Abstract
The sensitive telomeric repeat amplification protocol (TRAP) permits telomerase detection in mammalian cell and tissue extracts with very low telomerase activity levels. Unfortunately, conventional TRAP assays require complex post-amplification procedures, such as polyacrylamide gel electrophoresis and densitometry, to measure telomerase products. Therefore, a real-time quantitative TRAP assay (RQ-TRAP) was optimized in the present study and evaluated in comparison with a commercially available quantitative TRAP kit and by monitoring telomerase activity in human hepatocyte cultures, human hepatoma cell lines and telomerase reconstitution experiments. The novel real-time telomerase detection method has many advantages. Other than sample extraction and real-time cycling, no additional time-consuming steps have to be performed for telomerase quantification; reliable and linear telomerase quantification is possible down to single-cell dilutions without the interference of primer-dimer artifacts, and the costs are less. Moreover, the precision is similar to other amplification-based telomerase quantification assays and the results are comparable to data obtained with two commercially available assays. The closed-tube system reduces the risk of carryover contamination and supports high throughput. In conclusion, RQ-TRAP provides a new tool for the rapid and reliable quantification of telomerase activity.
INTRODUCTION
Telomeres, specific functional structures at the ends of eukaryotic chromosomes, are indispensable for chromosome protection and integrity (1). In proliferating cells lacking telomerase activity, telomeres progressively shorten with every cell division due to the end-replication problem and replication-associated erosion (2). Eventually, when telomeres are shortened and no longer protective, cells exit the cell cycle and enter a non-replicative state termed senescence (3). Telomerase, a unique ribonucleoprotein with reverse transcriptase activity, catalyzes the de novo addition of telomeric repeat sequences onto the eroding chromosome ends and thereby counterbalances telomere-dependent replicative aging (4). Telomerase is repressed in most human somatic cells, but reactivated in more than 80% of all human cancers (5). Because of its involvement in carcinogenesis, telomerase is a promising candidate as both tumor marker and therapeutic target for telomerase inhibitors or antisense constructs (6). In addition, telomerase reactivation was employed to enable human fibroblasts (7), human mammary epithelial cells (8), and human keratinocytes (9), among other cell types, to proliferate without restriction and to bypass the Hayflick limit (10).
The introduction of the sensitive PCR-based telomeric repeat amplification protocol (TRAP) to detect telomerase activity (11) has been quintessential for the evaluation of the role of telomerase in tumor development and cell immortalization. The original two-step TRAP assay utilizes a radiolabeled primer amenable to elongation with telomeric repeat sequences produced by functional telomerase enzyme extracted from cells or tissues. Because of the extremely low telomerase activity in mammalian cells, the TRAP assay includes a specific amplification protocol using a telomeric reverse primer and PCR technology. In the conventional TRAP assay, polyacrylamide gel electrophoresis and autoradiography are performed to visualize the typical six-base product ladder and to quantitate telomerase activity following densitometry. Since the introduction of the TRAP assay, various modifications have been suggested, especially to improve quantification and to simplify the time-consuming post-PCR steps. For example, combining the TRAP assay with a post-PCR hybridization protection protocol employing a chemiluminescent probe (12), or measuring the amount of double-stranded DNA generated in the TRAP assay with a fluorescent dye (13), were both proposed as rapid non-isotopic quantification methods. Using a different approach, energy transfer primers that emit fluorescence only upon incorporation into PCR products were applied to develop a closed-tube telomerase detection system (14). Alternatively, telomerase products were amplified in the presence of [3H]dTTP and the radioactivity of amplicons was measured following membrane filtration (15) to circumvent the need for gel analysis. Recently, a sensitive enzymatic luminometric inorganic pyrophosphate detection assay was introduced to quantitate the amount of inorganic pyrophosphate generated during the amplification of telomerase products as a measurement of telomerase activity (16). Two commercially available TRAP-based kits (see Materials and Methods) utilize biotinylated primers and enzyme-linked immunosorbent assays (ELISA) for post-PCR analysis (17). To improve linearity, the TRAP-ELISAs include competitive internal standards that also control for the presence of PCR inhibitors. These end-point PCR techniques have provided an interesting evolution of the conventional TRAP assay; however, they require complex multi-step procedures. Finally, direct telomerase activity assays without amplification of telomerase products have also been improved, but are generally less sensitive than PCR-based protocols (18,19).
To overcome some of the inherent problems of end-point PCR that make the accurate measurement of telomerase activity difficult, and to avoid post-PCR analysis steps, a real-time quantitative TRAP (RQ-TRAP) assay has been proposed (20). This assay employs a complex PCR master mix without eliminating primer-dimer artifacts that interfere with correct data interpretation. In the present study, we optimized and validated a simplified RQ-TRAP assay for the rapid quantification of telomerase activity. Our results demonstrate that a simple RQ-TRAP assay provides accurate telomerase activity data and is very effective in monitoring telomerase activity in cultured cells.
MATERIALS AND METHODS
Cell lines and cultures
The modified human embryonic kidney cell line 293T (21), a generous gift from Dr R. A. Weinberg, Whitehead Institute of Biomedical Research, Cambridge, Massachusetts, was cultured in minimum essential medium Eagle (Sigma-Aldrich). The human hepatoma cell lines Hep G2, acquired from the American Type Culture Collection (www.atcc.org), and HLE, HLF and HuH 7, purchased from the Japanese Coll ection of Research Bioresources (www.cellbank.nihs.go.jp), were cultured as recommended (see websites). Human fetal hepatocytes (HFH) were procured by Dr S. Gupta, Marion Bessin Liver Research Center, Albert Einstein College of Medicine, Bronx, New York, under approval from the institutional Committee on Clinical Investigations. Human adult and pediatric hepatocytes (HPH) were freshly isolated by the Liver Tissue Procurement and Distribution System, Minneapolis, Minnesota. All human hepatocyte cultures were maintained in a basic growth medium and cultured on tissue culture plastic as described (22). Human fetal pancreatic cells (HFP) were kindly provided by F. Wu, University of California, Davis Medical Center, Sacramento, California, and grown in RPMI-1640 medium (Invitrogen). Culture media were supplemented with 10% inactivated fetal calf serum (Invitrogen) and antibiotics. Cells were cultured in a humidified 5% CO2 atmosphere.
Sample extraction and inactivation for telomerase activity assays
Samples for telomerase activity assays were extracted following standard protocols (11). Briefly, cell pellets were resuspended in 3-[(3-cholamidopropyl)-dimethylammonio]-1-propane-sulfonate lysis buffer (1000 cells/µl) and incubated for 30 min on ice. After centrifugation at 16 000 g for 20 min at 4°C, aliquots of the supernatant were rapidly frozen and stored at –80°C. Protein concentration of extracts was determined with the DC Protein Assay (Bio-Rad). Telomerase was inactivated by digestion with 1 µg/µl DNase-free ribonuclease A (Sigma-Aldrich) for 20 min at 37°C.
TRAP-ELISA assays
Two commercially available TRAP-ELISA kits were utilized, the qualitative TRAPeze ELISA Telomerase Detection Kit (Intergen) and the quantitative TeloTAGGG Telomerase PCR ELISAPLUS (Roche Molecular Biochemicals). For maximum sensitivity, the TRAPeze ELISA was performed with 1 and 4 µg of cellular protein and 35 PCR cycles as outlined by the manufacturer. For the quantitative TeloTAGGG Telomerase PCR ELISAPLUS, cell extracts (1000 cells) were evaluated with an initial primer-elongation time of 10 min. Telomerase activity was calculated as suggested in the kit’s manual and compared with a control template of 0.1 amol telomeric repeats, representing a relative telomerase activity (RTA) of 100. Inactivated samples and lysis buffer served as negative controls.
RQ-TRAP
The SYBR Green RQ-TRAP assay was conducted with cell extracts (1000 cells), 0.1 µg of telomerase primer TS, and 0.05 µg of anchored return primer ACX, in 25 µl with SYBR Green PCR Master Mix (Applied Biosystems). Primer sequences were as described by Kim and Wu (23). Using the ABI Prism 7700 thermal cycler (Applied Biosystems), samples were incubated for 20 min at 25°C and amplified in 35 PCR cycles with 30 s at 95°C and 90 s at 60°C (two-step PCR). The threshold cycle values (Ct) were determined from semi-log amplification plots (log increase in fluorescence versus cycle number) and compared with standard curves generated from serial dilutions of telomerase-positive 293T cell extracts (1000, 100, 10, 1 cell). The default setting for the amplification threshold was 10 standard deviations above the mean background fluorescence. Standards, inactivated samples and lysis buffer were assayed anew on every plate. Each sample was analyzed at least in duplicates. Telomerase activity was expressed relative to 293T cells (RTA), i.e. the percentage of telomerase activity compared with 293T cells.
Telomerase reconstitution
Control amphotropic retroviral vectors and vectors delivering human telomerase reverse transcriptase (hTERT) were produced by cotransfecting 293T cells with either pBabe-puro or pBabe-puro-hTERT and the packaging plasmid pCL-Ampho (Imgenex) using Fugene 6 (Roche Molecular Biochemicals). The pBabe-puro plasmids were kindly provided by Dr R. A. Weinberg (see above). Viral supernatants were harvested 2 and 3 days after transfection and purified for immediate transduction in the presence of 8 µg/ml hexadimethrine bromide (Sigma-Aldrich). Cells were transduced with full-strength viral supernatants (∼20 000 transducing units/ml) for 3 h daily on 2–4 consecutive days, corresponding to a total multiplicity of infection of 0.5–1.5. Following transduction, cells were selected for 7–10 days with puromycin dihydrochloride (Sigma-Aldrich), using 0.75 µM for HFH, 2 µM for HPH and 1 µM for HFP.
Transgene expression analysis
Total RNA (100 ng), extracted with RNeasy Mini Kits (Qiagen), was examined with the LightCycler TeloTAGGG hTERT Quantification Kit (Roche Molecular Biochemicals). As suggested by the manufacturer, hTERT expression was normalized to the housekeeping gene porphobilinogen deaminase (PBGD) and expressed as hTERT copies per PBGD copy.
Statistical analysis
All values are represented as means ± SEM of the indicated number of measurements. Linear regression was assessed with the ‘least squares’ method and the coefficient of determination (R2) was calculated to evaluate strength of correlation. An ANOVA test and t-test statistics (two-tailed) were employed to determine significance, requiring P < 0.05 (95% confidence level) for statistical significance.
RESULTS
Optimization of the RQ-TRAP assay
Besides primers, nucleotides and Taq polymerase, the TRAP reaction mixture contains tris(hydroxymethyl)aminomethane buffer, MgCl2 and KCl. To ensure optimal reaction conditions for both telomerase and Taq polymerase activities, some investigators include, as originally suggested by Kim et al. (11), ethylene glycol-bis(β-aminoethylether)-N,N,N′,N′-tetraacetic acid (EGTA), T4 gene protein (13,20), bovine serum albumin (16) and polyoxyethylenesorbitan monolaurate (14). We compared a reaction mixture previously employed for a RQ-TRAP assay including T4 gene protein and EGTA (20) with a simple reaction mixture based on a commercially available SYBR Green PCR mix and without additional enhancers. Serial dilutions of Hep G2 cell extracts (1, 0.1, 0.01, 0.001 µg protein) were analyzed with both TRAP mixtures by real-time quantitative PCR and compared. The performance of both TRAP mixtures was similar, with linear regression values (R2) between 0.976 and 0.997 for the mixture containing T4 gene protein and EGTA (n = 3), and 0.968 and 0.994 for the simple SYBR Green mixture (n = 3). Ct values were identical, except for higher Ct values obtained with the complex reaction mixture and protein amounts of 0.01 µg or less. The data clearly demonstrates that T4 gene protein and EGTA are not essential for the TRAP assay and that a simple PCR master mix provides equally efficient telomere primer elongation and amplification.
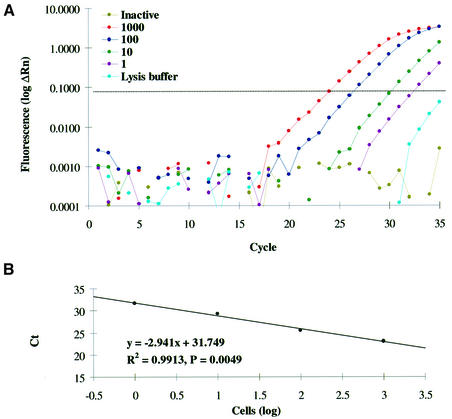
We conducted a series of systematic experiments with different primer concentrations and annealing temperatures in order to eliminate primer-dimer formation. Primer-dimer amplification was monitored by real-time quantitative SYBR Green PCR as described, using lysis buffer and inactivated cell extracts as no template controls. Amplification of primer-dimers interfered with telomerase detection in less than 10 telomerase-positive Hep G2 cells if the annealing temperature was below 55°C and 1.0 µg of both primers were used in a 25 µl reaction volume. In contrast, primer-dimer formation occurred only sporadically if the annealing temperature was raised to 60°C and the amount of anchored reverse primer ACX was decreased to 0.05 µg per reaction. Using these conditions and a two-step PCR protocol, primer-dimer formation was negligible and, if primer-dimer artifacts appeared, could be managed by raising the amplification threshold manually above the level of primer-dimer signals (Fig. 1A).
Figure 1.
A representative 293T-based standard curve. (A) Cell-number-dependent amplification is illustrated for the RQ-TRAP by charting cycle number versus log increase in fluorescence (log ΔRn) for different 293T cell quantities, inactivated cell extract (1000 cells) and lysis buffer. The dotted line represents the threshold of significant amplification. (B) Amplification of a serial dilution of 293T cell extract is graphed as log number of cells versus Ct with linear trendline for regression analysis (ANOVA test).
Optimized RQ-TRAP conditions were employed to set up standard curves for serial dilutions of telomerase-positive 293T cell extracts (1000, 100, 10, 1 cell). Semi-log amplification plots revealed a cell-number-dependent amplification with no significant increase in fluorescence for inactivated samples (1000 cells) and lysis buffer (Fig. 1A). A significant and strong linear relationship (ANOVA test) was confirmed by plotting the Ct values versus the log number of analyzed cells (Fig. 1B). Inhibition (increased Ct values) was observed if 5000 or more cells were analyzed. Using RQ-TRAP, telomerase activity was detected in single-cell samples confirming the high sensitivity of TRAP assays. However, results for such highly diluted samples were less reproducible, possibly due to random variations during sampling and dilution (Poisson error). With R2 between 0.976 and 0.994 (n = 5), the 293T-based standard curve provided a convenient and reliable tool to quantitate telomerase activity by cell-to-cell comparison, without the need to determine protein concentrations and to prepare standards with known amounts of telomeric repeats.
Linearity and accuracy of the RQ-TRAP assay
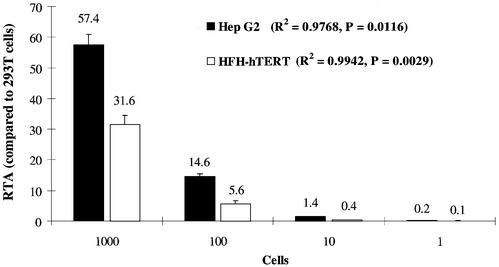
To investigate accuracy and linearity of the modified RQ-TRAP assay, serial dilutions of Hep G2 and telomerase-reconstituted HFH (HFH-hTERT) cell extracts (1000, 100, 10, 1 cell) were analyzed in four separate experiments in duplicates. The RTA values (percentage of telomerase activity in comparison with 293T cells) were pooled for each dilution (Fig. 2). There was a significant and strong linear relationship (ANOVA test) between the number of analyzed cells and RTA values for both Hep G2 and HFH-hTERT, with R2 values of 0.9768 and 0.9942, respectively. The coefficient of variation (CV) was determined for each dilution as an indicator of inter-assay variation. CV values were ∼12% for 1000 and 100 Hep G2 cells, and 16.3% for 10 analyzed cells. Higher CVs were observed for HFH-hTERT, with 18.9% for 1000 cells, 41.1% for 100 cells and 42.9% for 10 cells. As expected, the accuracy decreased if highly diluted samples (10 or less cells) were assayed. Therefore, telomerase activity measurements with the RQ-TRAP assay were performed with extracts from 1000 cells.
Figure 2.
Linearity of RQ-TRAP for different cell lines. Serial dilutions of Hep G2 and HFH-hTERT cell extracts were analyzed in replicates. Shown are the pooled results (n = 8) summarizing telomerase activity as a percentage of 293T cell telomerase activity (RTA). The error bars represent SEM. ANOVA test results are included in the figure.
Evaluation of the RQ-TRAP in comparison with TRAP-ELISA kits
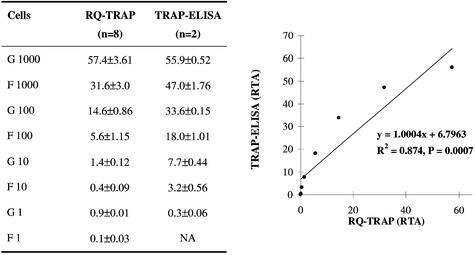
To evaluate the consistency of the new RQ-TRAP assay with commercially available quantitative TRAP kits, serial dilutions of Hep G2 and HFH-hTERT cell extracts (1000, 100, 10, 1 cell) were evaluated with the quantitative TeloTAGGG Telomerase PCR ELISAPLUS, and the results were compared with telomerase activity measurements obtained with the RQ-TRAP (Fig. 3). Statistical analysis revealed a highly significant (t-test, P = 0.0007) and strong (R2 = 0.874) correlation between the two assays. Considering the complex multi-step procedure of both assays, these findings indicate a high degree of consistency between the RQ-TRAP and commercially available quantitative TRAP kits.
Figure 3.
Correlation between RQ-TRAP and TRAP-ELISA. Serial dilutions of Hep G2 (G) and HFH-hTERT (F) cell extracts were analyzed with the TeloTAGGG Telomerase PCR ELISAPLUS. The data were compared with results obtained with the RQ-TRAP (table on the left) and graphed to visualize the correlation of the two assays. A linear trendline was added for correlation analysis. NA, no amplification.
The RQ-TRAP assay was utilized to quantitate telomerase activity in human fetal, human pediatric and human adult hepatocyte cultures, as well as in various human hepatoma cell lines. RQ-TRAP results were comparable with measurements obtained with the quantitative TRAP-ELISA (Table 1). Low RQ-TRAP results were validated with the TRAPeze ELISA. Telomerase activity was not detected in the two human hepatocyte samples 13/01 and 26/02 using the quantitative TRAP-ELISA (Table 1). In contrast, all human hepatocyte cultures were telomerase positive by RQ-TRAP and TRAPeze ELISA, although at very low levels.
Table 1. Telomerase activity in human hepatocytes and hepatoma cell lines.
| Cells | TRAP-ELISAa | RQ-TRAP | TRAPezeb |
|---|---|---|---|
| Human hepatocytes | |||
| Fetal-04/00 | 0.3 ± 0.16 (n = 4) | 0.5 ± 0.14 (n = 4) | + (n = 2) |
| Fetal-10/01 | 0.9 ± 0.53 (n = 4) | 1.1 ± 0.30 (n = 4) | + (n = 2) |
| Pediatric-11/01 | 0.2 ± 0.15 (n = 2) | 1.2 ± 0.11 (n = 4) | + (n = 2) |
| Pediatric-27/01 | 0.0 ± 0.01 (n = 2) | 1.8 ± 0.15 (n = 4) | + (n = 2) |
| Pediatric-13/01 | 0.0 ± 0.00 (n = 2) | 1.7 ± 0.15 (n = 2) | + (n = 2) |
| Pediatric-26/02 | 0.0 ± 0.00 (n = 4) | 0.4 ± 0.10 (n = 2) | + (n = 2) |
| Adult-23/00 | 1.0 ± 0.85 (n = 2) | 1.0 ± 0.38 (n = 4) | + (n = 2) |
| Adult-09/01 | 0.2 ± 0.10 (n = 2) | 0.8 ± 0.09 (n = 4) | + (n = 2) |
| Hepatoma cell lines | |||
| Hep G2 | 51.2 ± 3.40 (n = 4) | 57.4 ± 3.63 (n = 8) | ND |
| HLE | 35.5 ± 8.11 (n = 4) | 22.7 ± 1.02 (n = 6) | ND |
| HLF | 40.3 ± 7.57 (n = 4) | 36.8 ± 2.81 (n = 6) | ND |
| HuH 7 | 44.3 ± 6.75 (n = 4) | 37.3 ± 0.43 (n = 6) | ND |
All values are expressed as means ± SEM of the indicated number of measurements. Telomerase activity is tabulated as RTA per 1000 cells. ND, not determined.
aThe quantitative TeloTAGGG Telomerase PCR ELISAPLUS was employed to quantitate telomerase activity.
bLow RQ-TRAP results were confirmed with the qualitative TRAPeze ELISA Telomerase Detection Kit.
The RQ-TRAP assay was also employed to monitor telomerase reactivation following hTERT transduction. Human cells with low endogenous telomerase activity (HFH, HPH, HFP) were transduced with the catalytic telomerase subunit hTERT. Successful transduction was demonstrated by hTERT expression analysis. Transduced cultures exhibited high hTERT copy numbers and showed highly significant increases in telomerase activity (Table 2). In contrast, hTERT expression was not detected in untransduced cultures and in cultures transduced with the control vector without hTERT. Telomerase quantification in untransduced and transduced cultures of the same cell type demonstrated that the RQ-TRAP assay measured telomerase activity and not a cell-type related artifact.
Table 2. Telomerase reconstitution.
| Cells | hTERT copiesa | TRAP-ELISAb | RQ-TRAP |
|---|---|---|---|
| Fetal hepatocytes | |||
| HFH | NA | 0.9 ± 0.53 (n = 4) | 0.5 ± 0.14 (n = 4) |
| HFH-hTERT | 17.1 ± 4.13 (n = 6) | 28.8 ± 3.35 (n = 4)c | 30.3 ± 3.16 (n = 4)c |
| Pediatric hepatocytes | |||
| HPH | NA | 0.0 ± 0.01 (n = 4) | 1.8 ± 0.15 (n = 4) |
| HPH-hTERT | 55.2 ± 17.01 (n = 6) | 26.1 ± 4.07 (n = 4)c | 25.3 ± 6.86 (n = 4)c |
| Fetal pancreatic cells | |||
| HFP | NA | 0.0 ± 0.01 (n = 4) | 1.0 ± 0.34 (n = 4) |
| HFP-hTERT | 11.9 ± 4.84 (n = 2) | 67.7 ± 9.07 (n = 6)c | 52.4 ± 0.99 (n = 6)c |
All values are expressed as means ± SEM of the indicated number of measurements. Telomerase activity is tabulated as RTA per 1000 cells. NA, no amplification.
ahTERT expression was measured by real-time quantitative RT–PCR and copy numbers of hTERT were normalized to PBGD as housekeeping gene.
bThe quantitative TeloTAGGG Telomerase PCR ELISAPLUS was employed to quantitate telomerase activity.
cSignificant difference from untransduced control cultures (t-test, P < 0.05).
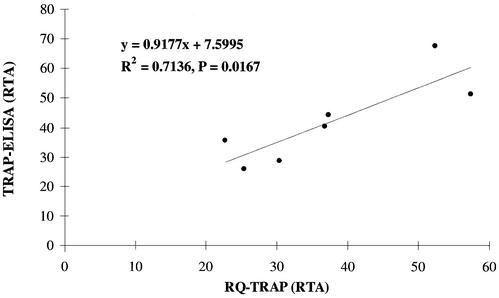
All telomerase activity measurements with the quantitative TRAP-ELISA and the RQ-TRAP obtained for telomerase-positive hepatoma cell lines (Table 1) and telomerase-reconstituted human cells (Table 2) were compared by charting the data pairs in an x–y scatter plot (Fig. 4). Correlation analysis confirmed a significant correlation (t-test, P = 0.0167) between the quantitative TRAP-ELISA and RQ-TRAP assay, with a correlation of 0.7136.
Figure 4.
Comparison of RQ-TRAP and TRAP-ELISA results. RTA measurements with the RQ-TRAP and quantitative TRAP-ELISA for various cell samples with high telomerase activity (n = 7) are graphed in an x–y scatter plot to visualize the correlation of the data obtained with both methods. Correlation analysis results are listed in the graph.
DISCUSSION
The goal of the present study was to quantitate telomerase activity employing real-time PCR in order to eliminate tedious and time-consuming post-synthetic procedures, such as polyacrylamide gel electrophoresis or ELISA, and to modify cycling conditions for improved sensitivity and linearity. Our RQ-TRAP method is based on the TRAP assay introduced by Kim et al. (11), and utilizes the refined anchored reverse primer ACX to reduce primer-dimer formation and to prevent 3′ elongation of telomerase products (23). Thermal cycling conditions (primer concentrations, annealing temperature, template amount) were optimized to ensure highly specific and linear amplification of telomerase products. The data demonstrates that our RQ-TRAP assay permits the rapid and reliable quantification of telomerase activity in cell culture samples without primer-dimer interference. The real-time, closed-tube protocol obviates the need for post-PCR steps, reduces the danger of carryover contamination, and permits sensitive and linear telomerase detection down to single-cell samples. In comparison, the observed linear range of telomerase activity for the TRAP-ELISA assays was from 250 to 5000 cells (17).
Our RQ-TRAP assay is highly linear, as shown for three different cell lines. RQ-TRAP inter-assay variation with a CV of 12% for 1000 analyzed cells was identical to the precision reported for other TRAP assays, including TRAP-ELISA kits (13,17). Moreover, this variation is consistent with the complex telomerase incubation and amplification procedure. Quantitative data generated with the RQ-TRAP for serial dilutions of a telomerase-positive hepatoma cell line (Hep G2) and telomerase reconstituted primary human hepatocytes (HFH-hTERT) correlated significantly with measurements obtained with the commercially available TeloTAGGG Telomerase PCR ELISAPLUS. Considering that the quantitative TRAP-ELISA utilizes a single telomeric repeat control for quantification, whereas telomerase activity measurements with the RQ-TRAP are based on serial dilutions of 293T cell extracts, it was not surprising that the degree of correlation between the two assays was lower (R2 = 0.7136) for samples with high telomerase activity (hepatoma cell lines and telomerase-reconstituted cells) compared with serial dilutions of cell extracts (R2 = 0.874). This lower but significant correlation for the high telomerase activity range might be explained by the large standard errors of the quantitative TRAP-ELISA for samples with high telomerase activity (see Table 1), which signifies the inherent problems of end-point PCR for accurate and reliable quantification of samples with high template concentrations. We believe that our RQ-TRAP assay, which surmounts the problems of end-point PCR by employing real-time quantitative PCR technology, generates more accurate and reproducible, and thus more reliable, telomerase activity data.
The RQ-TRAP assay takes less than 3 h, including sample extraction, with less than 20 min of hands-on time; a considerable improvement compared with TRAP-ELISA assays and other TRAP modifications. This makes the RQ-TRAP simpler and faster, allowing higher throughput and more convenient data generation. Thus, the assay will be beneficial in screening and monitoring cell cultures for telomerase activity, e.g. in immortalization experiments or stem cell differentiation projects.
Finally, using our RQ-TRAP assay, low telomerase activity levels were detected in two human hepatocyte samples that were evaluated as telomerase negative by the quantitative TRAP-ELISA. The presence of telomerase activity in these two samples was confirmed with the very sensitive but qualitative TRAPeze ELISA hit. Therefore, our findings also illustrate the increased sensitivity and reliability of the optimized RQ-TRAP method.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
The authors would like to thank Dr R. A. Weinberg, Whitehead Institute of Biomedical Research, Cambridge, Massachusetts, for supplying plasmids and cells, and Dr S. Gupta, Marion Bessin Liver Research Center, Albert Einstein College of Medicine, Bronx, New York, and F. Wu, University of California, Davis Medical Center, Sacramento, California, for providing primary human cell cultures. The study was supported by grants from the Studienstiftung des deutschen Volkes, Germany (to H.W.), the American Liver Foundation (to H.W.), the Alpha One Foundation (to M.Z.), and in part by Transplant Hope, University of California, Davis Medical Center (to M.Z.).
REFERENCES
- 1.Collins K. (2000) Mammalian telomeres and telomerase. Curr. Opin. Cell Biol., 12, 378–383. [DOI] [PubMed] [Google Scholar]
- 2.Allsopp R.C., Vaziri,H., Patterson,C., Goldstein,S., Younglai,E.V., Futcher,A.B., Greider,C.W. and Harley,C.B. (1992) Telomere length predicts replicative capacity of human fibroblasts. Proc. Natl Acad. Sci. USA, 89, 10114–10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campisi J. (1997) The biology of replicative senescence. Eur. J. Cancer, 33, 703–709. [DOI] [PubMed] [Google Scholar]
- 4.Greider C.W. and Blackburn,E.H. (1985) Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell, 43, 405–413. [DOI] [PubMed] [Google Scholar]
- 5.Shay J.W. and Bacchetti,S. (1997) A survey of telomerase activity in human cancer. Eur. J. Cancer, 33, 787–791. [DOI] [PubMed] [Google Scholar]
- 6.Davis A.J. and Siu,L.L. (2000) Telomerase: therapeutic potential in cancer. Cancer Invest., 18, 269–277. [DOI] [PubMed] [Google Scholar]
- 7.Bodnar A.G., Ouellette,M., Frolkis,M., Holt,S.E., Chiu,C.P., Morin,G.B., Harley,C.B., Shay,J.W., Lichtsteiner,S. and Wright,W.E. (1998) Extension of life-span by introduction of telomerase into normal human cells. Science, 279, 349–352. [DOI] [PubMed] [Google Scholar]
- 8.Wang J., Xie,L.Y., Allan,S., Beach,D. and Hannon,G.J. (1998) Myc activates telomerase. Genes Dev., 12, 1769–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dickson M.A., Hahn,W.C., Ino,Y., Ronfard,V., Wu,J.Y., Weinberg,R.A., Louis,D.N., Li,F.P. and Rheinwald,J.G. (2000) Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol. Cell. Biol., 20, 1436–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayflick L. and Moorhead,P.S. (1961) The serial cultivation of human diploid cell strains. Exp. Cell Res., 25, 585–621. [DOI] [PubMed] [Google Scholar]
- 11.Kim N.W., Piatyszek,M.A., Prowse,K.R., Harley,C.B., West,M.D., Ho,P.L., Coviello,G.M., Wright,W.E., Weinrich,S.L. and Shay,J.W. (1994) Specific association of human telomerase activity with immortal cells and cancer. Science, 266, 2011–2015. [DOI] [PubMed] [Google Scholar]
- 12.Hirose M., Abe-Hashimoto,J., Ogura,K., Tahara,H., Ide,T. and Yoshimura,T. (1997) A rapid, useful and quantitative method to measure telomerase activity by hybridization protection assay connected with a telomeric repeat amplification protocol. J. Cancer Res. Clin. Oncol., 123, 337–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gelmini S., Caldini,A., Becherini,L., Capaccioli,S., Pazzagli,M. and Orlando,C. (1998) Rapid, quantitative nonisotopic assay for telomerase activity in human tumors. Clin. Chem., 44, 2133–2138. [PubMed] [Google Scholar]
- 14.Uehara H., Nardone,G., Nazarenko,I. and Hohman,R.J. (1999) Detection of telomerase activity utilizing energy transfer primers: comparison with gel- and ELISA-based detection. Biotechniques, 26, 552–558. [DOI] [PubMed] [Google Scholar]
- 15.Szatmari I., Tokes,S., Dunn,C.B., Bardos,T.J. and Aradi,J. (2000) Modified telomeric repeat amplification protocol: a quantitative radioactive assay for telomerase without using electrophoresis. Anal. Biochem., 282, 80–88. [DOI] [PubMed] [Google Scholar]
- 16.Xu S., He,M., Yu,H., Cai,X., Tan,X., Lu,B. and Shu,B. (2001) A quantitative method to measure telomerase activity by bioluminescence connected with telomeric repeat amplification protocol. Anal. Biochem., 299, 188–193. [DOI] [PubMed] [Google Scholar]
- 17.Wu Y.Y., Hruszkewycz,A.M., Delgado,R.M., Yang,A., Vortmeyer,A.O., Moon,Y.W., Weil,R.J., Zhuang,Z. and Remaley,A.T. (2000) Limitations on the quantitative determination of telomerase activity by the electrophoretic and ELISA based TRAP assays. Clin. Chim. Acta, 293, 199–212. [DOI] [PubMed] [Google Scholar]
- 18.Francis R. and Friedman,S.H. (2002) A rapid direct telomerase assay method using 96-well streptavidin plates. Biotechniques, 32, 1154–1156, 1158,, 1160. [DOI] [PubMed] [Google Scholar]
- 19.Sun D., Hurley,L.H. and Von Hoff,D.D. (1998) Telomerase assay using biotinylated-primer extension and magnetic separation of the products. Biotechniques, 25, 1046–1051. [DOI] [PubMed] [Google Scholar]
- 20.Hou M., Xu,D., Bjorkholm,M. and Gruber,A. (2001) Real-time quantitative telomeric repeat amplification protocol assay for the detection of telomerase activity. Clin. Chem., 47, 519–524. [PubMed] [Google Scholar]
- 21.Counter C.M., Hahn,W.C., Wei,W., Caddle,S.D., Beijersbergen,R.L., Lansdorp,P.M., Sedivy,J.M. and Weinberg,R.A. (1998) Dissociation among in vitro telomerase activity, telomere maintenance and cellular immortalization. Proc. Natl Acad. Sci. USA, 95, 14723–14728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malhi H., Irani,A.N., Gagandeep,S. and Gupta,S. (2002) Isolation of human progenitor liver epithelial cells with extensive replication capacity and differentiation into mature hepatocytes. J. Cell Sci., 115, 2679–2688. [DOI] [PubMed] [Google Scholar]
- 23.Kim N.W. and Wu,F. (1997) Advances in quantification and characterization of telomerase activity by the telomeric repeat amplification protocol (TRAP). Nucleic Acids Res., 25, 2595–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.