain a lot of information about the conservation patterns and statistical properties of protein families. As a result, the sensitivity of methods which use profiles/HMMs in the database search is shown to be several times higher compared to the pairwise methods (4,11).
The aforementioned profile search methods compare a single test sequence to a template profile, which in some cases can result in missing some weak sequence similarities. For example, if the test sequence and its related sequences are distant from the template family, the template profile would not be sensitive enough to recognize the test sequence as belonging to the same template family. To increase the radius of detection of diverse family members one would want to explore the sequence space around the test sequence and compare two groups of sequences or two profiles to each other.
Several methods have been reported lately which align two protein family models with each other. For example, in the progressive and iterative multiple alignment methods two groups of sequences have been compared using the weighted ‘sum of pairs’ score (12,13). This measure can be pretty successful in aligning two closely related families, but fails in other cases since it does not take into account the statistical properties of the sequence groups. More sophisticated measures of comparing two profiles or HMMs have been proposed recently and proved to be successful in detecting weak similarities between conserved protein regions or for the classification of signal peptides (14,15). In the most recent papers the relative success of profile–profile alignment methods over sequence–profile alignment methods has been reported (16,17). For example, Yona et al. (17) used information theory to derive the profile–profile similarity score and found that the relative improvement of their profile–profile comparison method with respect to PSI-BLAST is almost the same as the improvement of PSI-BLAST compared to BLAST in detecting the SCOP family relationships within one superfamily.
Despite the fact that profile–profile comparison methods have been used successfully in genome annotation, fold recognition and protein classification (18–20), there has been a lack of evidence that profile–profile scoring schemes outperform sequence–profile scoring schemes. To prove the latter, one would need to make a direct comparison of two scoring schemes using the same alignment algorithm, set of parameters and protein family models. This work introduces a new core-based profile–profile alignment method, which is tested with different similarity measures for comparing two columns of profiles. The performance of the method is evaluated with respect to sensitivity and specificity of the database search and the accuracy of obtained alignments using the benchmark of structurally similar protein domains or domains from the same SCOP classification level. Finally, the test procedure enables the systematic analysis of the difference in performance produced by including the test profile term in the similarity measure and examines the factors this difference can be attributed to.
MATERIALS AND METHODS
A benchmark for comparison of different score functions
The essential idea in designing the benchmark is to measure the ability of different score functions to find evolutionary relationships described by SCOP or correctly identify similar structures defined by the VAST algorithm. To obtain a representative set of protein domains, first a non-redundant set of 1310 domains has been selected by single linkage clustering based on a BLAST P value of 10e–7 or less (21). Domain boundaries identified using a compactness algorithm (22) are taken from MMDB (23), which is distributed with ENTREZ (http://www.ncbi.nlm.nih.gov/Entrez/).
Each selected domain in the list of 1310 domains (list1310) has at least five structure neighbors and its domain definition agrees with the SCOP domain definition to a threshold of 80% mutual overlap. The list of structure neighbors is distributed with Entrez and structure–structure alignments in this set are computed by the VAST algorithm (24,25) based on complete chains or domains. To select a subset of test protein sequences for the experiments, the domains formed from only a one chain continuous segment are selected from list1310 ensuring that these test domains have at least one VAST neighbor and at least one member from the same SCOP family level in list1310. Trying not to use any information about domain boundaries, the full-length chain sequences are extracted from the corresponding test domains, which are guaranteed not to exceed a length of 250 residues for the purpose of speeding up the search process. As a result, the test set has been reduced to the 47 test sequences listed in the legend to Figure 1. According to the SCOP classification (26) the structures of domains derived from 47 test protein chains spanned four different classes, 36 folds and 41 superfamilies and by this criterion can be considered a diverse sample of protein domains. Test sequences can be found at http://www.ncbi.nlm.nih.gov/Entrez/ and query/template alignments can be obtained upon request.
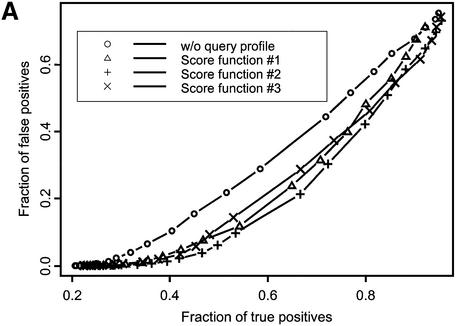
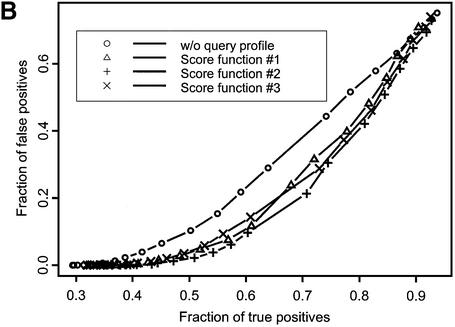
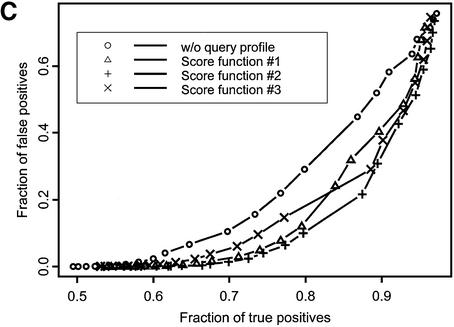
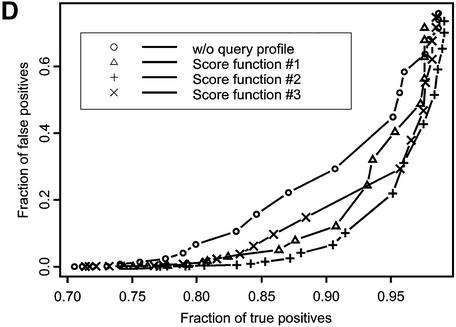
Figure 1.
ROC curves plotted for recognition of VAST neighbors (A) and SCOP domains at the fold (B), superfamily (C) and family (D) levels of classification. Different symbols denote various score functions (see Materials and Methods). Sequences in the test set are listed by their PDB code (lower case) and chain identifier (upper case): 1auoA, 1duzB, 1bftA, 1dbtA, 1bd8, 1a66A, 1aohB, 1exg, 1efiD, 1bqqT, 2occB, 1ctqA, 3pcgA, 1alvA, 2cut, 1ccwA, 1ad6, 1bt7, 1qa7B, 3nul, 1ecpA, 1a7gE, 1audA, 1aihA, 1qreA, 1huw, 1rcb, 1cewI, 1xnb, 1asu, 1czpA, 1guaB, 1aonO, 1cizA, 1tnrA, 1b9lA, 1rtm1, 1prtB, 1cnuA, 1c0gS,1nedA, 1aoiE, 1b5m, 4rhn, 1cjwA, 1dgwA, 3fib.
Fold recognition sensitivity and alignment accuracy
To find all relationships between 47 test sequences and their homologous (as defined by SCOP) or structurally similar (as defined by VAST) domains in list1310, each test sequence was compared against the database of 1310 domain structures using the core-based alignment algorithm (27,28). The true positive rate was calculated as the number of true positives found during the search above some Z-score threshold divided by the overall number of true positives. True positives here were defined as domains (out of list1310) with the same fold/superfamily/family or VAST assignments as a test domain. In cases where the test chain was composed of more than one domain (three of 47 test cases), the true positives for this test entry were identified as true positives for all domains included in the chain.
The false positive rate was calculated similarly to the number of false positives found divided by the overall number of false assignments (database size minus number of true positives). The Z-score was measured in terms of the momenta of the score distribution of random sequences with a given composition as a difference between the obtained alignment score and the expected score expressed in units of standard deviation of the random score distribution.
To compare the search sensitivity of different methods, another measure, the truncated receiver operating characteristic (ROC), has been used as well (29,30). ROCn was calculated as the sum of the number of true positives found at 1, 2, 3, …, n false positive levels (ti) divided by the overall number of true positives in the database (T) (30): ROCn = (ΣI = 1,…, n ti)/nT. The distribution of ROC values has been shown to be approximately normal and its standard deviation can be calculated analytically as σ2(Rn) = (ΣI = 1,…, n (tn + 1 – ti)2)/n2T2 (30).
The accuracy of obtained alignments, namely accuracy of molecular models of the test domains implied by the alignments of their sequences to the structures of the identified template domains, was calculated using the contact specificity. Contact specificity is defined as the fraction of correctly predicted non-local residue contacts in the molecular model which are also present in the experimental structure of the test domain (31): ACSpc = Ncp/Np. Here Ncp is the number of correctly predicted contacts for residues separated along the chain by at least five peptide bonds and having Cα atoms <8 Å apart. Np is the total number of non-local contacts in the predicted model. The measure we use in the alignment accuracy evaluation has been applied previously in the context of CASP structure prediction evaluations (31–34) and is based on correct prediction of residue–residue contacts, not on the comparison to a ‘true’ structure–structure alignment.
Alignment algorithm
Each test sequence was aligned to core elements of the template structure using the core-based alignment method described previously (27,28). Core elements of template domains were defined as continuous segments that were structurally conserved within a given family of domains (21,35). In general, core elements could be defined from multiple sequence alignments as continuous segments spanning only residues aligned in all sequences of the alignment. Loop lengths were constrained to disallow models with too few loop residues to span the distance between sequentially adjacent core elements.
Alternative alignments of each core segment were sampled by the Gibbs sampling algorithm. In this procedure the alignments of the center positions of core elements were sampled iteratively in the field of other core elements with fixed positions by using different types of score functions (see next section). The alignment of the center position of each core element was followed by the recruitment of additional residues at the N- or C-termini. Alignments were optimized using the simulated annealing schedule, which included 50 random alignment starts with 40 iterations of center point and end-point sampling, and each iteration in turn called for 10 cycles of center point alignments and 10 cycles of end-point refinement. Sampling was done iteratively until convergence with respect to recurrence of top-score alignments, which determined the overall run times. It should be noted that the algorithm is not related to any dynamic programming techniques and does not require gap penalties or ‘frozen approximation’ since the number of alignment variables is small enough to allow Gibbs sampling of alternative alignments with the direct evaluation of the overall alignment score. The core-based alignment algorithm has been tested in the CASP prediction experiment and proved to be successful in predicting the fold recognition targets (35).
Score functions
The scoring scheme for sequence–profile matches was the same as one described previously (28): it represented the difference between the score of the native test sequence aligned with a given template PSSM (Ssp) and the expected score obtained for random shuffles of the aligned residues in the test sequence (S0) (ΔSsp = Ssp – S0), where
Here, ai is the amino acid type in position i of the test sequence aligned with position j of the template and Wi,j are elements of the template PSSM. The shift factor (S0) represents a permuted sequence reference state and corrects for the test sequence composition (36).
Unlike the sequence–profile score, which involves the summation over the elements of the template PSSM, the profile–profile matching score should rather compare two aligned columns of profiles to each other. Three different measures for comparing two columns of profiles have been used in the study: two of them (correlation coefficient and dot product) are taken from pure mathematical considerations and the other uses some biological intuition trying to correctly encode the conservation patterns and variability in protein families.
In the simplest case the profile–profile matching score (Spp) for a given alignment i→j can be calculated as a dot product between a vector of observed frequencies and a vector of PSSM weights (score function 1):
Here Fi is the column of observed frequencies in the test profile and Wj is the corresponding column of PSSM weights of the template profile. It should be noted that in the limiting case of one sequence in the test profile or when all sequences in the test profile are identical to each other, this score will be reduced to the sequence–profile score. The expected profile– profile matching score is estimated for randomly permuted columns in the aligned region of the test profile similar to equation 1 and the difference between observed and expected matching scores is used in the alignment procedure (ΔSpp = Spp – S0). Calculating the reference state score directly in the profile–profile matching score allows us to avoid the optimization of the shift parameter, which effects the sensitivity of any chosen scoring scheme.
The second measure takes into account the ‘thickness’ or degree of divergence of both the test and template profiles (score function 2) (adopted with modification from a personal communication with John Spouge and Stephen Altschul):
Here, ni and nj are the numbers of independent observations or different amino acid types in columns i and j of the test and template profiles, respectively, which represent the measures of the diversity within the columns.
Pearson’s correlation coefficient between two columns of test and template profiles can be used as well (score function 3):
where Wi and Wj are vectors of PSSM scores for columns i and j, respectively. Scores constructed this way span values from –1, when there is a negative correlation between Wi and Wj, to 1 for columns with identical position-specific scores for each amino acid.
The profile–profile matching scores Spp calculated according to equations 2 and 3 will reduce to the corresponding elements of the amino acid substitution matrix in the case where the test and template profiles consist of identical sequences or nothing has been aligned to the test or template sequences in positions i and j. Test and template PSSMs were built by running PSI-BLAST 2.2.1 for five iterations with the default parameters and reporting matches which crossed the threshold of an E-value of 0.001 (http://www.ncbi.nlm.nih.gov/BLAST/).
RESULTS
Recognition sensitivity depends on the score function used
Figure 1 shows the fraction of true positive relationships detected with the different types of score functions plotted at various levels of false positive rate. Ideally, one would want to find the maximum number of true positives at a given false positive rate, so curves yielding better performance would lie farther to the right lower corner of the plot. There are two patterns apparent from this figure. First, the ROC curve corresponding to the original sequence–profile matching term lies above other ROC curves, where the test profile is used in the score calculation. For example, at the 1% level of false positives the original sequence–profile score function would yield 0.35, 0.58, 0.75 and 0.26 fractions of true positives for SCOP fold, superfamily and family levels and VAST neighbors recognition, respectively. At the same time, yielding the best performance, score function 2 would result in 0.46, 0.69, 0.84 and 0.37 fractions of true positives for the same levels of similarity, showing an increase of ∼15–30% with respect to the original sequence–profile score function.
The ROC100 statistic provides us with a quantitative measure to compare different scoring schemes. As can be seen from Table 1, the ROC100 statistic increases when the test profile is used in the calculation of the score for all types of profile–profile score functions. For example, in the case of score function 2, SCOP fold recognition improves from 0.269 ± 0.008 for the sequence–profile scoring scheme to 0.373 ± 0.010 (28% improvement), which demonstrates the statistically significant difference between various methods. Obviously, there is much room for improvement in profile– profile alignment methods since ROC100 values are not close to 1 even for the recognition at the SCOP family level.
Table 1. ROC100 values (mean values and their standard deviations) calculated with various score functions are listed for different SCOP categories and VAST structure neighbors.
| Without test profile | Score function | |||
|---|---|---|---|---|
| 1 | 2 | 3 | ||
| Vast neighbors | 0.197 ± 0.009 | 0.278 ± 0.013 | 0.296 ± 0.013 | 0.313 ± 0.012 |
| SCOP fold | 0.269 ± 0.008 | 0.349 ± 0.011 | 0.373 ± 0.010 | 0.364 ± 0.008 |
| SCOP superfamily | 0.456 ± 0.008 | 0.577 ± 0.010 | 0.600 ± 0.009 | 0.503 ± 0.008 |
| SCOP family | 0.694 ± 0.005 | 0.770 ± 0.007 | 0.800 ± 0.006 | 0.722 ± 0.004 |
It is clear from Table 1 that the Pearson correlation coefficient used as a similarity measure between two columns gives slightly higher ROC100 estimates for the recognition of VAST neighbors compared to other score functions. It can be explained by the fact that the Pearson correlation coefficient score function emphasizes not only similarities, but also encodes negative propensities of amino acids to be in a particular position. It can be crucial for detecting subtle similarities between, for example, non-homologous structure neighbors, which have different patterns of functionally conserved columns in their profiles. Moreover, from the same table we can see that the increase in sensitivity of the profile–profile method with respect to the sequence–profile alignment method is greatest for detecting VAST structure neighbors. This result is consistent with the previous observation by Yona and Levitt (17) that the position-specific weights of different amino acid types correlate very well with their secondary structure propensities for a given position and therefore structural similarities would be more easily recognized by the method based on the comparison of two profiles.
Recognition sensitivity depends on the test family diversity and level of detected similarity
A second pattern apparent from Figure 1 and Table 1 is that the recognition sensitivity strongly depends on the level of detected SCOP similarity, ranging from 0.46 in the case of the SCOP fold recognition to 0.84 for SCOP family recognition at a practical level of 1% false positives (for score function 2). The sensitivity of detecting structure neighbors is even smaller, ∼0.37, which shows that the SCOP fold level is defined more conservatively compared to VAST neighbors covering a rather broad range of structural similarities. It should be noted that test and template profiles were constructed by the PSI-BLAST algorithm, which can miss similarities based purely on a structural comparison, where there are no clear sequence motifs. It is consistent with the previous observations that only a small fraction of structure neighbors or folds can be detected by sequence comparison methods (3,37–39).
The sensitivity of the profile, or maximum radius of detection of remote family members, has been shown to depend on the diversity of the sequences included in the multiple alignment/profile (16,40,41). Profiles derived from the alignments of closely related sequences usually perform as well as pairwise alignment methods in the database search. At the same time, as more and more diverse sequences are included in the alignment, the low alignment accuracy can become an issue and dilute the information content of the profile.
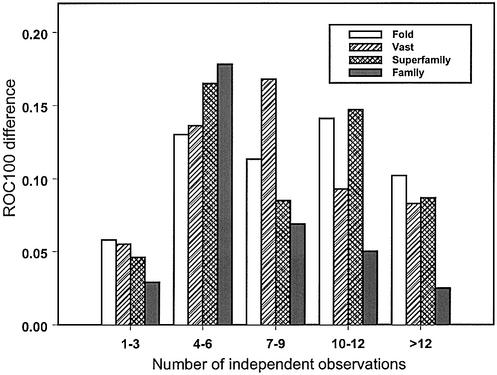
The same pattern can be observed for the improvement of the ROC100 statistic upon including the test profile term (Fig. 2). If the test family is not very diverse (between one and three of the number of different amino acid types per column), the improvement is not significant. The same is true for very diverse non-informative profiles, where the number of independent observations is >12. As can be seen from Figure 2, at around four to six amino acid types per column, the recognition improvement is high enough for all levels of the SCOP hierarchy, as well as structure neighbors, which is consistent with the optimal range of diversity reported earlier (41). More precisely, the desirable range of diversity depends on the level of similarity to be recognized. For example, similarity at the SCOP family level is more easily detected when the informativeness of the profile is not too low and diversity is not too high, whereas families encompassing more diverse members are more suitable for detecting the similarity at the SCOP superfamily, fold and VAST levels.
Figure 2.
Difference in recognition of VAST structure neighbors or SCOP fold/superfamily/family categories between sequence–profile and profile– profile alignment methods is plotted against the diversity of the test profile. Diversity is measured as the average number of independent observations (average number of different amino acid types per column of the alignment). Improvement in recognition is calculated as the average difference in ROC100 statistics between the profile–profile alignment method with scorefunction 2 and the sequence–profile alignment method.
Improvement in alignment accuracy
To assess different score functions for their ability to produce accurate alignments, the accuracies of models predicted by obtained sequence–profile or profile–profile alignments have been calculated. Table 2 shows the contact specificity values for models based on the detected domains from the same SCOP fold category averaged over different bins of sequence similarity. As can be seen from this table, alignment accuracy strongly depends on the sequence similarity between the test and template domains, which is consistent with the previous observations that accuracy of the alignments decreases with the evolutionary distance (39,42). In our case, contact specificity ranges from ∼0.20 for distantly related pairs to ∼0.90 for pairs with >40% sequence identity.
Table 2. Average contact specificity of alignments between test sequences and template domains detected with Z-score > 7 from the same fold SCOP category is shown for each bin of similarity between test and template domains.
| Identity (%) | Without test profile | Score function | ||
|---|---|---|---|---|
| 1 | 2 | 3 | ||
| 0–5 | 0.19 | 0.26 | 0.16 | 0.29 |
| 5–10 | 0.26 | 0.33 | 0.31 | 0.34 |
| 10–15 | 0.31 | 0.33 | 0.33 | 0.38 |
| 15–20 | 0.34 | 0.39 | 0.40 | 0.40 |
| 20–25 | 0.50 | 0.54 | 0.53 | 0.51 |
| 25–30 | 0.63 | 0.58 | 0.57 | 0.60 |
| 30–40 | 0.74 | 0.75 | 0.75 | 0.66 |
| >40 | 0.79 | 0.79 | 0.77 | 0.85 |
Similarity between test and template domains is calculated as the average percent identity in structure–structure alignments, given that the test and template structures are found to be VAST neighbors. For the purposes of comparison between different score functions contact specificity was averaged only over those test–template pairs found by all four types of score functions.
To compare the alignment accuracy of models produced with different score functions, contact specificity values are listed for SCOP folds found by all four tested score functions (Table 2). In other words, additional folds found by profile– profile score functions are not considered in this analysis. It is clear from this table that the alignment accuracy improves when test profile is used in the score calculation. Below 25% of sequence identity between the test and template domains, the improvement is maximum and models obtained with profile–profile score functions are up to 30% more accurate than the models obtained with the original sequence–profile score function. The same pattern is observed with another measure of alignment accuracy, RMSD (not shown). Above this level of similarity all four score functions and, in general, all sequence alignment methods yield accurate alignments.
Analysis of the obtained results showed that the profile– profile alignment algorithm can sometimes detect relationships between different SCOP families that cannot be easily inferred from the conventional sequence analysis. For example, tryptophan biosynthesis enzyme (2tsyA) and OMP-decarboxylase (1dbtA) belong to two different families of the same SCOP superfamily of ribulose phosphate-binding barrels. Although these proteins are very structurally similar and carry the same type of enzymatic activity (lyases), they act on different substrates (EC 4.2.1.20 and EC 4.1.1.23) and do not share significant sequence similarity (11% sequence identity). In order to detect interlinks between these two families, a CD search versus a non-redundant sequence database (43) has been performed and it has been found that sequences detected by both families constitute <1% of all detected sequences. In contrast, the profile–profile alignment method finds the 1dbtA–2tsyA pair with a very high Z-score equal to 14 and reasonable alignment quality of ∼41% of contact specificity or 6 A of RMSD. Interestingly enough, the original sequence–profile method is not capable of obtaining high quality alignment between these two proteins yielding 14% of contact specificity and 14 A of RMSD.
DISCUSSION
Several decades ago it was observed that protein sequences can be clustered into well-defined protein superfamilies of related sequences and that this sequence hierarchy may reflect the evolutionary history of contemporary proteins (44). With the rapid increase in the number of protein sequences and thoroughly characterized protein families, it has been shown that statistical properties of protein families are very important in most cases in establishing the correct evolutionary connections between sequences in the sequence space (8,11). This paper presents a next step in this direction, namely the algorithm, which is based on comparing the statistical properties of two protein families. The described profile–profile comparison algorithm is capable of detecting weak similarities between protein families, which cannot be found by sequence–profile alignment methods. It has been shown that the degree of improvement can be very significant not only in terms of increased sensitivity, but also in terms of higher accuracy of obtained alignments.
The size and boundaries of protein families in the sequence space vary to a great extent from one family to another and different profile search strategies would be useful in identifying sequence relationships in each particular case. Among the factors in defining the optimal strategy, the similarity between the test and template sequences and diversity of protein families are shown to be the most important ones. In agreement with this observation, the score function, which explicitly takes into account the diversity of test and template families, is found to yield somewhat better results compared to other profile–profile comparison score functions. Finally, analysis of different strategies for comparing two profiles presented in this paper can be of practical use for protein annotation and classification as well as supplementing the existing sequence alignment methods.
Acknowledgments
ACKNOWLEDGEMENTS
I thank Stephen Bryant, John Spouge and Stephen Altschul for helpful discussions and suggestions and Yuri Wolf and Benjamin Shoemaker for critically reading the manuscript. I also thank Jie Chen for providing me with the data about SCOP/MMDB domain overlap and the Intramural Research Program for support.
REFERENCES
- 1.Pearson W.R. and Lipman,D.J. (1988) Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA, 85, 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 3.Park J., Teichmann,S.A., Hubbard,T. and Chothia,C. (1997) Intermediate sequences increase the detection of homology between sequences. J. Mol. Biol., 273, 349–354. [DOI] [PubMed] [Google Scholar]
- 4.Salamov A.A., Suwa,M., Orengo,C.A. and Swindells,M.B. (1999) Combining sensitive database searches with multiple intermediates to detect distant homologues. Protein Eng., 12, 95–100. [DOI] [PubMed] [Google Scholar]
- 5.Gribskov M., McLachlan,A.D. and Eisenberg,D. (1987) Profile analysis: detection of distantly related proteins. Proc. Natl Acad. Sci. USA, 84, 4355–4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eddy S.R. (1996) Hidden Markov models. Curr. Opin. Struct. Biol., 6, 361–365. [DOI] [PubMed] [Google Scholar]
- 7.Hughey R. and Krogh,A. (1996) Hidden Markov models for sequence analysis: extension and analysis of the basic method. Comput. Appl. Biosci., 12, 95–107. [DOI] [PubMed] [Google Scholar]
- 8.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karplus K., Sjolander,K., Barrett,C., Cline,M., Haussler,D., Hughey,R., Holm,L. and Sander,C. (1997) Predicting protein structure using hidden Markov models. Proteins, (suppl. 1), 134–139. [DOI] [PubMed] [Google Scholar]
- 10.Neuwald A.F., Liu,J.S., Lipman,D.J. and Lawrence,C.E. (1997) Extracting protein alignment models from the sequence database. Nucleic Acids Res., 25, 1665–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park J., Karplus,K., Barrett,C., Hughey,R., Haussler,D., Hubbard,T. and Chothia,C. (1998) Sequence comparisons using multiple sequences detect three times as many remote homologues as pairwise methods. J. Mol. Biol., 284, 1201–1210. [DOI] [PubMed] [Google Scholar]
- 12.Gotoh O. (1993) Optimal alignment between groups of sequences and its application to multiple sequence alignment. Comput. Appl. Biosci., 9, 361–370. [DOI] [PubMed] [Google Scholar]
- 13.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pietrokovski S. (1996) Searching databases of conserved sequence regions by aligning protein multiple-alignments. Nucleic Acids Res., 24, 3836–3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lyngso R.B., Pedersen,C.N. and Nielsen,H. (1999) Metrics and similarity measures for hidden Markov models. Proc. Int. Conf. Intell. Syst. Mol. Biol., 178–186. [PubMed] [Google Scholar]
- 16.Rychlewski L., Jaroszewski,L., Li,W. and Godzik,A. (2000) Comparison of sequence profiles. Strategies for structural predictions using sequence information. Protein Sci., 9, 232–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yona G. and Levitt,M. (2002) Within the twilight zone: a sensitive profile-profile comparison tool based on information theory. J. Mol. Biol., 315, 1257–1275. [DOI] [PubMed] [Google Scholar]
- 18.Pawlowski K., Zhang,B., Rychlewski,L. and Godzik,A. (1999) The Helicobacter pylori genome: from sequence analysis to structural and functional predictions. Proteins, 36, 20–30. [PubMed] [Google Scholar]
- 19.Henikoff J.G., Greene,E.A., Pietrokovski,S. and Henikoff,S. (2000) Increased coverage of protein families with the blocks database servers. Nucleic Acids Res., 28, 228–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pawlowski K., Rychlewski,L., Zhang,B. and Godzik,A. (2001) Fold predictions for bacterial genomes. J. Struct. Biol., 134, 219–231. [DOI] [PubMed] [Google Scholar]
- 21.Matsuo Y. and Bryant,S.H. (1999) Identification of homologous core structures. Proteins, 35, 70–79. [PubMed] [Google Scholar]
- 22.Holm L. and Sander,C. (1994) Parser for protein folding units. Proteins, 19, 256–268. [DOI] [PubMed] [Google Scholar]
- 23.Marchler-Bauer A., Addess,K.J., Chappey,C., Geer,L., Madej,T., Matsuo,Y., Wang,Y. and Bryant,S.H. (1999) MMDB: Entrez’s 3D structure database. Nucleic Acids Res., 27, 240–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gibrat J.F., Madej,T. and Bryant,S.H. (1996) Surprising similarities in structure comparison. Curr. Opin. Struct. Biol., 6, 377–385. [DOI] [PubMed] [Google Scholar]
- 25.Madej T., Gibrat,J.F. and Bryant,S.H. (1995) Threading a database of protein cores. Proteins, 23, 356–369. [DOI] [PubMed] [Google Scholar]
- 26.Murzin A.G., Brenner,S.E., Hubbard,T. and Chothia,C. (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol., 247, 536–540. [DOI] [PubMed] [Google Scholar]
- 27.Bryant S.H. (1996) Evaluation of threading specificity and accuracy. Proteins, 26, 172–185. [DOI] [PubMed] [Google Scholar]
- 28.Panchenko A.R., Marchler-Bauer,A. and Bryant,S.H. (2000) Combination of threading potentials and sequence profiles improves fold recognition. J. Mol. Biol., 296, 1319–1331. [DOI] [PubMed] [Google Scholar]
- 29.Gribskov M. and Robinson,N.L. (1996) Use of receiver operating characteristic (ROC) analysis to evaluate sequence matching. Comput. Chem., 20, 25–33. [DOI] [PubMed] [Google Scholar]
- 30.Schaffer A.A., Aravind,L., Madden,T.L., Shavirin,S., Spouge,J.L., Wolf,Y.I., Koonin,E.V. and Altschul,S.F. (2001) Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res., 29, 2994–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marchler-Bauer A. and Bryant,S.H. (1997) Measures of threading specificity and accuracy. Proteins, (suppl. 1), 74–82. [DOI] [PubMed] [Google Scholar]
- 32.Levitt M. (1997) Competitive assessment of protein fold recognition and alignment accuracy. Proteins, (suppl. 1), 92–104. [DOI] [PubMed] [Google Scholar]
- 33.Moult J., Hubbard,T., Fidelis,K. and Pedersen,J.T. (1999) Critical assessment of methods of protein structure prediction (CASP): round III. Proteins, (suppl. 3), 2–6. [PubMed] [Google Scholar]
- 34.Marchler-Bauer A., Levitt,M. and Bryant,S.H. (1997) A retrospective analysis of CASP2 threading predictions. Proteins, (suppl. 1), 83–91. [DOI] [PubMed] [Google Scholar]
- 35.Panchenko A., Marchler-Bauer,A. and Bryant,S.H. (1999) Threading with explicit models for evolutionary conservation of structure and sequence. Proteins, 37, 133–140. [DOI] [PubMed] [Google Scholar]
- 36.Bryant S.H. and Altschul,S.F. (1995) Statistics of sequence-structure threading. Curr. Opin. Struct. Biol., 5, 236–244. [DOI] [PubMed] [Google Scholar]
- 37.Brenner S.E., Chothia,C. and Hubbard,T.J. (1998) Assessing sequence comparison methods with reliable structurally identified distant evolutionary relationships. Proc. Natl Acad. Sci. USA, 95, 6073–6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gerstein M. (1998) Measurement of the effectiveness of transitive sequence comparison, through a third ‘intermediate’ sequence. Bioinformatics, 14, 707–714. [DOI] [PubMed] [Google Scholar]
- 39.Sauder J.M., Arthur,J.W. and Dunbrack,R.L.,Jr (2000) Large-scale comparison of protein sequence alignment algorithms with structure alignments. Proteins Struct. Funct. Genet., 40, 6–22. [DOI] [PubMed] [Google Scholar]
- 40.Aravind L. and Koonin,E.V. (1999) Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J. Mol. Biol., 287, 1023–1040. [DOI] [PubMed] [Google Scholar]
- 41.Panchenko A. and Bryant,S. (2002) A comparison of position-specific score matrices based on sequence and structure alignments. Protein Sci., 11, 361–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson J.D., Plewniak,F. and Poch,O. (1999) A comprehensive comparison of multiple sequence alignment programs. Nucleic Acids Res., 27, 2682–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marchler-Bauer A., Panchenko,A., Shoemaker,B., Thiessen,P., Geer,L. and Bryant,S. (2002) CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res., 30, 281–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dayhoff M.O. (1976) The origin and evolution of protein superfamilies. Fed. Proc., 35, 2132–2138. [PubMed] [Google Scholar]