Abstract
It is generally believed that bryophytes are the earliest land plants. However, the phylogenetic relationships among bryophytes, including mosses, liverworts and hornworts, are not clearly resolved. To obtain more information on the earliest land plants, we determined the complete nucleotide sequence of the chloroplast genome from the hornwort Anthoceros formosae. The circular double-stranded DNA of 161 162 bp is the largest genome ever reported among land plant chloroplasts. It contains 76 protein, 32 tRNA and 4 rRNA genes and 10 open reading frames (ORFs), which are identical with the chloroplast genome of the other green plants analyzed. The major difference is a larger inverted repeat than that of the liverwort Marchantia, Anthoceros contains an excess of ndhB and rps7 genes and the 3′ exon of rps12. The genes matK and rps15, commonly found in the chloroplast genomes of land plants, are pseudogenes. The intron of rrn23 is the first finding in the known chloroplast genomes of land plants. A striking feature of the hornwort chloroplast is that more than half of the protein-coding genes have nonsense codons, which are converted into sense codons by RNA editing. Maximum-likelihood (ML) analysis, based on 11 518 amino acid sites of 52 proteins encoded in the chloroplast genomes of the green plants, placed liverworts as the sister to all other land plants.
INTRODUCTION
Approaches based on morphological, physiological and molecular data in extant plants have identified bryophytes as the earliest land plants (1,2). However, the exact phylogeny remains unresolved, especially with regard to which group of bryophytes represents the earliest form of land plants. Much morphological and molecular data (3), including sequences of nuclear small subunit rRNA (4), chloroplast rbcL (5) and rRNA internal transcribed spacers (6), and mitochondrial cox3 (7) and small subunit rRNA (8), support paraphyly of the bryophytes (9,10). However, these studies have not resolved the issue of the basal-most lineage of land plants in spite of the use of genes from all three genomes. Phylogenetic analysis based on chloroplast genes and nuclear rRNA genes (11) and cladistic analyses based on male gametogenesis (3,12) suggest hornworts to be the first. However, the fossil evidence (13) and intron structures of mitochondrial genomes (10,14) support liverworts as the earliest land plants.
The complete nucleotide sequences of many chloroplast genomes have been determined and this information has contributed to phylogenetic analyses. The angiosperms Nicotiana (15), Oryza (16), Zea (17) and Spinacia (18), the gymnosperm Pinus (19), the bryophyte liverwort Marchantia (20), the green alga Chlorella (21) and the prasinophytes Nephroselmis (22) and Mesostigma (23), all of which belong to the green plant lineage, have been studied. The genome of the charophyte Chaetosphaeridium, which is thought of as a direct ancestor of land plants, was recently published (24). In addition, studies on the whisk fern Psilotum (accession no. AP004638; T.Wakasugi, A.Nishikawa, K.Yamada and M.Sugiura, in preparation) and the moss Physcomitrella (C.Sugiura, Y.Kobayashi, S.Aoki, C.Sugita and M.Sugita, in preparation) will soon be published. The missing data on the chloroplast genome with which to identify the earliest land plants could come from hornworts and early land plant lycopods. Therefore, we have determined the complete nucleotide sequence of the hornwort Anthoceros formosae chloroplast genome. Here we discuss the earliest land plants inferred from the entire structure of chloroplast genomes.
MATERIALS AND METHODS
Total DNA was isolated from A.formosae as described previously (25) and used as a template for PCR. Thalli of the hornwort grown on 1/2 KnopII-agar medium were homogenized in a buffer containing 50 mM Tris–HCl pH 8.0, 10 mM EDTA, 20% sucrose, 5 mM 2-mercaptoethanol and 0.1% BSA with a Waring blender. The homogenate was passed through cheesecloth and spun at 1000 g for 10 s to remove unbroken cells. The chloroplast-rich fraction was precipitated from the supernatant by centrifugation at 3000 g for 10 min. Chloroplast DNA was solubilized in an extraction buffer containing 100 mM Tris–HCl pH 8.0, 50 mM EDTA, 500 mM NaCl, 10 mM 2-mercaptoethanol and 1% SDS and extracted by a standard method with phenol/chloroform/isoamyl alcohol (25:24:1). Chloroplast DNA was amplified using a PCR amplification kit (Takara, Osaka, Japan) with appropriate primers. The primers (accession no. AB086179) were designed from the partial sequences of several clones of an Anthoceros chloroplast DNA library which were analyzed previously (unpublished). The amplified DNA was ligated into Charomid vector (Nippon Gene) as described (25) and the resulting 18 clones were confirmed to cover the entire chloroplast genome. They were subcloned into Bluescript vector (Stratagene) to analyze their nucleotide sequences. Nucleotide sequences were analyzed with the Prism Dye Terminator Cycle sequencing kit (Applied Biosystems) on a DNA sequencer (Model 310; Applied Biosystems). The resulting sequences were treated with Genetyx-Mac v.8.5 and Genetyx-Mac/Atsq v.3.0 software (SDC, Tokyo). Nucleotide sequences for every coding region including pseudogenes and open reading frames (ORFs) were corrected by direct sequencing of re-amplified A.formosae DNA. These regions were amplified with the primers (AB086179), which were designed from the genomic sequence of 20–50 nt upstream and downstream of the coding region, so as to be able to sequence the entire coding region. Amplified regions were directly analyzed with the Prism Dye Terminator Cycle sequencing kit (Applied Biosystems). Additional primers within the coding regions were also designed from the genomic sequence.
Total RNA was isolated from A.formosae using CTAB (26) with a slight modification and cDNA for chloroplast transcripts was synthesized as described previously (25). Nucleotide sequence for every coding region of cDNA including pseudogenes and ORFs was amplified and sequenced as described above.
Phylogenetic analysis was carried out using 11 518 amino acid sites of 52 genes. They were atpA, atpB, atpE, atpF, atpH, atpI, clpP, petA, petB, petD, petG, psaA, psaB, psaC, psaI, psaJ, psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbL, psbM, psbN, psbT, psbZ, rbcL, rpl2, rpl14, rpl16, rpl20, rpl23, rpl32, rpl36, rpoA, rpoB, rpoC1, rpoC2, rps2, rps3, rps4, rps7, rps8, rps11, rps12, rps14, rps18 and rps19 from the genomes of A.formosae (AB086179), Marchantia polymorpha (20), Chlorella vulgaris (21), Chaetosphaeridium globosum (24), Nicotiana tabacum (15), Pinus thunbergii (19) and Psilotum nudum (T.Wakasugi, A.Nishikawa, K.Yamada and M.Sugiura, unpublished results). The amino acid sequences of Anthoceros were obtained from cDNA sequences (accession nos AB087416–AB087494) because it contained a large number of editing sites. The amino acid sequences were aligned using CLUSTAL W, v.1.81 (27). Maximum likelihood (ML) analysis was performed by PROTML in the MOLPHY v.2.3b3 package (28). Using constraint phylogeny within vascular plants [Psilotum (Pinus, Nicotiana)], the trees were examined for their approximate likelihoods under the JTT-F model (29) with the ‘-e’ option. Local bootstrap probability was estimated using the resampling of estimated log likelihood (RELL) method (30,31) with the best tree.
RESULTS
Overall genome organization
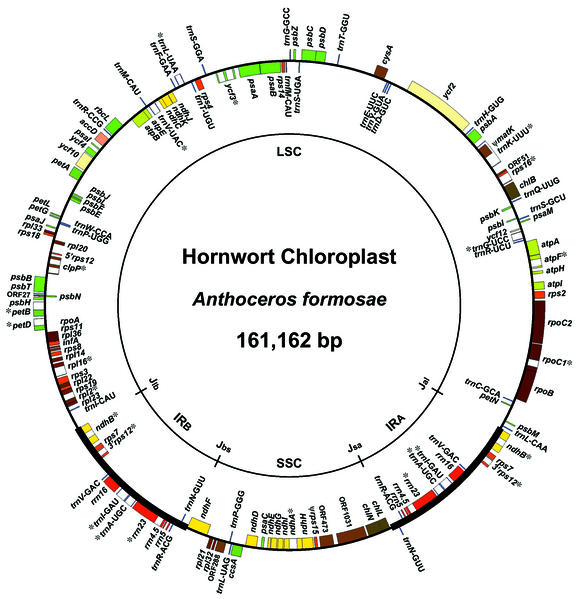
The circular chloroplast DNA of A.formosae is 161 162 bp long and divided into two regions by a pair of inverted repeat regions (IR) of 15 744 bp each. The large single copy region and small single copy region are 107 503 and 22 171 bp, respectively (Fig. 1). This is the largest genome ever reported among land plant chloroplasts, though the gene contents and arrangement are quite similar to those of the liverwort M.polymorpha (20). The major difference is a larger IR than that of Marchantia, Anthoceros contains an excess of ndhB and rps7 genes and the 3′ exon of rps12. Identified genes and conserved ORFs (ycfs) are listed in Table 1. We have found 112 genes including 76 protein coding genes, four rRNA genes and 32 tRNA genes. In addition, we have found 10 ORFs and two pseudogenes. Each ORF shows 60–70% identity with that of Marchantia. ORF1031 and ORF473 are homologous to the 5′ and 3′ regions of the ycf1 of Nicotiana, respectively. There is a large intergenic spacer between the cysA and trnT-GGU and we have detected a transcript longer than 770 nt. However, the transcript did not show any ORF form where no RNA editing was detected.
Figure 1.
Gene map of the A.formosae chloroplast genome. Genes shown inside the circle are transcribed clockwise and genes on the outside are transcribed counter-clockwise. Genes with related functions are shown in different colors. Asterisks denote split genes.
Table 1. Gene list of Anthoceros formosae chloroplast DNA.
| RNA genes | |||||
| Ribosomal RNAs | |||||
| 23S rDNA* ×2 | 16S rDNA ×2 | 5S rDNA ×2 | 4.5S rDNA ×2 | ||
| Transfer RNAs | |||||
| trnA(UGC)* ×2 | trnC(GCA) | trnD(GUC) | trnE(UUC) | trnF(GAA) | trnG(GCC) |
| trnG(UCC)* | trnH(GUG) | trnI(CAU) | trnI(GAU)* ×2 | trnK(UUU)* | trnL(CAA) ×2 |
| trnL(UAA)* | trnL(UAG) | trnfM(CAU) | trnM(CAU) | trnN(GUU) ×2 | trnP(GGG) |
| trnP(UGG) | trnQ(UUG) | trnR(ACG) ×2 | trnR(CCG) | trnR(UCU) | trnS(GGA) |
| trnS(GCU) | trnS(UGA) | trnT(GGU) | trnT(UGU) | trnV(GAC) ×2 | trnV(UAC)* |
| trnW(CCA) | trnY(GUA) | ||||
| Protein genes | |||||
| Photosynthesis | |||||
| Photosystem I | |||||
| psaA | psaB | psaC | psaI | psaJ | psaM |
| Photosystem II | |||||
| psbA | psbB | psbC | psbD | psbE | psbF |
| psbH | psbI | psbJ | psbK | psbL | psbM |
| psbN | psbT | psbZ | |||
| Cytochrome | |||||
| petA | petB* | petD* | petG | petL | petN |
| ATP synthase | |||||
| atpA | atpB | atpE | atpF* | atpH | atpI |
| Rubisco | |||||
| rbcL | |||||
| Chlorophyll biosynthesis | |||||
| chlB | chlL | chlN | |||
| NADH dehydrogenase | |||||
| ndhA* | ndhB* ×2 | ndhC | ndhD | ndhE | ndhF |
| ndhG | ndhH | ndhI | ndhJ | ndhK | |
| Ribosomal proteins | |||||
| Large subunits | |||||
| rpl2* | rpl14 | rpl16* | rpl20 | rpl21 | rpl22 |
| rpl23 | rpl32 | rpl33 | rpl36 | ||
| Small subunits | |||||
| rps2 | rps3 | rps4 | rps7 ×2 | rps8 | rps11 |
| rps12** | rps14 | ψrps15 | rps16 | rps18 | rps19 |
| Transcription/translation | |||||
| RNA polymerase | |||||
| rpoA | rpoB | rpoC1* | rpoC2 | ||
| Translation factor | |||||
| infA | |||||
| Miscellaneous proteins | |||||
| accD | ccsA | clpP** | cysA | ψmatK | |
| Conserved ORFs | |||||
| ORF27 | ORF33 (ycf12) | ORF51 | ORF170 (ycf3)** | ORF184 (ycf4) | ORF288 |
| ORF473 | ORF508 (ycf10) | ORF1031 | ORF2392 (ycf2) |
Pseudogenes are marked ‘ψ’. Hypothetical conserved frames are shown with ‘ycf’. Asterisks indicate number of introns in the gene. Two copy genes are shown with ‘×2’.
The differences in genes in the chloroplast genomes of Anthoceros and Marchantia are summarized in Table 2. The intron of rrn23 in Anthoceros is the first finding in the known chloroplast genomes of land plants. Two introns of ycf3 are commonly present in the chloroplast genomes of land plants, except Marchantia, in which only the first intron is found. The second intron is present in Chaetosphaeridium. ycf66 is absent in Anthoceros but present in the other bryophytes Marchantia and Physcomitrella. The gene trnP-GGG is present in Anthoceros, but is absent in Physcomitrella and is a pseudogene in Marchantia. The genes matK and rps15 are commonly present in all the known chloroplast genomes of the land plants and Chaetosphaeridium, but are pseudogenes in Anthoceros. The overall GC content of the genome is 32.9%, which is closer to that of the algae Chlorella (31.6%) (21), Odentella (31.8%) (32) and Porphyra (33%) (33) rather than Chaetosphaeridium (29.6%) (24) and the land plants Marchantia (28.8%) (34) and angiosperms (38–39%) (15–17).
Table 2. Differences in genes of chloroplast genomes.
| rrn23 intron | ycf3 intron | ycf66 | trnP-GGG | matK | rps15 | |
|---|---|---|---|---|---|---|
| Nicotiana (15) | – | + + | – | – | + | + |
| Pinus (19) | – | + + | – | + | + | + |
| Psilotuma | – | + + | – | + | + | + |
| Physcomitrellab | – | + + | + | – | + | + |
| Anthocerosc | + | + + | – | + | ψ | ψ |
| Marchantia (20) | – | + – | + | ψ | + | + |
| Chaetosphaeridium (24) | – | – + | + | + | + | + |
| Chlorella (21) | + | – – | – | – | – | – |
The presence (+) or absence (–) of each molecular character, and pseudogene (ψ) are shown. References are shown in parentheses.
aAP004638, T.Wakasugi, A.Nishikawa, K.Yamada and M.Sugiura, in preparation.
bC.Sugiura, Y.Kobayashi, S.Aoki, C.Sugita and M.Sugita, in preparation.
cThis paper.
RNA editing
A number of peculiar gene structures have been found in the chloroplast genome of Anthoceros, as described in our previous papers (25,35). Therefore RNA editing events have been systematically investigated in every transcript from the Anthoceros chloroplasts, except for some tRNAs. In total, 507 C→U and 432 U→C conversions have been identified in the transcript of 68 genes and eight ORFs. The unusual initiation codon ACG in atpB, atpH, petA, cysA and ccsA is converted into the usual initiation codon AUG by RNA editing of C→U. 164 nonsense codons of UGA, UAA and UAG in the 52 protein-coding genes and the seven ORFs are also converted into sense codons of CGA, CAA and CAG, respectively, by U→C conversion. The anticodon UUC in trnK is expectedly converted into UUU. The conversion of the anticodon by RNA editing is the first finding in plants, though RNA editing in the stem of tRNA has been known in plant mitochondria (36–38). Not all the nonsense codons within putative coding regions, however, are recovered by RNA editing. Several nonsense codons found in rps15- and matK-like sequences are not edited, though their transcripts are detected. Therefore we denote them as pseudogenes. Details of RNA editing will be provided in subsequent publications.
Phylogenetic analysis
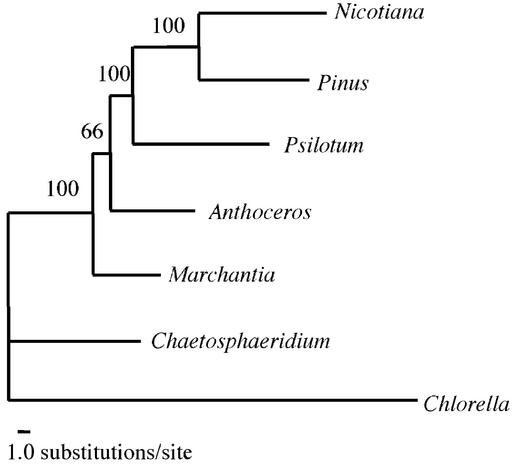
Phylogenetic relationship among the early land plants was inferred from the ML analysis based on 11 518 amino acid sites of 52 genes (Fig. 2). The tree of which the relative bootstrap probability was 75.3% indicated that Marchantia was sister to the other land plants, including Anthoceros. Local bootstrap probabilities of those branches supporting this phylogenetic relationship were quite high (100%), though the divergence between Marchantia and Anthoceros was 66%. In the second greatest-likelihood tree, Marchantia and Anthoceros formed a clade with a relative probability of 24.5%.
Figure 2.
The ML tree based on amino acid sequences of chloroplast genes. The horizontal length of each branch is proportional to the number of amino acid substitutions estimated by the ML method based on the JTT-F model. The number on each branch is the local bootstrap probability (%) estimated by the RELL method.
DISCUSSION
RNA editing is required in all major lineages of land plants except in complex thalloid liverworts (7,39). No editing, however, is required in algae (39,40), suggesting that liverworts are the earliest land plants. Extensive RNA editing has been observed in the hornwort A.formosae (25,35), suggesting that hornworts were the first to acquire RNA editing. However, simple leafy liverworts contain the editing mechanism (39,41). Thus, assuming liverworts in total as a monophyletic group (4,42), RNA editing itself is not a phylogenetically informative character for the plants discussed here.
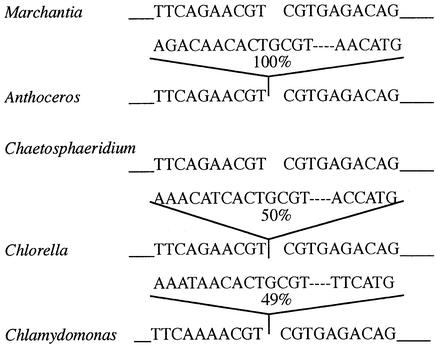
We have found an intron of rrn23 in Anthoceros. The intron is located in the homologous site of the charophytes Chlorella (21) and Chlamydomonas (43), but there is no intron in any known chloroplast genomes of land plants and Chaetosphaeridium. The sequence and location of them suggest that the intron of Anthoceros was derived from early charophytes, which was lost from coleochaetales and the common ancestor of the other land plants (Fig. 3), suggesting hornworts to be the earliest land plants. This view is supported by cladistic analyses based on male gametogenesis (3,12) and on ultrastructural data (42), and phylogenetic analyses based on nuclear small subunit rDNA (4), mitochondrial cox3 (7) and nad5 (44), mitochndrial small subunit rDNA (8) and a multigene data set (11,45). However, ycf66 is absent in the chloroplast genome of Anthoceros, as it is in the known vascular plants, but is present in those of the other bryophytes, suggesting that hornworts are the sister to vascular plants.
Figure 3.
Occurrence of the rrn23 intron. The intron of rrn23 has been found in Anthoceros and Chlorella (21) but no intron has been found in Marchantia (20) and Chaetosphaeridium (24). The sequences surrounding the intron show the position corresponding to intron 15 of Chlamydomonas (43). The identity (%) of each intron sequence to that of Anthoceros is shown.
ML analysis based on amino acid sequences of chloroplast genomes showed that liverworts were sister to the other land plants and bryophytes were monophyletic with relative bootstrap probabilities of 75.3 and 24.5%, respectively. The former, ‘liverworts-basal topology’, was supported from the distribution of mitochondrial introns (10,14) and from the fossil evidence (13). Although the latter is contradictory with the view that bryophytes are palaphyletic (46), it cannot be ruled out from our present results. The complete chloroplast structures of the moss Physcomitrella and lycopods, which are the closest relatives to bryophytes within vascular plants, will give important information about the earliest land plants.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs Hasebe and Nishiyama of the National Institute for Basic Biology for the valuable suggestions for phylogenetic analysis, Drs Wakasugi and Yamada of Toyama University and Dr Sugita of Nagoya University for unpublished data, and Dr Tsudzuki of Aichi-Gakuin University for drawing the physical map. We very much thank Prof. K. Wada for introducing the study of the hornworts and Mr T. Ishii of Otsuka Pharmaceutical Co. for establishing the method of preparing hornwort DNA. This work was supported in part by a Grant-in-Aid from the Ministry of Education, Japan.
DDBJ/EMBL/GenBank accession no. AB086179
REFERENCES
- 1.Bremer K., Humphries,C.J., Mishler,B.D. and Churchill,S.P. (1987) On cladistic relationships in green plants. Taxon, 36, 339–349. [Google Scholar]
- 2.Kenrick P. and Crane,P.R. (1997) The origin and early evolution of plants on land. Nature, 389, 33–39. [Google Scholar]
- 3.Mishler B.D., Lewis,L.A., Buchheim,M.A., Renzaglia,K.S., Garbary,D.J., Delwiche,C.F., Zechman,F.W., Kantz,T.S. and Chapman,R.L. (1994) Phylogenetic relationships of the “green algae” and “bryophytes.” Ann. Mo. Bot. Gard., 81, 451–483. [Google Scholar]
- 4.Hedderson T.A., Chapman,R.L. and Rootes,W.L. (1996) Phylogenetic relationships of bryophytes inferred from nuclear-encoded rRNA gene sequences. Plant Syst. Evol., 200, 213–224. [Google Scholar]
- 5.Lewis L.A., Mishler,B.D. and Vilgalys,R. (1997) Phylogenetic relationships of the liverworts (Hepaticae), a basal embryophyte lineage, inferred from nucleotide sequence data of the chloroplast gene rbcL. Mol. Phylogenet. Evol., 7, 377–393. [DOI] [PubMed] [Google Scholar]
- 6.Samigullin T.H., Valiejo-Roman,K.M., Troitsky,A.V., Bobrova,V.K., Filin,V.R., Martin,W. and Antonov,A.S. (1998) Sequences of rDNA internal transcribed spacers from the chloroplast DNA of 26 bryophytes: properties and phylogenetic utility. FEBS Lett., 422, 47–51. [DOI] [PubMed] [Google Scholar]
- 7.Malek O., Laettig,K., Hiesel,R., Brennicke,A. and Knoop,V. (1996) RNA editing in bryophytes and a molecular phylogeny of land plants. EMBO J., 15, 1403–1411. [PMC free article] [PubMed] [Google Scholar]
- 8.Duff R.J. and Nickrent,D.L. (1999) Phylogenetic relationships of land plants using mitochondrial small-subunit rDNA sequences. Am. J. Bot., 86, 372–386. [PubMed] [Google Scholar]
- 9.Mishler B.D. and Churchill,S.P. (1984) A cladistic approach to the phylogeny of the “bryophytes.” Brittonia, 36, 406–424. [Google Scholar]
- 10.Pruchner D., Nassal,B., Schindler,M. and Knoop,V. (2001) Mosses share mitochondrial group II introns with flowering plants, not with liverworts. Mol. Genet. Genomics, 266, 608–613. [DOI] [PubMed] [Google Scholar]
- 11.Nishiyama T. and Kato,M. (1999) Molecular phylogenetic analysis among bryophytes and tracheophytes based on combined data of plastid coded genes and the 18S rRNA gene. Mol. Biol. Evol., 16, 1027–1036. [DOI] [PubMed] [Google Scholar]
- 12.Garbary D.J., Renzaglia,K.S. and Duckett,J.G. (1993) The phylogeny of land plants: a cladistic analysis based on male gametogenesis. Plant Syst. Evol., 188, 237–269. [Google Scholar]
- 13.Edwards D., Duckett,J.G. and Richardson,J.B. (1995) Hepatic characters in the earliest land plants. Nature, 374, 635–636. [Google Scholar]
- 14.Qiu Y.-L., Cho,Y., Cox,J.C. and Palmer,J.D. (1998) The gain of three mitochondrial introns identifies liverworts as the earliest land plants. Nature, 394, 671–674. [DOI] [PubMed] [Google Scholar]
- 15.Shinozaki K., Ohme,M., Tanaka,M., Wakasugi,T., Hayashida,N., Matsubayashi,T., Zaita,N., Chunwongse,J., Obokata,J., Yamaguchi-Shinozaki,K. et al. (1986) The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J., 5, 2043–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hiratsuka J., Shimada,H., Whittier,R., Ishibashi,T., Sakamoto,M., Mori,M., Kondo,C., Honji,Y., Sun,C.R., Meng,B.-Y. et al. (1989) The complete sequence of the rice (Oryza sativa) chloroplast genome: intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol. Gen. Genet., 217, 185–194. [DOI] [PubMed] [Google Scholar]
- 17.Maier R.M., Neckermann,K., Igloi,G.L. and Koessel,H. (1995) Complete sequence of the maize chloroplast genome: gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J. Mol. Biol., 251, 614–628. [DOI] [PubMed] [Google Scholar]
- 18.Schmitz-Linneweber C., Maier,R.M., Alcaraz,J.P., Cottet,A., Herrmann,R.G. and Mache,R. (2001) The plastid chromosome of spinach (Spinacia oleracea): complete nucleotide sequence and gene organization. Plant Mol. Biol., 45, 307–315. [DOI] [PubMed] [Google Scholar]
- 19.Wakasugi T., Tsudzuki,J., Ito,S., Nakashima,K., Tsudzuki,T. and Sugiura,M. (1994) Loss of all ndh genes as determined by sequencing the entire chloroplast genome of the black pine Pinus thunbergii. Proc. Natl Acad. Sci. USA, 91, 9794–9798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohyama K., Fukuzawa,H., Kohchi,T., Shirai,H., Sano,T., Sano,S., Umesono,K., Shiki,Y., Takeuchi,M., Chang,Z. et al. (1986) Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature, 322, 572–574. [Google Scholar]
- 21.Wakasugi T., Nagai,T., Kapoor,M., Sugita,M., Ito,M., Ito,S., Tsudzuki,J., Nakashima,K., Tsudzuki,T., Suzuki,Y. et al. (1997) Complete nucleotide sequence of the chloroplast genome from the green alga Chlorella vulgaris: the existence of genes possibly involved in chloroplast division. Proc. Natl Acad. Sci. USA, 94, 5967–5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turmel M., Otis,C. and Lemieux,C. (1999) The complete chloroplast DNA sequence of the green alga Nephroselmis olivacea: insights into the architecture of ancestral chloroplast genomes. Proc. Natl Acad. Sci. USA, 96, 10248–10253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemieux C., Otis,C. and Turmel,M. (2000) Ancestral chloroplast genome in Mesostigma viride reveals an early branch of green plant evolution. Nature, 403, 649–652. [DOI] [PubMed] [Google Scholar]
- 24.Turmel M., Otis,C. and Lemieux,C. (2002) The chloroplast and mitochondrial genome sequences of the charophyte Chaetosphaeridium globosum: insights into the timing of the events that restructured organelle DNAs within the green algal lineage that led to land plants. Proc. Natl Acad. Sci. USA, 99, 11275–11280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshinaga K., Iinuma,H., Masuzawa,T. and Uedal,K. (1996) Extensive RNA editing of U to C in addition to C to U substitution in the rbcL transcripts of hornwort chloroplasts and the origin of RNA editing in green plants. Nucleic Acids Res., 24, 1008–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gawel N.J. and Jarret,R.L. (1991) A modified CTAB DNA extraction procedure for Musa and Ipomoea. Plant Mol. Biol. Rep., 9, 262–266. [Google Scholar]
- 27.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adachi J. and Hasegawa,M. (1996) MOLPHY version2.3: programs for molecular phylogenetics based on maximum likelihood. Comput. Sci. Monogr., 28, 1–150. [Google Scholar]
- 29.Jones D.T., Taylor,W.R. and Thornton,J.M. (1992) The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci., 8, 275–282. [DOI] [PubMed] [Google Scholar]
- 30.Kishino H., Miyata,T. and Hasegawa,M. (1990) Maximum likelihood inference of protein phylogeny and the origin of chloroplasts. J. Mol. Evol., 30, 151–160. [Google Scholar]
- 31.Hasegawa M. and Kishino,H. (1994). Accuracies of the simple methods for estimating the bootstrap probability of a maximum-likelihood tree. Mol. Biol. Evol., 11, 142–145. [Google Scholar]
- 32.Kowallik K.V., Stoebe,B., Schaffran,I., Kroth-Pancic,P. and Freier,U. (1995) The chloroplast genome of a chlorophyll a+c-containing alga, Odontella sinensis. Plant Mol. Biol. Rep., 13, 336–342. [Google Scholar]
- 33.Reith M. and Munholland,J. (1995) Complete nucleotide sequence of the Porphyra purpurea chloroplast genome. Plant Mol. Biol. Rep., 13, 333–335. [Google Scholar]
- 34.Umesono K. and Ozeki,H. (1987) Chloroplast gene organization in plants. Trends Genet., 3, 281–287. [Google Scholar]
- 35.Yoshinaga K., Kakehi,T., Shima,Y., Iinuma,H., Masuzawa,T. and Ueno,M. (1997) Extensive RNA editing and possible double-stranded structures determining editing sites in the atpB transcripts of hornwort chloroplasts. Nucleic Acids Res., 25, 4830–4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marechal-Drouard L., Kumar,R., Remacle,C. and Small,I. (1996) RNA editing of larch mitochondrial tRNA(His) precursors is a prerequisite for processing. Nucleic Acids Res., 24, 3229–3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Binder S., Marchfelder,A. and Brennicke,A. (1994) RNA editing of tRNA(Phe) and tRNA(Cys) in mitochondria of Oenothera berteriana is initiated in precursor molecules. Mol. Gen. Genet., 244, 67–74. [DOI] [PubMed] [Google Scholar]
- 38.Fey J., Tomita,K., Bergdoll,M. and Marechal-Drouard,L. (2000) Evolutionary and functional aspects of C-to-U editing at position 28 of tRNA(Cys) (GCA) in plant mitochondria. RNA, 6, 470–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steinhauser S., Beckert,S., Capesius,I., Malek,O. and Knoop,V. (1999) Plant mitochondrial RNA editing. J. Mol. Evol., 48, 303–312. [DOI] [PubMed] [Google Scholar]
- 40.Hiesel R., Combettes,B. and Brennicke,A. (1994) Evidence for RNA editing in mitochondria of all major groups of land plants except the Bryophyta. Proc. Natl Acad. Sci. USA, 91, 629–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Freyer R., Kiefer-Meyer,M.C. and Koessel,H. (1997) Occurrence of plastid RNA editing in all major lineages of land plants. Proc. Natl Acad. Sci. USA, 94, 6285–6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garbary D.J. and Renzaglia,K.S. (1998) Bryophyte phylogeny and the evolution of land plants: evidence from development and ultrastucture. In Bates,J.W., Ashton,N.W. and Duckett,J.G. (eds), Bryology for the Twenty-first Century. Maney Publishing and British Bryological Society, Leeds, UK. pp. 45–63.
- 43.Turmel M., Gutell,R.R., Mercier,J.P., Otis,C. and Lemieux,C. (1993) Analysis of the chloroplast large subunit ribosomal RNA gene from 17 Chlamydomonas taxa. Three internal transcribed spacers and 12 group I intron insertion sites. J. Mol. Biol., 232, 446–467. [DOI] [PubMed] [Google Scholar]
- 44.Beckert S., Steinhauser,S., Muhle,H. and Knoop,V. (1999) A molecular phylogeny of bryophytes based on nucleotide sequences of the mitochondrial nad5 gene. Plant Syst. Evol., 218, 179–192. [Google Scholar]
- 45.Nickrent D.L., Parkinson,C.L., Palmer,J.D. and Duff,R.J. (2000) Multigene phylogeny of land plants with special reference to bryophytes and the earliest land plants. Mol. Biol. Evol., 17, 1885–1895. [DOI] [PubMed] [Google Scholar]
- 46.Qiu Y.-L. and Palmer,J.D. (1999) Phylogeny of early land plants: insights from genes and genomes. Trends Plant Sci., 4, 26–30. [DOI] [PubMed] [Google Scholar]