Abstract
Complete genomes of three closely related Gram-positive bacteria Streptococcus pyogenes, Strepto coccus pneumoniae and Lactococcus lactis are analyzed for abundances of short DNA sequence motifs (frequent words). The character and extent of frequent words are strikingly different among these genomes. The frequent words of S.pneumoniae split into three categories: parts of the previously characterized RUP and BOX repetitive elements and a 24 bp tandem repeat in the gene SP1772. The most abundant frequent words of L.lactis are all related to the 13 bp motif, WWNTTACTGACRR or its inverted complement YYGTCAGTAANWW. Distributional analysis of this motif, which we called highly repetitive motif (HRM), indicates its possible dual role. Frequent occurrences immediately downstream of genes suggest a possible role in transcription termination whereas spacings of consecutive HRMs consistent with the DNA helical period are indicative of a protein-binding site. Two regions of the L.lactis genome feature an intriguing pattern of several periodically occurring HRMs separated by precisely 59 bp. In a striking contrast to S.pneumoniae and L.lactis, S.pyogenes contains hardly any frequent words.
INTRODUCTION
Streptococci are low G+C, Gram-positive, non-motile bacteria. Some species cause disease in humans and animals while lactic streptococci are non-pathogenic and important in the dairy industry. The genomes of three different Streptococcus species were recently completely sequenced. Streptococcus pyogenes (1) is responsible for a wide variety of diseases in humans including scarlet fever, toxic shock syndrome and rheumatic fever. Streptococcus pneumoniae causes pneumonia, bacteremia and meningitis (2). In contrast, Lactococcus lactis is a non-pathogenic bacterium that lives on plants or in animals in nature and its cultures are employed in milk fermentation (3). All three genomes are roughly congruent in genome size (S.pyogenes 1.85 Mb, S.pneumoniae 2.16 Mb and L.lactis 2.37 Mb), and similar in genomic G+C content (38.5, 39.7 and 35.3%, respectively) and in genome signature (dinucleotide relative abundances) (4).
DNA repeats can be classified in three types. (i) Simple sequence repeats (SSRs), or microsatellites, composed of extensive tandem iterations of a short oligonucleotide. Some SSRs promote formation of non-canonical DNA structures (5,6) which can play regulatory roles in gene expression (7). In some pathogenic bacteria, SSRs induce mutations that can counter host defense mechanisms (8). (ii) The second class of repeats emphasizes frequent dispersed motifs typically up to ∼20 bp length. Such repeats are often associated with regulatory or structural elements. For example, uptake signal sequences are pronounced in Haemophilus influenzae (9,10), chi sites (which promote recombination in association with the RecBCD complex) and REP elements (repeated extragenic palindrome of unknown function) are highly abundant in Escherichia coli and Salmonella typhimurium (11–13), and highly iterated palindrome (HIP) sequences are very frequent and rather evenly dispersed in the Synechocystis genome (10,14,15). (iii) The third class features long repeats (typically hundreds of base pairs in length) of lower copy number which are unlikely to occur by chance. Repeats of this class often contain insertions and/or deletions. In bacteria, these repeats often consist of various kinds of mobile elements and families of duplicated genes. Ohno (16) has proposed that genomes have been amplified by extensive duplications of genes that subsequently diverge. In other cases, genomes can be reduced in size from their progenitors due to loss of genes as in Mycoplasma and Rickettsia genomes.
In this paper, we focus on the second class of repeats—highly repetitive oligonucleotide motifs of intermediate length. We analyze the genomes of S.pyogenes, S.pneumoniae and L.lactis for a proliferation of repetitive DNA sequence motifs (frequent words). Appropriate statistics for detecting frequent words in genomes with biased G+C content were proffered in Karlin et al. (10). The most frequent words in the S.pneumoniae genome are parts of previously characterized BOX and RUP (Repeat Unit of Pneumococcus) repeat families. The BOX elements, of unknown function and typically of 100–200 bp length, tend to occur in intergenic regions (17). The RUP repeats, generally 107 bp in length, are related to IS elements (18). In contrast, S.pyogenes lacks abundant frequent words. Most S.pyogenes frequent words occur <20 times and are parts of tRNA genes or IS elements. The L.lactis genome contains nearly 1000 iterations of a 13 bp highly repetitive motif (HRM). This motif differs from chi sites identified in L.lactis as the 7 bp motif GCGCGTG (19). The distribution of the HRMs relative to genes and spacings between consecutive HRMs are analyzed in this paper. We speculate that this motif serves a dual role as a transcription terminator and as a protein-binding site.
MATERIALS AND METHODS
DNA sequences
Annotated DNA sequences of complete genomes of L.lactis (3), S.pyogenes (1) and S.pneumoniae (2) where obtained from GenBank.
Frequent words (oligonucleotides)
We apply Poisson distribution approximations associated with generalized occupancy problems of balls-in-urns [see Karlin and Leung (20) for mathematical details]. For a sequence S of length L, there is a natural word size s determined by the inequalities:
As–1 ≤ L < As1
where A is the alphabet size (A = 4 for DNA, A = 20 for proteins). For the complete genomes of L.lactis, S.pneumoniae and S.pyogenes, the appropriate word size according to equation 1 is s = 11 bp. For each word w, the copy threshold rw is determined as the least integer satisfying the inequality:
exp(–pwL)(pwL)rw / rw! ≤ 1 / L2
where the word size s is the parameter of formula 1 and pw = fi1fi1i2...fis–1is with fi the frequency of nucleotide i in S and fij the transition frequency in S from nucleotide i to nucleotide j. A general feature of this formulation is that the lower the expected frequency of a word, the lower the cut-off required for it to be frequent. By equating the left side of the inequality 2 to 1 / L, at most one frequent word is expected in a random sequence (10).
Analysis of the distribution of frequent words
In probing the organization of a genome, the general problem arises of how to characterize anomalies in the spacings of markers in a long sequence of nucleotides or amino acids. These include properties of clumping (too many neighboring short spacings), overdispersion (too many long gaps between markers) and excessive regularity (too few short spacings and/or too few long gaps). Questions concerning the distribution (spacings) of a marker array can be approached by consideration of the cumulative lengths of r consecutive distances along the marker array where R(r)i is the distance (number of letters) between marker i and marker i + r designated r-scan lengths (10). The spans of the longest and shortest r-scans are useful statistics for detecting significant clumping, significant overdispersion or excessive regularity in the spacings of the marker. We compare the distribution of {R(r)i} calculated under a random model with the observed r-scan lengths. Let m*r = mini R(r)i, M*r = maxi R(r)i.
The theoretical probabilities for a randomly distributed marker array of n points obey the asymptotic relations (n → ∞):
Prob{m*r ≥ x / n(1 + 1 / r)} ≈ exp(–xr / r!)
Prob{M*r ≤ n–1[ln n + (r – 1)ln(ln n)]} ≈ exp[–e–x / (r – 1)!]
These formulas provide benchmarks as to whether the minimum and/or maximum spacing deviates significantly from randomness.
RESULTS
Lactococcus lactis
Frequent words. In the L.lactis genome, 4697 different oligonucleotides of length 11 bp qualify as frequent words by the criteria of formulas 1 and 2. The most over-represented frequent words match the consensus sequence WWNTTACTGACRR or its inverted complement YYGTCAGTAANWW [W = weak (A or T), R = purine, Y = pyrimidine and N = any base], labeled W and W′, respectively. Occurrences of W and W′ in the genome relative to genes and intergenic regions are summarized in Table 1. W and W′ occur 916 times in the genome and only 321 (35%) overlap genes. Considering that genes occupy 85% of the L.lactis DNA, this shows a strong tendency of W and W′ to occur in intergenic regions. Allowing for one error (a single nucleotide mismatch with the consensus sequence), W and W′ occur 2094 times and 1129 (54%) in genes. We call the W and W′ words the HRM.
Table 1. Counts of the words W and W′ in the L.lactis genome in genes and intergenic regions.
| Word | Count in genes | Count in intergenic regions |
|---|---|---|
| W and W′ | 321 | 595 |
| W and W′ allowing for one error | 1129 | 965 |
| Close dyad WW′ | 13 | 98 |
| Close dyad W′W | 5 | 33 |
13mers matching the consensus sequence WWNTTACTGACRR and its inverted complement YYGTCAGTAANWW are designated W and W′, respectively. Close dyads are defined as W followed by W′, or W′ followed by W, separated by ≤20 bp.
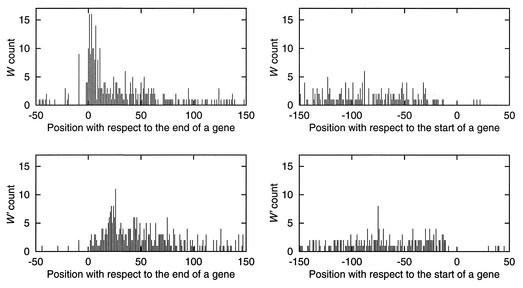
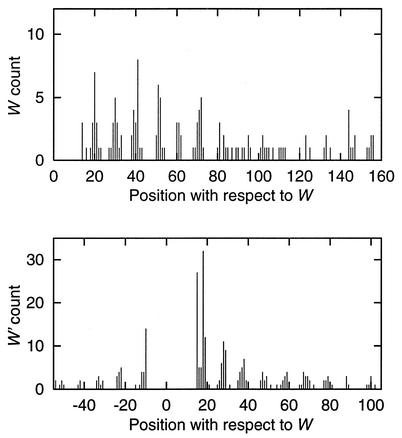
Distribution of HRMs. Figure 1 displays histograms of W and W′ counts in the vicinity of starts and stops of genes. W exhibits a sharp peak following the 3′ end of a gene within 12 bp downstream or overlapping a stop codon. W′ occurs predominantly ∼25 bp downstream of a stop codon. In contrast, neither W nor W′ exhibit a positional preference with respect to gene start sites. The W and W′ words often form close dyads, i.e. WW′ or W′W pairs separated by ≤20 bp. The WW′ dyad occurs 111 times, 98 of which are intergenic. The dyad W′W occurs 38 times and 33 are intergenic. Figure 2 shows counts of W (top) and W′ (bottom) at specific positions relative to another W nearby in the sequence. Both plots show a strong periodic pattern of ∼10 bp, similar to a helical period of DNA in a canonical B conformation (21,22).
Figure 1.
Histograms of the word W = WWNTTACTGACRR and W′ = YYGTCAGTAANWW counts at specific positions relative to 5′ and 3′ ends of genes. In the plots on the left, position 0 corresponds to the last base of the stop codon and positive coordinates indicate positions downstream of a gene. On the right, position 0 corresponds to the first base of the translation initiation codon and negative coordinates indicate positions upstream of a gene.
Figure 2.
Occurrence of the W = WWNTTACTGACRR and W′ = YYGTCAGTAANWW words at specific positions with respect to another W nearby in the genomic sequence. Position 0 signifies that the two words completely overlap. Positive and negative coordinates indicate positions downstream and upstream, respectively, from position 0. Only positive coordinates are shown in the top panel because the plot is symmetrical about 0.
r-Scan analyses (see Materials and Methods) of W and W′ spacings reveal several significant clusters (Table 2). The HRM clusters appear uncorrelated with gene function. Two segments, 757517–757889 and 1242384–1242839, stand out exhibiting a pronounced periodic pattern. The region 757517–757889 contains six W′ words between the genes yhfF and dnaB separated by gaps of exactly 59 bp. The region 1242384–1242839, located mainly inside the gene ymeA, contains seven W′ words (some with one error) also separated by gaps of 59 bp and the last W′ is followed by W constituting a 29 bp palindrome.
Table 2. Statistically significant clusters of HRMs in the L.lactis genome and the flanking genes.
| Cluster location | Number of markers | Descriptiona |
|---|---|---|
| 160191–160241 | 3 | ybfB+. –3. W. 6. W′. 6. W′. 116. ybfC+ |
| 165117–165229 | 4 | codY+. 295. W. 46. W′. 9. W. 6. W′. 0. gatC+ |
| 427668–427782 | 4 | yecE+. 1. W. 2. W′. 3. W′. 58. W′. 100. msmK+ |
| 524347–524396 | 3 | yfcF+. 0. W. 7. W. 4. W′. 6. yfcG– |
| 597537–597660 | 4 | yfjD+. –1. W. 39. W. 5. W′. 28. W. 21. yfjE– |
| 757517–757889 | 6 | yhfF+. 2. W′. 59. W′. 59. W′. 59. W′. 59. W′. 59. W′. 12. dnaB+ |
| 824270–824489 | 5 | ftsY+(168. W. 2. W′. 77. W. 38. W. 38. W. 990) |
| 1157481–1157558b | 4 | ylfH+. 2. W. 1. W′. 11. W. 11. W′. 21. ylfI+ |
| 1200263–1200313 | 3 | yljJ+. 31. W. 10. W. 2. W′. 136. als+ |
| 1242384–1242839b | 8 | ymeA–(30. W′. 60. W′. 59. W′. 59. W′. 59. W′. 59. W′. 18). 41. W′. –3. W. 39. leuC+ |
| 1346478–1346528 | 3 | yneH–. 87. W′. 9. W. 3. W′. 44. pabB– |
| 1698284–1698324 | 3 | pepQ+. 52. W′. 1. W′. 1. W′. 35. yqjD– |
| 1768471–1768639b | 5 | serS+. 6. W. 36. W. 6. W′. 49. W′. 13. W. 13. yrgH+ |
| 1857086–1857159 | 4 | ysfG–. 47. W′. 9. W. 8. W. 5. W′. 23. rpoC– |
aThe description of each cluster shows the gaps between the HRMs (W or W′) in the cluster and between the HRMs and the flanking genes. Segments inside genes are in parentheses; negative numbers indicate the word overlaps the start or end of the gene. Gene orientation is indicated by + (direct strand) or – (complementary strand).
bSome of the words in the cluster contain one error.
Other frequent words, besides W and W′, occur in ∼10 copies and are parts of IS elements.
Streptococcus pneumoniae
The genome of S.pneumoniae contains 2906 distinct oligonucleotides of length 11 bp qualifying as frequent words by formula 2 (see Materials and Methods). The most abundant frequent words are parts of an imperfect 24 bp tandem repeat found in 540 copies in the gene SP1772 (2). The SP1772 gene encodes a putative member of a cell wall surface anchor protein family. The 24 bp repeat translates into iterations of the amino acid sequence SASTSASA which comprise 4320 amino acids of the 4776 amino acid protein. Other frequent words are mostly parts of two families of repeats, RUP and BOX elements. The RUP element has been characterized as a derivative of an insertion sequence that could still be mobile (18).
Distribution of BOX repeats. The function of the BOX element (17) is unknown. The S.pneumoniae genome contains 127 BOX elements (2). One copy is part of a possible pseudogene SP0388 whereas the remaining 126 are intergenic. Among these 126 intergenic BOX elements, 47 are between genes encoded in the same strand as the BOX element and 30 are between genes encoded in the complementary strand to the BOX sequence. Forty BOX elements are between convergent genes whereas only nine are between divergent genes. Interestingly, 16 BOX elements occur in precisely the same position in a gene’s 3′-flanking region such that the first three bases of the BoxA consensus sequence (TAA) coincide with the gene’s stop codon. An additional 11 BOX elements start within 16 bp downstream of a stop codon. Twelve BOX elements are located next to putative pseudogenes (regions with similarity to known genes but containing frameshifts or in-frame stop codons), in addition to one BOX inside the pseudogene SP0388. Neither BOX nor RUP elements form significant clusters and both are randomly distributed as verified by r-scan statistics.
Streptococcus pyogenes
In contrast to the previous two genomes, S.pyogenes contains hardly any frequent words. There are 517 oligonucleotides of 11 bp length in the S.pyogenes genome that qualify as frequent words but none exceeds the copy number threshold rw (see formula 2 in Materials and Methods) by more than six copies. The most frequent words are parts of tRNA genes or IS elements.
DISCUSSION
With the frequent word analysis (10), we identified abundant oligonucleotide motifs in the genomes of Gram-positive bacteria S.pneumoniae, S.pyogenes and L.lactis. All three are roughly equivalent in genome size, and similar in G+C content and genome signature (Table 3). The dinucleotide relative abundance comparisons (4) characterize these genomes as closely related, at about the same level of similarity as between E.coli and Vibrio cholerae, or Mycobacterium tuberculosis and Mycobacterium leprae. Despite these taxonomically close relationships, the characters of frequent words contrast sharply among these genomes. The predominant frequent words of L.lactis are parts of the 13 bp HRM of the consensus sequence WWNTTACTGACRR and its inverted complement YYGTCAGTAANWW. The frequent words of S.pneumoniae divide into three categories. One set of frequent words derives from a 24 bp sequence iterated (with few errors) 540 times in tandem contained in the gene SP1772 (2). The frequent words of the second set are parts of the 107 bp RUP element (18). The last set of frequent words relates to the 100–200 bp BOX element of unknown function (17). Interestingly, S.pyogenes lacks frequent words.
Table 3. Comparisons of L.lactis, S.pneumoniae and S.pyogenes genomic characteristics.
| Characteristica | L.lactis | S.pneumoniae | S.pyogenes |
|---|---|---|---|
| Genome size | 2.37 Mb | 2.16 Mb | 1.85 Mb |
| No. of annotated ORFs | 2266 | 2094 | 1696 |
| G+C content | 35.5% | 39.7% | 38.5% |
| ρTA* | 0.67 | 0.72 | 0.77 |
| ρCG* | 0.77 | 0.69 | 0.71 |
| ρAA* or ρTT* | 1.24 | (1.15) | (1.17) |
| τGATC* | 0.37 | 0.58 | 0.75 |
| τCCGG* | (0.81) | 0.58 | 0.73 |
| τGGCC* | (0.79) | 0.73 | (0.86) |
aIncluded are genome signature extremes for di- (ρ* values) and tetranucleotides (τ* values) defined as ρ*XY = f*XY / (f*Xf*Y), where f*X is a frequency of nucleotide X in the sequence concatenated with its inverted complement sequence and f*XY is the corresponding frequency of dinucleotide XY. For tetranucleotides, τ*XYZW = (f*XYZWf*XYf*YZf*ZWf*XNZf*YNW f*XN1N2W) / (f*Xf*Yf*Zf*Wf*XYZf*YZWf*XYNWf*XNZW), N stands for any nucleotide (4,32). The values are significantly high if ρ*XY ≥ 1.23 and significantly low if ρ*XY ≤ 0.78. The same thresholds apply for τ* values. Values in parentheses are within the normal range.
The HRMs of L.lactis are mostly confined to intergenic regions. We propose that the HRM plays a dual role. Its frequent occurrence in the 3′-flanking region of many genes suggests a possible function in transcription termination (Fig. 1) analogous to Rho independent terminators of E.coli consisting of a short G+C-rich palindrome followed by an iteration of T (12). Palindromes promote formation of stem–loop structures and RNA polymerase pausing where the terminator may destabilize the interaction between the RNA polymerase and template DNA (23). The HRMs often occur as close dyad pairs (Table 1) which could form a stem–loop structure. In addition, 61 HRMs extend to the exact sequence TTTTACTGACAGAAA (the HRM part is underlined) which could possibly establish a stem–loop conformation.
The HRMs often occur in pairs, either in the same DNA strand (close pairs) or in opposite strands (close dyads). In both cases, the spacings between the HRMs are not random (Fig. 2). The peaks are separated by ∼10 bp, similar to the DNA helical period (21,22). Pairs of HRMs positioned at multiples of the DNA helical period ensure that the HRMs face the same side of the DNA double helix. We propose that the HRMs are binding sites for an unidentified protein or proteins that cooperatively bind DNA at neighboring HRM sites. This unusual helical phasing of the HRMs facilitates interaction between the protein molecules bound to adjacent HRMs or possibly with another protein. There are documented cases of a helical phasing of DNA-binding sites required to facilitate interaction between DNA-bound proteins. For example, the transcription activator CRP requires a binding site positioned at multiples of the helical period from the rest of the promoter and incorrect positioning of the binding site decreases promoter activity (24–26). We hypothesize that the HRMs, apart from their possible role as transcription terminators, also serve as binding sites for an unidentified DNA-binding protein contributing to transcriptional control, DNA organization, replication control or regulation of some other cellular process involving DNA.
The HRMs constitute several statistically significant clusters (Table 2). In particular, two regions stand out with several HRMs arranged periodically every 72 bp (13 bp HRMs separated by 59 bp gaps). The first such region is found between the genes yhfF (hypothetical protein) and dnaB (involved in replication initiation). The other region encompasses the complete length of the ORF ymeA and its 5′ flank. Notably, this region is proximal to the replication terminus near position 1 260 000 (3). Assuming an average DNA helical period of 10.4 bp per helical turn (21), the 72 bp periodicity positions the HRMs so that all face the same side of the DNA molecule and occur exactly seven helical turns apart. We speculate that these regions could play an important role in DNA organization, e.g. as attachment sites or as regulatory elements.
Lactococcus lactis chi sites were characterized as the motif GCGCGTG (19) but these were not detected by the method of frequent words. The chi 7mer occurs only 188 times, far fewer than the 8 bp chi sites of E.coli, counted 1009 times in the E.coli K-12 genome (27).
Many frequent words of the S.pneumoniae genome relate to the two previously characterized motifs RUP and BOX. The RUP repeats are 107 bp long and situated strictly in intergenic regions. They are similar to IS elements, specifically to the IS630 family. It was proposed that the mobility of the RUP elements is mediated by the IS630 transposase and the RUPs may facilitate genomic rearrangements contributing to genomic flexibility (18).
The function of the BOX repeat is unknown. It consists of three parts, BoxA, BoxB and BoxC. The BoxA consensus is 58 bp long but commonly truncated to ∼40 bp. BoxB is typically 43 bp long. The 50 bp BoxC is the most conserved part. The length of the complete BOX element varies because BoxB may occur in multiple copies or may be missing (17). It was originally speculated (17) that the BOX repeats were regulatory elements associated with virulence and/or competence but this was disputed when the complete genome became available (2). What does the BOX distribution indicate about its possible function? The BOX elements are rarely found between divergent genes but are often located proximal downstream of a gene. This has also been observed for BOX elements in the R6 strain of S.pneumoniae (28). Location in the 3′ flank of genes suggests a possible role in transcription termination, localization, or stability of the mRNA transcript. In fact, the BOX elements can form a stable secondary structure featuring multiple stem–loop arrangements (17). The S.pneumoniae ORF SP1772 contains a remarkable repeat of 24 bp tandemly iterated 540 times. This tandem repeat is unique among current complete bacterial genomes. It translates into iterations of the amino acid sequence SASTSASA (2). The hypothetical protein encoded by this gene is 4776 amino acids long and the repeat accounts for 4320 amino acids. The protein consists of a 400 amino acid N-terminal part, followed by the tandem repeat region and a 57 amino acid C-terminus which contains an unusual run of nine charged amino acids, KRRKRDEEE. It was annotated as a cell wall surface anchor protein based on a conserved 40 amino acid motif near the C-terminus. However, the 29 members of this family listed in the PFAM database (29) do not contain a serine-rich repeat resembling the one in SP1772. Only the putative surface protein SdrC of Staphylococcus aureus among the members of this family contains an iteration of the dipeptide SD extending over 170 amino acids (30). Tettelin et al. (2) suggested that the serines in SP1772 could be glycosylated producing a structure similar to mucins. Strikingly, the strain R6 of S.pneumoniae (28) does not possess a homolog of the SP1772 protein and lacks the associated tandem repeat. Since the R6 strain is non-pathogenic, it is intriguing to speculate about a possible role of the SP1772 protein in pathogenicity. Coincidentally, SP1772 is surrounded by a pair of transposon remnants ∼400 bp upstream and ∼4000 bp downstream whereas the stretch of genes upstream of SP1772 is conserved (including gene order) between the two strains. It is possible that the two transposons proximal to SP1772 were involved in a horizontal transfer event that brought in the SP1772 gene. Besides the tandem repeat in SP1772, the frequent words in the two strains are nearly identical.
The lack of frequent words in S.pyogenes is enigmatic in comparison to the many frequent words in S.pneumoniae and L.lactis but also in comparison with other prokaryotic genomes. In fact, among 46 complete genomes, only the obligate intracellular parasites Rickettsia prowazekii, Chlamydia trachomatis and Chlamydia multocida, in addition to S.pyogenes, contain no frequent words exceeding the copy threshold rw by at least 10 copies. The lack of highly repetitive frequent words is also valid for another S.pyogenes strain, MGAS8232, whose complete genome has been recently released (31).
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported in part by NIH grants 5R01GM10452-35 and 5R01HG00335-12.
REFERENCES
- 1.Ferretti J.J., McShan,W.M., Ajdic,D., Savic,D.J., Savic,G., Lyon,K., Primeaux,C., Sezate,S., Suvorov,A.N., Kenton,S. et al. (2001) Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl Acad. Sci. USA, 98, 4658–4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tettelin H., Nelson,K.E., Paulsen,I.T., Eisen,J.A., Read,T.D., Peterson,S., Heidelberg,J., DeBoy,R.T., Haft,D.H., Dodson,R.J. et al. (2001) Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science, 293, 498–506. [DOI] [PubMed] [Google Scholar]
- 3.Bolotin A., Wincker,P., Mauger,S., Jaillon,O., Malarme,K., Weissenbach,J., Ehrlich,S.D. and Sorokin,A. (2001) The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res., 11, 731–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell A., Mrázek,J. and Karlin,S. (1999) Genome signature comparisons among prokaryote, plasmid and mitochondrial DNA. Proc. Natl Acad. Sci. USA, 96, 9184–9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nordheim A. and Rich,A. (1983) The sequence (dC–dA)n × (dG–dT)n forms left-handed Z-DNA in negatively supercoiled plasmids. Proc. Natl Acad. Sci. USA, 80, 1821–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Htun H. and Dahlberg,J.E. (1989) Topology and formation of triple-stranded H-DNA. Science, 243, 1571–1576. [DOI] [PubMed] [Google Scholar]
- 7.van Holde K. and Zlatanova,J. (1994) Unusual DNA structures, chromatin and transcription. Bioessays, 16, 59–68. [DOI] [PubMed] [Google Scholar]
- 8.Moxon E.R., Rainey,P.B., Nowak,M.A. and Lenski,R.E. (1994) Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr. Biol., 4, 24–33. [DOI] [PubMed] [Google Scholar]
- 9.Smith H.O., Tomb,J.F., Dougherty,B.A., Fleischmann,R.D. and Venter,J.C. (1995) Frequency and distribution of DNA uptake signal sequences in the Haemophilus influenzae Rd genome. Science, 269, 538–540. [DOI] [PubMed] [Google Scholar]
- 10.Karlin S., Mrázek,J. and Campbell,A.M. (1996) Frequent oligonucleotides and peptides of the Haemophilus influenzae genome. Nucleic Acids Res., 24, 4263–4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krawiec S. and Riley,M. (1990) Organization of the bacterial chromosome. Microbiol. Rev., 54, 502–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blaisdell B.E., Rudd,K.E., Matin,A. and Karlin,S. (1993) Significant dispersed recurrent DNA sequences in the Escherichia coli genome. Several new groups. J. Mol. Biol., 229, 833–848. [DOI] [PubMed] [Google Scholar]
- 13.Stern M.J., Ames,G.F., Smith,N.H., Robinson,E.C. and Higgins,C.F. (1984) Repetitive extragenic palindromic sequences: a major component of the bacterial genome. Cell, 37, 1015–1026. [DOI] [PubMed] [Google Scholar]
- 14.Robinson N.J., Robinson,P.J., Gupta,A., Bleasby,A.J., Whitton,B.A. and Morby,A.P. (1995) Singular over-representation of an octameric palindrome, HIP1, in DNA from many cyanobacteria. Nucleic Acids Res., 23, 729–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mrázek J., Bhaya,D., Grossman,A.R. and Karlin,S. (2001) Highly expressed and alien genes of the Synechocystis genome. Nucleic Acids Res., 29, 1590–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohno S. (1999) Gene duplication and the uniqueness of vertebrate genomes circa 1970–1999. Semin. Cell. Dev. Biol., 10, 517–522. [DOI] [PubMed] [Google Scholar]
- 17.Martin B., Humbert,O., Camara,M., Guenzi,E., Walker,J., Mitchell,T., Andrew,P., Prudhomme,M., Alloing,G., Hakenbeck,R., Morrison,H.A., Boulnois,G.J. and Claverys,J.-P. (1992) A highly conserved repeated DNA element located in the chromosome of Streptococcus pneumoniae. Nucleic Acids Res., 20, 3479–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oggioni M.R. and Claverys,J.P. (1999) Repeated extragenic sequences in prokaryotic genomes: a proposal for the origin and dynamics of the RUP element in Streptococcus pneumoniae. Microbiology, 145, 2647–2653. [DOI] [PubMed] [Google Scholar]
- 19.Biswas I., Maguin,E., Ehrlich,S.D. and Gruss,A. (1995) A 7-base-pair sequence protects DNA from exonucleolytic degradation in Lactococcus lactis. Proc. Natl Acad. Sci. USA, 92, 2244–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karlin S. and Leung,M.-Y. (1991) Some limit theorems on distributional patterns of balls in urns. Ann. Appl. Prob., 1, 513–538. [Google Scholar]
- 21.Wang J.C. (1979) Helical repeat of DNA in solution. Proc. Natl Acad. Sci. USA, 76, 200–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhodes D. and Klug,A. (1980) Helical periodicity of DNA determined by enzyme digestion. Nature, 286, 573–578. [DOI] [PubMed] [Google Scholar]
- 23.Henkin T.M. (1996) Control of transcription termination in prokaryotes. Annu. Rev. Genet., 30, 35–57. [DOI] [PubMed] [Google Scholar]
- 24.Gaston K., Bell,A., Kolb,A., Buc,H. and Busby,S. (1990) Stringent spacing requirements for transcription activation by CRP. Cell, 62, 733–743. [DOI] [PubMed] [Google Scholar]
- 25.Ushida C. and Aiba,H. (1990) Helical phase dependent action of CRP: effect of the distance between the CRP site and the –35 region on promoter activity. Nucleic Acids Res., 18, 6325–6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merkel T.J., Dahl,J.L., Ebright,R.H. and Kadner,R.J. (1995) Transcription activation at the Escherichia coli uhpT promoter by the catabolite gene activator protein. J. Bacteriol., 177, 1712–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blattner F.R., Plunkett,G.,III, Bloch,C.A., Perna,N.T., Burland,V., Riley,M., Collado-Vides,J., Glasner,J.D., Rode,C.K., Mayhew,G.F. et al. (1997) The complete genome sequence of Escherichia coli K-12. Science, 277, 1453–1474. [DOI] [PubMed] [Google Scholar]
- 28.Hoskins J., Alborn,W.E.,Jr, Arnold,J., Blaszczak,L.C., Burgett,S., DeHoff,B.S., Estrem,S.T., Fritz,L., Fu,D.J., Fuller,W. et al. (2001) Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol., 183, 5709–5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bateman A., Birney,E., Durbin,R., Eddy,S.R., Howe,K.L. and Sonnhammer,E.L. (2000) The Pfam protein families database. Nucleic Acids Res., 28, 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Josefsson E., McCrea,K.W., Ni Eidhin,D., O’Connell,D., Cox,J., Hook,M. and Foster,T.J. (1998) Three new members of the serine–aspartate repeat protein multigene family of Staphylococcus aureus. Microbiology, 144, 3387–3395. [DOI] [PubMed] [Google Scholar]
- 31.Smoot J.C., Barbian,K.D., Van Gompel,J.J., Smoot,L.M., Chaussee,M.S., Sylva,G.L., Sturdevant,D.E., Ricklefs,S.M., Porcella,S.F., Parkins,L.D. et al. (2002) Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc. Natl Acad. Sci. USA, 99, 4668–4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karlin S., Mrázek,J. and Campbell,A.M. (1997) Compositional biases of bacterial genomes and evolutionary implications. J. Bacteriol., 179, 3899–3913. [DOI] [PMC free article] [PubMed] [Google Scholar]