Abstract
Recombinant baculoviruses have established themselves as a favoured technology for the high-level expression of recombinant proteins. The construction of recombinant viruses, however, is a time consuming step that restricts consideration of the technology for high throughput developments. Here we use a targeted gene knockout technology to inactivate an essential viral gene that lies adjacent to the locus used for recombination. Viral DNA prepared from the knockout fails to initiate an infection unless rescued by recombination with a baculovirus transfer vector. Modified viral DNA allows 100% recombinant virus formation, obviates the need for further virus purification and offers an efficient means of mass parallel recombinant formation.
INTRODUCTION
Baculoviruses have been exploited as expression vectors since 1983 (1). A highly expressed but redundant virus very late gene is replaced by a gene of interest to produce a recombinant virus that expresses a recombinant protein as part of the virus life cycle (2). Baculoviruses are robust, safe and have a good record of high-level expression, particularly of complex proteins [e.g. (3)]. The insect cell background carries out most of the post-translational modifications found in mammalian cells (4) and a wide variety of vectors and cloning strategies are available (5). These qualities have suggested recombinant baculoviruses as a favoured technology for the delivery of high throughput protein expression solutions aimed at whole genome coverage (6), particularly for the highly processed proteins whose expression in Escherichia coli is challenging. However, the formation of the recombinant virus is a significant bottleneck in the process. Recombinant formation is frequently done via genetic exchange in vivo followed by selection based on an altered phenotype (2) although other systems such as direct cloning into the viral genome (7), recombination in yeast (8) and transposition in E.coli (9) have also been used. Historically, the most direct technology, recombination within insect cells, has always led to a background of wild type virus in addition to the recombinant and this has necessitated multiple virus isolation steps to avoid eventual outgrowth by the wild type. This has been partly circumvented by the use of linearised viral DNA which cannot initiate a viral infection unless rescued by the recombination event (10). However, even with the use of linear DNA, background virus is a possibility and plaque assays to ensure 100% recombinant are advised. Clearly, if recombinant baculoviruses are to be a key technology in the generation of many thousands of recombinants, the generation of background virus, however low in frequency, needs to be avoided.
Here, using a recombinational cloning methodology that uses RecE and RecT gene products, ‘ET cloning’ (11), we describe a viral gene knockout that renders the virus non-viable unless rescued by recombinant formation. Recombin ation protocols are both shortened and made 100% efficient by this modification allowing true ‘walk away’ recombinant formation and the purposeful inclusion of mass baculovirus expression to high throughput protein expression strategies.
RESULTS AND DISCUSSION
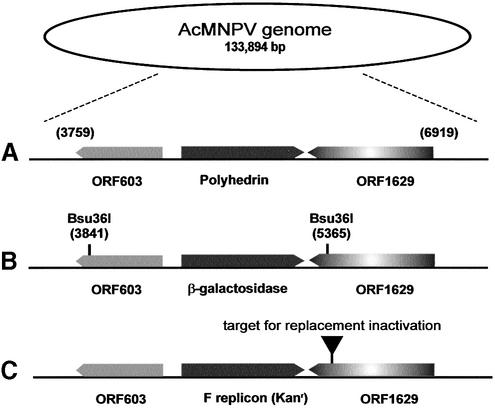
In principle there are three methods for the insertion of foreign genes into baculoviruses, recombination within insect cells, recombination in E.coli (or yeast) and ligation in vitro. Recombinant baculovirus formation requiring integration of a coding region into the genome before virus production [e.g. using site-specific transposition in E.coli (9,12)] may require multiple cloning steps for each separate coding region and for some purposes, such as the creation of expression libraries, this is inherently difficult. For this reason we adopted the traditional recombination process in insect cells as the basis for the modification in recombination in our experiments. In this technology, viral DNA is used in a co-transfection experiment with a transfer vector which encodes the desired coding region, flanked by the baculovirus late transcription signals and sufficient baculovirus DNA to allow efficient recombination with the viral genome (2,12). Although recombination can occur at several loci in the viral genome, such as the p10 site (13), the locus of recombination used in most experiments is the polyhedrin gene which maps to 4520–5258 on the 133 894 bp complete genome (Fig. 1A). Kitts and Possee (10) described the introduction of point mutations into the wild type baculovirus genome that generated novel sites for the infrequent cutting restriction enzyme Bsu36I at positions 3841 and 5365. These sites flank the polyhedrin locus in the upstream and downstream genes ORF603 (3759–4365) and ORF1629 (5287–6919), respectively (Fig. 1B). Cleavage with Bsu36I necessarily truncates both ORFs and, as ORF1629 encodes an essential gene product involved either in nucleocapsid packaging (14,15) or modification of the virion RNA polymerase (16), a viable genome can only be formed if the truncation is bridged and repaired by recombination with a suitable transfer vector or by a sufficiently long PCR product (2,12,17). Background wild type virus results either from incompletely digested viral genome present in the transfecting DNA or by the re-ligation in vivo of the two Bsu36I fragments removed by restriction. As the principle of inactivation of ORF1629 is a highly effective selection against background virus we sought to mimic the effect of the restriction site introduction described by genetic inactivation of ORF1629 (Fig. 1C).
Figure 1.
Comparative maps of the baculovirus polyhedrin locus. (A) The polyhedrin locus and surrounding genes on the wild type AcMNPV (accession no. L22858) genome. (B) The intermediate viral derivative BacPak6 showing the position of Bsu36I sites introduced to allow genome linearisation prior to transfection (10). Note, Bsu36I sites present in the β-galactosidase gene are not shown. (C) The AcMNPV bacmid used for insertional mutagenesis of ORF1629.
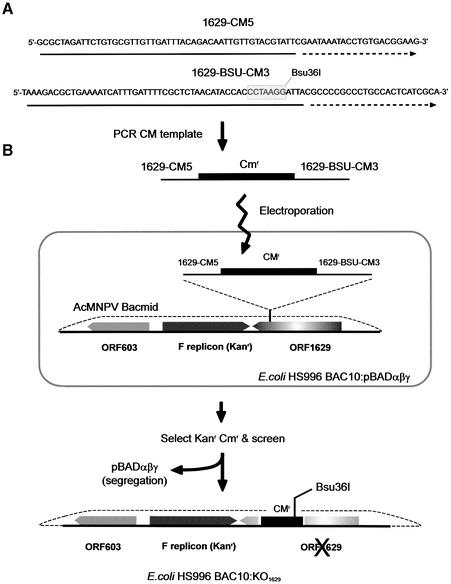
As ORF1629 interruption in vivo would render the virus non-viable, genetic knockout was done in a bacmid version of the baculovirus genome held in E.coli. To do this we established ET cloning (11) for the baculovirus genome. Escherichia coli:BAC10, containing the infectious baculovirus genome in an F factor based bacmid replicon, was sourced and transformed into the ET host HS996 (selecting KanR). Subsequently, an ET competent strain was generated by transformation of HS996:BAC10 by pBADαβγ (selecting ApR) to produce the ET recipient strain HS996:BAC10: pBADαβγ (ApR KanR). To provide the marker for gene knockout, the chloramphenicol acetyl transferase gene was amplified to include ∼50 bp 5′ extensions homologous to the baculovirus genome at two adjacent sites within ORF1629 (Fig. 2). Although adjacent, the baculovirus sequences present on oligonucleotides did not adjoin so that inactivation of ORF1629 could not revert by fragment excision. In addition, a single Bsu36I site was introduced to allow linearisation of the genome as linear DNA is more efficient than circular for recombination (10). The amplified fragment was transformed into HS996:BAC10:pBADαβγ, selection made for chloramphenicol resistance and colonies screened by amplification of the ORF1629 locus. A strain of HS996:BAC10 (CmR KanR), with the correct genetic profile, was termed BAC10:KO1629 and prepared by large-scale bacmid protocols and cleaved by Bsu36I for use in transfection. The entire knockout procedure is summarised (Fig. 2).
Figure 2.
Procedure for knockout of ORF1629. (A) The oligonucleotides used for knockout with salient features indicated. Solid underlining indicates sequence homology of oligonucleotides 1629 CM5 and 1629 BSU CM3 to AcMNPV nucleotides 5291–5339 and 5359–5413, respectively. Dotted underlining indicates homology to chloramphenicol acetyl transferase. The introduced Bsu36I site is indicated. (B) Summary of the steps involved in the ET cloning strategy resulting in creation of Bac10:KO1629.
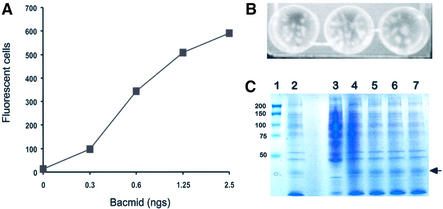
To assess BAC10:KO1629 as a substrate for recombination with a transfer vector and the subsequent generation of recombinant viruses, Spodoptera frugiperda (Sf9) cells were transfected with mixtures of linearised BAC10:KO1629 DNA and pVSVGTMGFP, a transfer vector encoding GFP (18) following a standard transfection protocol using lipofectamine (19). Cells were harvested at 2 days post transfection and analysed by fluorescence-activated cell scan (FACS). At this time, GFP fluorescence is a measure of primary transfectants in which transfer vector and bacmid have successfully recombined but have not yet initiated secondary virus amplification. A linear dose response curve of fluorescence with DNA concentration was obtained for linearised bacmid over the DNA concentration range used (up to ∼5 ng of DNA per 106 cells giving ∼15% of total cells transfected) for the transfection conditions in use (Fig. 3A). In the absence of transfer vector, saturating amounts of bacmid DNA, either linearised or supercoiled, failed to generate infectious virus on repeated transfection. Virus present in the supernatant of productive transfections was plaque assayed and individual plaque examined for fluorescence. All plaques showed fluorescence (n > 1000 of which ∼90 individual plaques are shown in Fig. 3B) and infected cells within a plaque exhibited fluorescence in the Golgi and on the cell surface typical of the secreted and anchored form of GFP encoded by the transfer vector used (18). From these data we conclude that the gene knockout of ORF1629 in bacmid BAC10:KO1629 is genetically stable and is unable to initiate virus infection unless rescued by recombination with an appropriate transfer vector. In practice, this means that BAC10:KO1629 viral DNA produces 100% recombinant virus following transfection. Although the formation of the recombinant virus uses modified viral DNA, the eventual genetic structure of the virus is the same as that produced by traditional methods. As expected therefore, examination of the expression level of several viruses expressing GFP generated using BAC10: KO1629 showed no change when compared with viruses produced by alternate methodologies (Fig. 3C). In addition, no instability or reduction in virus titre has been observed with any recombinant produced by transfection using BAC10:KO1629.
Figure 3.
Transfection dose response curve and characterisation of recombinants. (A) Bac10:KO1629 preparations were quantitated by OD280 and introduced into Sf9 cells with saturating amounts (500 ng) of transfer vector using lipofectamine. Cells were harvested at 2 days post infection, resuspended in Facsflow (Beckton Dickenson) and the number of fluorescent cells determined. Data are number of GFP positive cells per 5000. Transfection with vector only gave no fluorescence and transfection by Bac10:KO1629 only gave no virus recovery. (B) Plaque assay of progeny recombinant baculoviruses expressing GFP. Three dishes with well isolated plaques were photographed following illumination by UV light transmitted by a UV/blue plate converter (UVP, Upland CA) and photographed through an orange filter. Examination of multiple dishes failed to reveal any non-fluorescent plaques. (C) GFP (arrowed) expression levels of four recombinant viruses (lanes 4–7) produced using modified bacmid DNA were compared with GFP expressed by a recombinant made using non-modified viral DNA (lane 2). Marker proteins with the molecular mass (in kDa) shown are in lane 1 and a non-infected cell extract in lane 3. No reduction in expression level was apparent between lanes 4–7 and lane 2.
Since its inception in 1983 (1), the formation of recombinant baculoviruses has improved incrementally from the original technology, where loss of the polyhedron inclusion bodies was the sought phenotype, through the use of more easily detected markers such as β-galactosidase (20,21), to systems with inherently low backgrounds (10) or those which pre-incorporate the gene to be expressed into the baculovirus genome before virus formation (7,9). The former technologies all rely on individual virus isolation, usually through plaque assay, before expansion of the recombinant virus. Although limited work can be done without plaque assay, even low level contamination with wild type virus will result in outgrowth over a period of time with an apparent and concomitant lowering of expression yield (22). In the latter technologies, which rely on site-specific recombination in E.coli, only recombinant genomes are produced for transfection. However, a separate transposition event is required for the creation of each recombinant viral genome making work with large numbers, such as libraries, problematic. In the modification described here, we sought to retain the original technology (recombination in insect cells) but to modify the viral genome such that background virus was impossible. The necessity of recombinant virus isolation following transfection would then be eliminated. To do this, ORF1629 adjacent to the polyhedrin locus was inactivated by site-specific insertion into a bacmid version of the genome [see also (23,24)]. The bacmid source of the genome also allows large amounts of viral DNA for transfection to be produced easily. The knockout strategy included the elimination of reversion and the provision of linear viral DNA, preferable for transfection purposes.
Renewed interest in mass protein expression has followed developments in structural genomics [for overviews see (25,26)]. Systems for such applications need to be simple to operate, robust and productive. Expression in E.coli fulfils these requirements and, for ORFs of microbial origin or for ORFs that encode relatively small and simple (not glycosylated, not heavily disulphide bonded) proteins of eukaryotic origin this has proved efficient [e.g. (27)]. Similarly, in vitro systems have also been adapted to allow mass multiple protein expression (28). However, for large complex proteins, particularly glycoproteins, or for proteins whose activity depends on secondary modification, expression in a higher eukaryotic cell is desirable (29). Recombinant baculoviruses have the potential to fulfil this ‘second tier’ expression requirement as they are robust, high yielding and carry out the majority of protein secondary modifications that have been examined (2,4). A major practical restriction however has been the necessity of virology technique, particularly plaque assays. The modification described here removes this requirement as, with recombinants formed using the modified bacmid described, no background virus can be produced. Indeed, as both bacmid and transfer DNA can be prepared easily in quantity, large volumes of recombinant baculovirus and/or infected cells can be achieved directly without any isolation of the virus through large-scale DNA transfection as has been described for mammalian cells (30).
Acknowledgments
ACKNOWLEDGEMENTS
We thank R. D. Possee, Oxford, for helpful discussion. The work was supported by the Biotechnology and Biological Sciences Research Council, UK.
REFERENCES
- 1.Smith G.E., Summers,M.D. and Fraser,M.J. (1983) Production of human beta interferon in insect cells infected with a baculovirus expression vector. Mol. Cell. Biol., 3, 2156–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Reilly D.R., Miller,L.K. and Luckow,V.A. (1992) Baculovirus Expression Vectors: a Laboratory Manual. W.H.Freeman and Company, New York.
- 3.Stern L.J. and Wiley,D.C. (1992) The human class II MHC protein HLA-DR1 assembles as empty alpha beta heterodimers in the absence of antigenic peptide. Cell, 68, 465–477. [DOI] [PubMed] [Google Scholar]
- 4.Luckow V.A. and Summers,M.D. (1988) Trends in the development of baculovirus expression vectors. Biotechnology, 6, 47–55. [Google Scholar]
- 5.Jones I. and Morikawa,Y. (1996) Baculovirus vectors for expression in insect cells. Curr. Opin. Biotechnol., 7, 512–516. [DOI] [PubMed] [Google Scholar]
- 6.Albala J.S., Franke,K., McConnell,I.R., Pak,K.L., Folta,P.A., Rubinfeld,B., Davies,A.H., Lennon,G.G. and Clark,R. (2000) From genes to proteins: high-throughput expression and purification of the human proteome. J. Cell. Biochem., 80, 187–191. [DOI] [PubMed] [Google Scholar]
- 7.Ernst W.J., Grabherr,R.M. and Katinger,H.W. (1994) Direct cloning into the Autographa californica nuclear polyhedrosis virus for generation of recombinant baculoviruses. Nucleic Acids Res., 22, 2855–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel G., Nasmyth,K. and Jones,N. (1992) A new method for the isolation of recombinant baculovirus. Nucleic Acids Res., 20, 97–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luckow V.A., Lee,S.C., Barry,G.F. and Olins,P.O. (1993) Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J. Virol., 67, 4566–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kitts P.A. and Possee,R.D. (1993) A method for producing recombinant baculovirus expression vectors at high frequency. Biotechniques, 14, 810–817. [PubMed] [Google Scholar]
- 11.Muyrers J.P., Zhang,Y., Testa,G. and Stewart,A.F. (1999) Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res., 27, 1555–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patil M., Chan-Fook,C., Jones,I.M. and Chapple,S.D. (2001) Generation of baculovirus recombinants using PCR-amplified fragments. Biotechniques, 30, 1212–1214, 1216. [DOI] [PubMed] [Google Scholar]
- 13.Martens J.W., van Oers,M.M., van de Bilt,B.D., Oudshoorn,P. and Vlak,J.M. (1995) Development of a baculovirus vector that facilitates the generation of p10-based recombinants. J. Virol. Methods, 52, 15–19. [DOI] [PubMed] [Google Scholar]
- 14.Pham D.Q., Hice,R.H., Sivasubramanian,N. and Federici,B.A. (1993) The 1629-bp open reading frame of the Autographa californica multinucleocapsid nuclear polyhedrosis virus encodes a virion structural protein. Gene, 137, 275–280. [DOI] [PubMed] [Google Scholar]
- 15.Vialard J.E. and Richardson,C.D. (1993) The 1,629-nucleotide open reading frame located downstream of the Autographa californica nuclear polyhedrosis virus polyhedrin gene encodes a nucleocapsid-associated phosphoprotein. J. Virol., 67, 5859–5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iorio C., Vialard,J.E., McCracken,S., Lagace,M. and Richardson,C.D. (1998) The late expression factors 8 and 9 and possibly the phosphoprotein p78/83 of Autographa californica multicapsid nucleopolyhedrovirus are components of the virus-induced RNA polymerase. Intervirology, 41, 35–46. [DOI] [PubMed] [Google Scholar]
- 17.Monsma S.A., Oomens,A.G. and Blissard,G.W. (1996) The GP64 envelope fusion protein is an essential baculovirus protein required for cell-to-cell transmission of infection. J. Virol., 70, 4607–4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chapple S.D. and Jones,I.M. (2002) Non-polar distribution of green fluorescent protein on the surface of Autographa californica nucleopolyhedrovirus using a heterologous membrane anchor. J. Biotechnol., 95, 269–275. [DOI] [PubMed] [Google Scholar]
- 19.Keith M.B., Farrell,P.J., Iatrou,K. and Behie,L.A. (2000) Use of flow cytometry to rapidly optimize the transfection of animal cells. Biotechniques, 28, 148–154. [DOI] [PubMed] [Google Scholar]
- 20.Lalumiere M. and Richardson,C.D. (1995) Production of recombinant baculoviruses using rapid screening vectors that contain the gene for beta-galactosidase. Methods Mol. Biol., 39, Human Press, 161–177. [DOI] [PubMed] [Google Scholar]
- 21.Richardson C.D. (ed.) (1995) Baculovirus expression protocols. Methods in Molecular Biology, vol 39, Humana Press, Totowa, NJ.
- 22.van Lier F.L., van Duijnhoven,G.C., de Vaan,M.M., Vlak,J.M. and Tramper,J. (1994) Continuous beta-galactosidase production in insect cells with a p10 gene based baculovirus vector in a two-stage bioreactor system. Biotechnol. Prog., 10, 60–64. [DOI] [PubMed] [Google Scholar]
- 23.Hou S., Chen,X., Wang,H., Tao,M. and Hu,Z. (2002) Efficient method to generate homologous recombinant baculovirus genomes in E.coli. Biotechniques, 32, 783–784, 786,788. [DOI] [PubMed] [Google Scholar]
- 24.Lin G. and Blissard,G.W. (2002) Analysis of an Autographa californica nucleopolyhedrovirus lef-11 knockout: LEF-11 is essential for viral DNA replication. J. Virol., 76, 2770–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campbell I.D. (2002) Timeline: The march of structural biology. Nature Rev. Mol. Cell Biol., 3, 377–381. [DOI] [PubMed] [Google Scholar]
- 26.Stevens R.C., Yokoyama,S. and Wilson,I.A. (2001) Global efforts in structural genomics. Science, 294, 89–92. [DOI] [PubMed] [Google Scholar]
- 27.Buchanan S.G., Sauder,J.M. and Harris,T. (2002) The promise of structural genomics in the discovery of new antimicrobial agents. Curr. Pharm. Des., 8, 1173–1188. [DOI] [PubMed] [Google Scholar]
- 28.Rungpragayphan S., Kawarasaki,Y., Imaeda,T., Kohda,K., Nakano,H. and Yamane,T. (2002) High-throughput, cloning-independent protein library construction by combining single-molecule DNA amplification with in vitro expression. J. Mol. Biol., 318, 395–405. [DOI] [PubMed] [Google Scholar]
- 29.Gilbert M. and Albala,J.S. (2002) Accelerating code to function: sizing up the protein production line. Curr. Opin. Chem. Biol., 6, 102–105. [DOI] [PubMed] [Google Scholar]
- 30.Wurm F. and Bernard,A. (1999) Large-scale transient expression in mammalian cells for recombinant protein production. Curr. Opin. Biotechnol., 10, 156–159. [DOI] [PubMed] [Google Scholar]