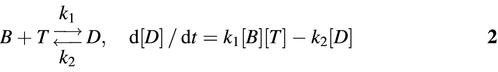Abstract
Molecular beacons are oligonucleotide probes capable of forming a stem–loop hairpin structure with a reporter dye at one end and a quencher at the other end. Conventional molecular beacons are designed with a target-binding domain flanked by two complementary short arm sequences that are independent of the target sequence. Here we report the design of shared-stem molecular beacons with one arm participating in both stem formation when the beacon is closed and target hybridization when it is open. We performed a systematic study to compare the behavior of conventional and shared-stem molecular beacons by conducting thermodynamic and kinetic analyses. Shared-stem molecular beacons form more stable duplexes with target molecules than conventional molecular beacons; however, conventional molecular beacons may discriminate between targets with a higher specificity. For both conventional and shared-stem molecular beacons, increasing stem length enhanced the ability to differentiate between wild-type and mutant targets over a wider range of temperatures. Interestingly, probe–target hybridization kinetics were similar for both classes of molecular beacons and were influenced primarily by the length and sequence of the stem. These findings should enable better design of molecular beacons for various applications.
INTRODUCTION
Molecular beacons are dual-labeled oligonucleotide probes that fluoresce upon hybridization with a complementary target sequence (1). The oligonucleotide is labeled at one end with a fluorescent reporter dye and at the opposite end with a fluorescence quencher. Molecular beacons are designed to form a stem–loop hairpin structure in the absence of target, forcing the fluorescence reporter group in proximity with the quencher group. In this conformation, fluorescence is quenched. In the presence of a complementary target molecule, the molecular beacon opens due to the formation of the more stable probe–target duplex, increasing distance between the reporter and quencher, and restoring fluorescence. The competing reaction between hairpin formation and target hybridization improves specificity of molecular beacons compared with linear probes, while the transition between quenched and fluorescent states allows for the differentiation between bound and unbound probes (2). Molecular beacons are capable of having >200-fold increase in fluorescence intensity upon hybridization (1).
The unique target recognition and signal transduction capabilities of molecular beacons have led to their application in many biochemical and biological assays including quantitative PCR (3,4), protein–DNA interactions (5,6), multiplex genetic analysis (7,8), and the detection of mRNA in living cells (9–11). The thermodynamic and kinetic properties of a molecular beacon are dependent on its structure and sequence in complex ways (2,12). Moreover, the signal-to-background ratio in target detection is dependent not only on design (length and sequence of the stem and probe) but also on the quality of oligonucleotide synthesis and purification (13,14) and the assay conditions employed. Molecular beacons must be carefully designed for each application in order to achieve optimal performance: precise knowledge of how different design parameters influence performance is critical in exploiting the greatest benefit from the use of molecular beacon probes.
Here we describe a design variant for molecular beacons where one arm of the stem participates in either hairpin formation or target hybridization, which we call ‘shared-stem’ molecular beacons. In contrast, conventional molecular beacons are designed such that the loop sequence is complementary to the target while the stem sequences are self-complementary but unrelated to the target sequence. This new design may offer certain advantages over conventional molecular beacon design, especially in two-probe fluorescence resonance energy transfer (FRET) assays (15–19). We have examined the thermodynamic and kinetic properties of both shared-stem and conventional molecular beacons and made a systematic comparison between them. In particular, we have quantified the changes in enthalpy and entropy upon the formation of probe–target duplexes as determined by the probe and stem lengths. We have also studied how the melting behavior, specificity and hybridization on-rate depend on the stem length of molecular beacons. The implications of these findings are discussed.
MATERIALS AND METHODS
Oligonucleotide synthesis
Oligonucleotide probes and targets were synthesized using standard phosphoramidite chemistry on an Applied Biosystems model 394 automated DNA synthesizer (Foster City, CA). Molecular beacons were purified using a two-step reverse phase (RP) plus ion-exchange (IE) high performance liquid chromatography (HPLC) on a Waters Model 600E HPLC system (Millipore Corp., Milford, MA). For RP-HPLC purification, oligonucleotides were loaded on a Hamilton PRP-1 column and eluted with a linear 5–50% acetonitrile gradient in 0.1 M triethyl-ammonium acetate pH 7.2 over 40 min. The oligonucleotides were additionally purified by IE-HPLC using a Source™ column (Amersham Pharmacia Biotech, Piscataway, NJ) and eluted with a linear 0–50% 1 M LiCl gradient in 0.1 M Tris pH 8.0 over 40 min. Unmodified (target) oligonucleotides were purified using polyacrylamide gel electrophoresis. All oligonucleotides were synthesized at Integrated DNA Technologies, Inc. (Coralville, IA).
Molecular beacon design
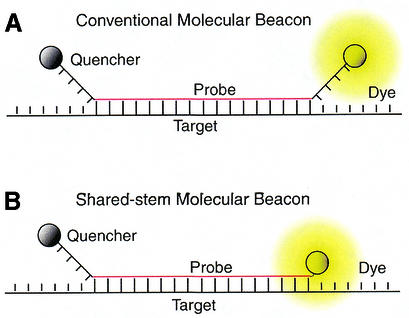
Two types of molecular beacons were designed and synthesized; both contain target-specific probe sequence complementary in antisense orientation to exon 6 of the human GAPDH gene, a Cy3 fluorophore at the 5′ end, and a dabcyl quencher at the 3′ end. As illustrated in Figure 1A, one type follows the conventional design of molecular beacons in that the target-specific probe domain was centrally positioned between two complementary arms that form the stem; the sequence of these arms were independent of the target sequence. Shared-stem molecular beacons, on the other hand, were designed to have one arm of the stem complementary to the target sequence, as shown schematically in Figure 1B. In both cases, the probe length Lp is defined as the portion of the molecular beacon that is complementary to the target, and the stem length Ls is the number of bases of each complementary arm. The probe length was chosen as Lp = 19 bases for all the molecular beacons studied (see Table 1). In general, the probe length should be chosen to avoid the potential of forming secondary structure within the loop of the molecular beacon (http://www.bioinfo.math.rpi.edu/∼mfold), and to achieve an optimal combination of specificity, kinetic rate and melting behavior.
Figure 1.
Alternative molecular beacon designs. (A) Conventional molecular beacons have stem sequences that are independent of the target sequence. (B) Shared-stem molecular beacons are designed such that one arm of the stem participates in both hairpin formation and target hybridization.
Table 1. The design of probes and target oligonucleotides.
| Name | Sequence (5′–3′) | Notes |
|---|---|---|
| Shared-stem 19/4a | Cy3-GAGTCCTTCCACGATACCActc-Dabcyl | Probe 19/Stem 4 |
| Shared-stem 19/5a | Cy3-GAGTCCTTCCACGATACCAgactc-Dabcyl | Probe 19/Stem 5 |
| Shared-stem 19/7a | Cy3-GAGTCCTTCCACGATACCAggactc-Dabcyl | Probe 19/Stem 7 |
| Conventional 19/4a | Cy3-cctcGAGTCCTTCCACGATACCAgagg-Dabcyl | Probe 19/Stem 4 |
| Conventional 19/5a | Cy3-ctgacGAGTCCTTCCACGATACCAgtcag-Dabcyl | Probe 19/Stem 5 |
| Conventional 19/6a | Cy3-ctgagcGAGTCCTTCCACGATACCAgctca-Dabcyl | Probe 19/Stem 6 |
| Target WTb | ACTTTGGTATCGTGGAAGGACTCATGA | Perfect match |
| Target Ab | ACTTTGGTATCGTGGAAGGAaTCATGA | Single mismatch |
| Target Bb | ACTTTGGTATCGTaGAAGGACTCATGA | Single mismatch |
| Target Cb | ACTTTGGTATCGTaGAAGGAaTCATGA | Double mismatch |
| Target Db | ACTTTGGTATCGTaaAAGGACTCATGA | Double mismatch |
aMolecular beacons: lower case, bases added to create stem domains; upper case, probe–target hybridizing domains; upper case bold, bases participating in both stem hairpin and target binding.
bTargets: underscore, 19-base sequence complementary to beacons; lower case bold, mismatch bases in targets.
Conventional molecular beacons were synthesized with Ls = 4, 5 and 6 bases. The shared-stem molecular beacons were synthesized with Ls = 4, 5 and 7 bases. As illustrated by Figure 2A, a 6-base stem could not be synthesized because the shared-stem molecular beacon sequence is constrained, i.e. part of the arm sequence that makes up the 6-base stem is predetermined since the 5′ end of the shared-stem molecular beacon must complement the target sequence and the 3′ stem is created solely to complement the 5′-stem sequence. This inadvertently forces an additional base pairing in the stem. It should be noted that the stem sequence of a shared-stem molecular beacon is not adjustable since one arm of the stem is designed to complement the target. This limitation often precludes the design of certain stem/probe length combinations, as demonstrated in Figure 2B for a molecular beacon with a probe length of 18 bases and a stem length of 4 bases. Five target oligonucleotides were also synthesized, one wild-type and four with mismatches at assorted locations, as shown in Table 1.
Figure 2.
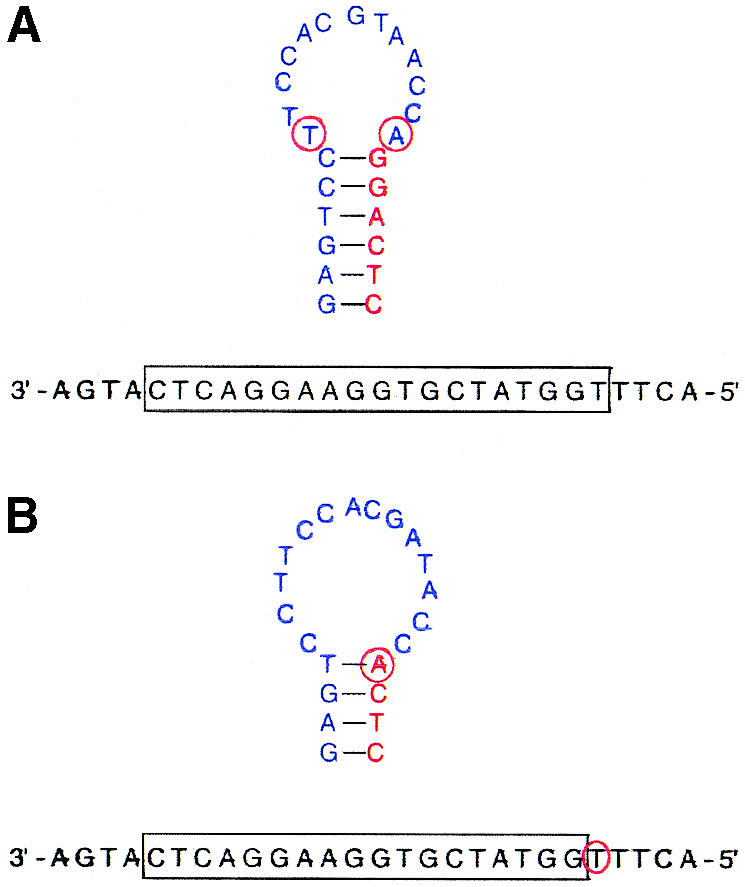
Examples of the design constraint of shared-stem molecular beacons with certain stem/probe combinations. (A) The design of a molecular beacon with a probe length of 19 bases and a stem length of 6 bases inadvertently resulted in additional bases participating in stem formation (red circles). (B) The design of a molecular beacon with a probe length of 18 bases and a stem length of 4 bases inadvertently resulted in an additional base participating in target hybridization (red circle).
Equilibrium analysis
Molecular beacons in the presence of target were assumed to exist mainly in three phases: (i) as a duplex with target, (ii) as a stem–loop hairpin, and (iii) in random coil conformation. It should be noted, however, that under certain circumstances other states of the molecular beacon may exist and contribute to thermodynamic equilibrium, including unintended hybrids of the beacon and target. Dissociation constants describing the transition between these phases were determined by analyzing the thermal denaturation profile of molecular beacons in the presence and absence of target (2). Denaturation profiles were obtained by recording the fluorescence intensity of a 50 µl solution containing 200 nM of molecular beacon in the presence of 0–20 µM of target at temperatures ranging from 5 to 95°C. Specifically, the temperature of the hybridization solution was brought to 95°C and reduced by 1°C increments to 5°C. The temperature was then raised with 1°C increments back to 95°C to ensure that the solution reached equilibrium and no hysteresis had occurred. The temperature was held at each temperature increment for 10 min and fluorescence was measured for the final 30 s. The fluorescence intensity of each test solution was adjusted to correct for the intrinsic variance of fluorescence over temperature. Each thermal denaturation assay was performed in hybridization buffer containing 10 mM Tris, 50 mM KCl and 5 mM MgCl2.
The fluorescence intensity data describing the thermal denaturation profile of each molecular beacon and molecular beacon–target duplex was used to determine the respective dissociation constant as described in Bonnet et al. (2). Specifically, dissociation constants K12 characterizing the transition between phase 1 (bound to target) and phase 2 (closed beacon) of molecular beacons were obtained for all beacon–target pairs and for all molecular beacons in the absence of target. Furthermore, the dissociation constants K12 were used to determine the changes in enthalpy (ΔH12) and entropy (ΔS12) associated with each beacon–target duplex. The errors calculated for the thermodynamic parameters signify a 95% confidence interval.
Molecular beacon specificity
The fraction of molecular beacons bound to target, α, was calculated for each molecular beacon–target pair as a function of temperature. All calculations utilized the thermodynamic parameters, enthalpy change ΔH12 and entropy change ΔS12, obtained from the thermal denaturation profiles for each beacon–target duplex:
α / [(1 – α)(η – α)B̂0] = e(–ΔH12 / Rθ) + (ΔS12 / R)1
where θ is the temperature in Kelvin, R is the gas constant, η = T0/B0, B̂0 = B0 / c0, T0 and B0 are, respectively, initial concentration of target and beacons, and c0 is the unit concentration, 1 M (20). The value of α was calculated for each molecular beacon–target pair as a function of temperature for samples containing B0 = 200 nM of molecular beacon and T0 = 400 nM of target. The melting temperature θm is defined as the temperature at which half of the molecular beacons are bound to target, i.e. α = 0.5.
Kinetic analysis
A SPEX fluorolog-2 spectrofluorometer with an SFA-20 rapid kinetics stopped-flow accessory and a temperature/trigger module (SFA-12) was used to measure molecular beacon– target binding kinetics. Specifically, the fluorescence intensity emitted from a rapidly mixed solution containing 250 nM molecular beacons and 2.5 µM targets was recorded over time for each molecular beacon–target pair. The hybridization reaction was assumed to obey the second order reaction kinetics:
where [B], [T] and [D] are the concentrations of unbound molecular beacon, unbound target and molecular beacon– target duplex, respectively; k1 is the on-rate and k2 the off-rate of molecular beacon–target hybridization. The exact solution of equation 2 gives:
1 – {[D(t)] / [Deq]} = e–Δk1t[1 – λ{[D(t)] / [Deq]}]3
where Δ = 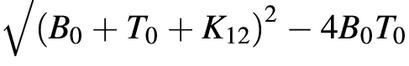 , [Deq] = (B0 + T0 + K12 – Δ) / 2, λ = [Deq]2 / B0T0, and K12 = k2 / k1 is the dissociation constant discussed above. Since the concentration of molecular beacon–target duplex is unknown at any given time, it was assumed that [F(t) – F0] / (Feq – F0) = [D(t)] / [Deq] where F(t) is the fluorescence intensity at time t, F0 is the initial fluorescence intensity, and Feq is the fluorescence intensity as t → ∞. In order to obtain the on-rate k1 based on the fluorescence measurement, two different curve-fitting schemes were used. The first utilized a least-squares method by fitting a straight line to a logarithmic form of equation 3:
, [Deq] = (B0 + T0 + K12 – Δ) / 2, λ = [Deq]2 / B0T0, and K12 = k2 / k1 is the dissociation constant discussed above. Since the concentration of molecular beacon–target duplex is unknown at any given time, it was assumed that [F(t) – F0] / (Feq – F0) = [D(t)] / [Deq] where F(t) is the fluorescence intensity at time t, F0 is the initial fluorescence intensity, and Feq is the fluorescence intensity as t → ∞. In order to obtain the on-rate k1 based on the fluorescence measurement, two different curve-fitting schemes were used. The first utilized a least-squares method by fitting a straight line to a logarithmic form of equation 3:
(1 / Δ)ln(1 – {[F(t) – F0] / (Feq – F0)}) = (1 / Δ)ln(1 – λ{[F(t) – F0] / [Feq – F0]}) – k1t4
with a slope equal to k1. Alternatively, a non-linear least-squares method was used to determine the value of k1 from equation 3 directly. The results obtained using these two approaches were compared.
RESULTS
Thermal analysis
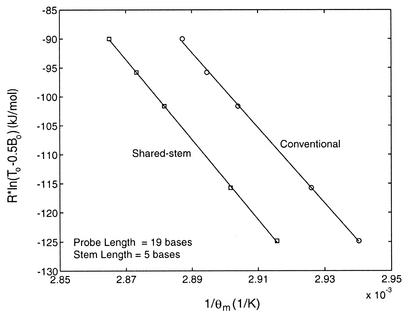
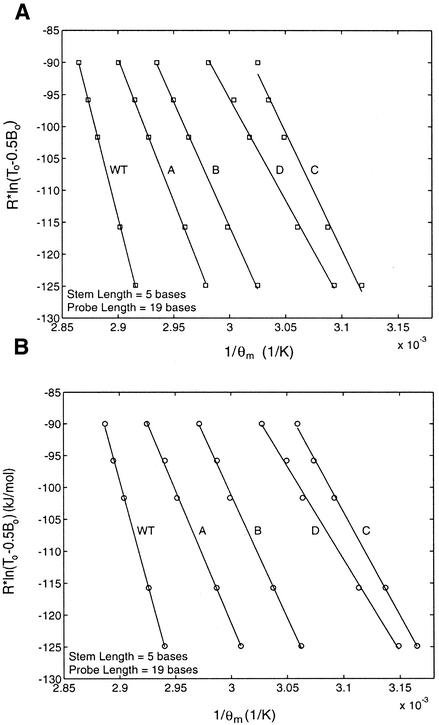
To better understand how the performance of shared-stem molecular beacons differs from that of the conventional molecular beacons, the thermodynamic parameters of these two types of molecular beacon were obtained and compared. In particular, the enthalpy and entropy changes ΔH12 and ΔS12 describing the phase transition between bound-to-target and stem–loop conformations were determined for conventional and shared-stem molecular beacons using van’t Hoff plots. As demonstrated in Figure 3, these plots display the inverse of melting temperature 1 / θm as determined by Rln(T0 – 0.5B0) shown as the ordinate. Since at melting temperature,
Figure 3.
Comparison of the melting temperature of shared-stem and conventional molecular beacons as determined by the initial concentrations of probe and target. By fitting the data with a straight line, changes in enthalpy (slope of the fitted line) and entropy (y-intercept) characterizing the phase transition between bound-to-target and stem–loop conformations of a molecular beacon were obtained.
Rln(T0 – 0.5B0) = –ΔH12(1 / θm) + ΔS125
the slope of the fitted straight line of each curve in Figure 3 represents the enthalpy change –ΔH12 and the y-intercept represents the entropy change ΔS12. It was found that, in general, the shared-stem molecular beacons have a higher melting temperature, i.e. they form more stable probe–target duplexes than conventional molecular beacons. The changes of enthalpy and entropy for all the molecular beacon–target combinations tested are summarized in Table 2. To minimize the number of independent variables involved in controlling probe–target hybridization, all the molecular beacons were designed to have identical probe sequences. Furthermore, for molecular beacons with a stem length of 5 bases and a probe length of 19 bases, the stem sequence of the conventional molecular beacons was chosen such that energetically the stem was similar to that of the shared-stem molecular beacons. The free energy changes were calculated using nearest neighbor approximations (21).
Table 2. Changes in enthalpy and entropy of conventional and shared-stem molecular beacons in the presence of target.
| Target | Probe length | Conventional molecular beacons | Shared-stem molecular beacons | ||||
|---|---|---|---|---|---|---|---|
| Stem length | ΔH (kJ/mol) | ΔS (J/mol·K) | Stem length | ΔH (kJ/mol) | ΔS (J/mol·K) | ||
| WT | 19 | 4 | 823 ± 168 | 2281 ± 489 | 4 | 862 ± 116 | 2383 ± 336 |
| A | 19 | 4 | 577 ± 62 | 1595 ± 184 | 4 | 708 ± 36 | 1967 ± 106 |
| B | 19 | 4 | 527 ± 27 | 1471 ± 79 | 4 | 586 ± 54 | 1628 ± 161 |
| C | 19 | 4 | 472 ± 23 | 1336 ± 70 | 4 | 478 ± 49 | 1340 ± 148 |
| D | 19 | 4 | 480 ± 38 | 1352 ± 115 | 4 | 521 ± 87 | 1461 ± 262 |
| WT | 19 | 5 | 649 ± 28 | 1784 ± 83 | 5 | 690 ± 16 | 1887 ± 46 |
| A | 19 | 5 | 418 ± 23 | 1133 ± 70 | 5 | 446 ± 20 | 1205 ± 59 |
| B | 19 | 5 | 385 ± 24 | 1055 ± 73 | 5 | 391 ± 24 | 1057 ± 72 |
| C | 19 | 5 | 324 ± 17 | 901 ± 54 | 5 | 369 ± 57 | 1025 ± 175 |
| D | 19 | 5 | 291 ± 25 | 790 ± 76 | 5 | 319 ± 28 | 861 ± 85 |
| WT | 19 | 6 | 467 ± 16 | 1265 ± 48 | 7 | 413 ± 10 | 1096 ± 28 |
| A | 19 | 6 | 404 ± 25 | 1105 ± 76 | 7 | 370 ± 13 | 998 ± 38 |
| B | 19 | 6 | 380 ± 26 | 1055 ± 79 | 7 | 351 ± 39 | 956 ± 117 |
| C | 19 | 6 | 367 ± 19 | 1058 ± 61 | 7 | 260 ± 30 | 707 ± 93 |
| D | 19 | 6 | 373 ± 29 | 1065 ± 91 | 7 | 245 ± 25 | 653 ± 79 |
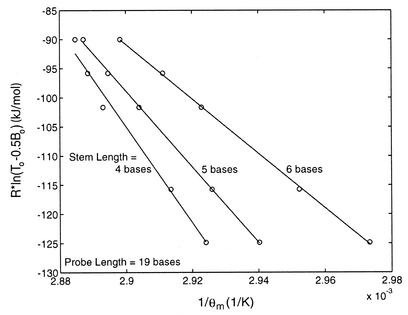
The difference in thermodynamic behavior between conventional and shared-stem molecular beacons can be understood in terms of the ability of the flanking arms to interact with each other. With shared-stem molecular beacons, once part of the stem (one arm) is bound to the target, it is less likely to interact with its complementary arm, resulting in a more stable probe–target duplex. In contrast, the arms of a conventional molecular beacon do not bind to the target and are thus more likely to interact with each other as driven by thermal energy, increasing the tendency of forming a closed molecular beacon by dissociating from the target. Not surprisingly, the stem length of a molecular beacon influenced the equilibrium state of both the shared-stem and conventional beacons in the presence of target. As shown in Figure 4, as the stem length was increased from 4 to 6 bases, conventional molecular beacons were found to dissociate from target molecules more readily. A very similar trend was true for shared-stem molecular beacons (data not shown). This indicates that hybridization is less favorable for molecular beacons with longer stems.
Figure 4.
Determination of the changes in enthalpy (slope of the fitted line) and entropy (y-intercept) characterizing the phase transition between bound-to-target and stem–loop conformations for conventional molecular beacons. A similar trend was found for shared-stem molecular beacons.
The changes in enthalpy and entropy that control the dissociation of conventional and shared-stem molecular beacons from targets with mismatches were also determined (see Table 2). It was found that shared-stem molecular beacons formed more stable duplexes with each of the target molecules tested. However, as displayed in Figure 5, when point mutations were present in the target oligonucleotide, both types of molecular beacons dissociated from their targets more readily. The magnitude of change depended on both the number of mismatches and their location. Compared with the wild-type target, a point mutation near the center of the probe-binding domain (Target B) was found to have a larger effect on molecular beacon dissociation than a mutation near the end of the probe-binding domain (Target A). As expected, two point mutations (Targets C and D) on a target had a more profound effect on the dissociation of molecular beacons from targets than that with one point mutation.
Figure 5.
Determination of the changes in enthalpy (slope of the fitted line) and entropy (y-intercept) characterizing the phase transition between bound-to-target and stem–loop conformations for (A) shared-stem and (B) conventional molecular beacons interacting with wild-type and mutant targets.
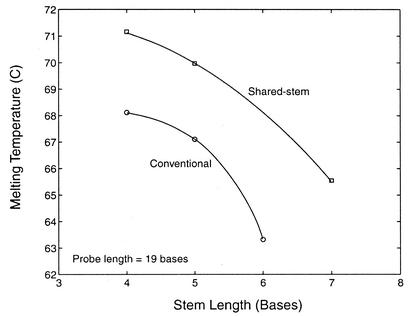
Melting temperature
To further elucidate the effect of molecular beacon structure on the stability of the probe–target duplex, the melting temperatures θm for conventional and shared-stem molecular beacons with a probe length of 19 bases and stem lengths ranging from 4 to 7 bases were compared, as shown in Figure 6. It was found that conventional molecular beacons had lower melting temperatures than shared-stem molecular beacons for each of the stem lengths considered; however, both types of molecular beacon exhibited similar trends. Specifically, the melting temperature progressively decreased as the stem length increased. In fact, it appears that the melting temperature would be quite low for conventional molecular beacons with a probe length of 19 bases and a stem length of 7 bases or greater. This may be because that with long free arms of the stem a bound molecular beacon is very easy to dissociate from the target and form a stable hairpin structure even at low temperatures.
Figure 6.
Comparison of melting temperatures as a function of stem length for conventional and shared-stem molecular beacons in the presence of wild-type target.
Molecular beacon specificity
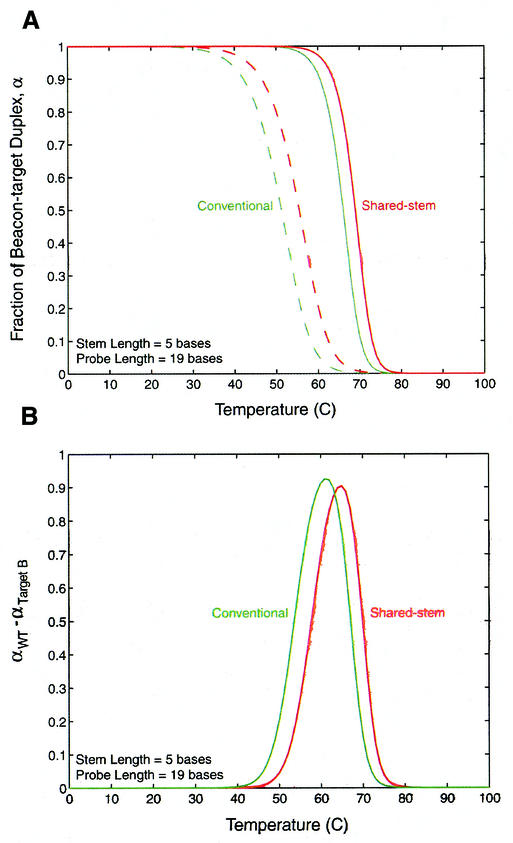
Melting curves that display the fraction of molecular beacons in duplex form, α, as a function of temperature were obtained for each molecular beacon and probe–target pair. As demonstrated in Figure 7A, the difference in melting temperature θm (i.e. temperature at α = 0.5) between beacon–wild-type-target and beacon–mutant-target duplexes was found to be slightly larger for conventional molecular beacons compared with corresponding shared-stem molecular beacons. Furthermore, the difference in the fraction of molecular beacons bound to wild-type target and mutant target, αWT – αTarget B, as a function of temperature was found to be similar for conventional and shared-stem molecular beacons, although the maximum value of αWT – αTarget B is slightly higher for the former, as shown in Figure 7B. The conventional molecular beacons were also found to maintain a value of αWT – αTarget B > 0 over a slightly broader range of temperatures than shared-stem molecular beacons, but again the difference is very small. This implies that the conventional molecular beacons may exhibit only a slightly higher specificity than shared-stem molecular beacons.
Figure 7.
Melting behavior of conventional and shared-stem molecular beacons with a 19-base probe and a five-base stem. (A) Melting curves for conventional (green) and shared-stem (red) molecular beacons in the presence of wild-type (solid line) and mutant (dashed line) target. (B) The difference in the fraction of conventional (green) or shared-stem (red) molecular beacons bound to wild-type and mutant targets.
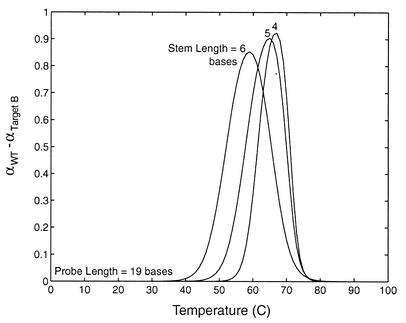
The effect of stem length on molecular beacon specificity was also found to be similar for conventional and shared-stem molecular beacons. Specifically, the curves in Figure 8 demonstrate that, as the stem length is increased the heightened competition between a unimolecular reaction and bimolecular hybridization broadens the transition between bound and unbound states. This results in an improved ability to discriminate between targets over a wider range of temperature but lowers the maximum difference in the fraction of beacons bound to wild-type and mutant targets.
Figure 8.
The difference in the fraction of beacons bound to wild-type and mutant targets for shared-stem molecular beacons with stem lengths of 4, 5 and 6 bases. The same trend is true for conventional molecular beacons.
Kinetic analysis
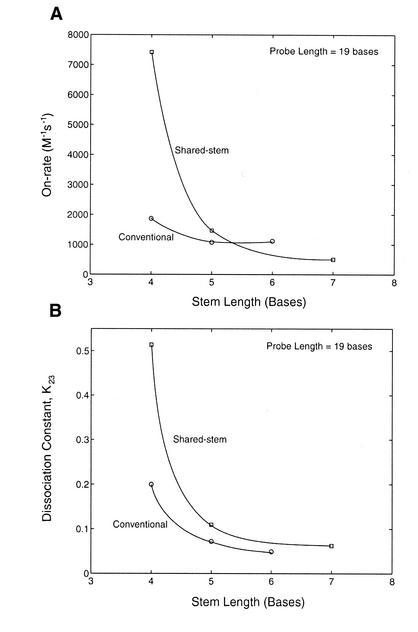
The on-rate of shared-stem and conventional molecular beacons hybridized to wild-type target as a function of stem length is displayed in Figure 9A. It is seen that for shared-stem molecular beacons, an increase in stem length from 4 to 5 bases induced a 5-fold reduction in its on-rate, which was further reduced by 3-fold when the stem length was increased from 5 to 7 bases. In contrast, the on-rate of conventional molecular beacons only decreased slightly when the stem length was increased from 4 to 6 bases. It is interesting to note that, with stem lengths of 5 bases or larger, the shared-stem and conventional molecular beacons have on-rates differing only by less than a factor of 2. However, the shared-stem molecular beacons with a 4-base stem hybridized to wild-type targets four times faster than the corresponding conventional molecular beacons. This large difference in the rate of hybridization most likely resulted from the variations in the stability of the hairpin structure. To further illustrate this, the dissociation constants K23 of the conventional and shared-stem molecular beacons in the absence of target are shown in Figure 9B. Interestingly, there seems to be a clear correlation between the on-rate of beacon hybridization and the stability of the stem–loop structure. This is understandable since K23 represents the transition from hairpin (phase 2) to random coiled (phase 3) conformations of molecular beacons, and a higher K23 implies that the molecular beacons are easier to open.
Figure 9.
Comparison between conventional and shared-stem molecular beacons on (A) the on-rate of hybridization with target and (B) the dissociation constant without target (i.e. the transition between stem–loop hairpin and random-coiled beacons) for molecular beacons with a 19-base probe length and various stem lengths.
DISCUSSION
Molecular beacons have become a very useful tool for many homogeneous single-stranded nucleic acid detection assays due to their ability to differentiate between bound and unbound states and their improved specificity over linear probes. However, to optimize the performance of molecular beacons for different applications, it is necessary to understand their structure–function relationships. This may become critical in certain assays since the addition (or deletion) of just a single nucleotide to the stem can dramatically change the behavior of molecular beacons. Here we describe a new design of molecular beacons, i.e. the shared-stem molecular beacons, of which the stem-arm nearest the reporter dye participates in both hairpin formation and target hybridization. In contrast to conventional molecular beacons whose stems are independent of the target sequence and thus can freely rotate around the probe–target duplex, this new design helps immobilize the fluorophores of molecular beacons when they hybridize to the target, which is desirable when two molecular beacons are used in a FRET assay (19). Specifically, with shared-stem molecular beacons, there is a better control of the distance between the donor dye on one beacon and the acceptor dye on the other beacon, since the rotational motion of the fluorophore-attached stem-arm is constrained, as illustrated in Figure 1B. To facilitate the design and application of, and to reveal the differences between, shared-stem and conventional molecular beacons, we performed a systematic study of the thermodynamic and kinetic parameters that control the hybridization of these molecular beacons with complementary and mismatched targets.
In general, it was found that compared with shared-stem molecular beacons, conventional molecular beacons form less stable duplexes with single-stranded nucleic acid targets but have a slightly improved ability to discriminate between wild-type and mutant targets. The difference in the duplex stability may be understood by considering the thermal-driven interactions between the two stem-forming arms after the molecular beacon hybridized to a target molecule. Unlike linear oligonucleotide probes, a molecular beacon can have two stable conformations: bound to target, and as a stem–loop hairpin. These two stable states compete with each other, giving rise to an improved specificity. The additional freedom inherent in both arms of conventional molecular beacon increases the likelihood that, when a beacon–target complex is partially denatured due to thermal fluctuations, these arms will have a higher probability of encountering each other, allowing the molecular beacon to dissociate from the target. There might exist other explanations for the difference in the stability of probe–target duplexes of conventional and shared-stem molecular beacons and this possibility needs to be further explored.
The reduced stability for conventional beacon hybrids also corresponds to a smaller value in the free energy difference between bound and unbound states of the probe–target duplex. The change in free energy due to any mismatch between the probe and target, therefore, will have a more profound effect on the preference of the stem–loop hairpin conformation of the conventional molecular beacons, leading to an improved ability to differentiate between wild-type and mutated targets. However, this improvement was found to be marginal.
With any given probe length and sequence, the hybridization kinetics of molecular beacons appears to be primarily dependent on the length and sequence of the stem, regardless of whether they are designed in the conventional or shared-stem configuration. Both types of molecular beacon exhibited comparable hybridization rates when the dissociation constants describing the thermal fluctuation induced opening of the stem–loop structure, K23, were similar. When the difference in K23 for the shared-stem and conventional molecular beacons was increased, so was the difference in the hybridization on-rate.
In addition to the above-mentioned differences in the behavior of shared-stem and conventional molecular beacons, it is worth mentioning that, with the same probe length (i.e. bases that hybridize to the target) of a conventional beacon, a shared-stem molecular beacon has a shorter loop length, thus reducing the potential for secondary structure within the loop portion of the beacon. Furthermore, the choice of the stem length is independent of the probe length for conventional molecular beacons, whereas there are certain constraints on the stem-length and probe-length combinations in designing the share-stem molecular beacons. This, together with the dependence of the thermodynamic and kinetic properties on the probe and stem lengths demonstrated in this study, should be considered in the design of molecular beacons for specific applications.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Andrei Laikhter and Erin Edgar for assistance with oligonucleotide synthesis. This work was supported in part by the Wallace H. Coulter Foundation and by a Cutting Edge Research Award from Georgia Institute of Technology.
REFERENCES
- 1.Tyagi S. and Kramer,F.R. (1996) Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol., 14, 303–308. [DOI] [PubMed] [Google Scholar]
- 2.Bonnet G., Tyagi,S., Libchaber,A. and Kramer,F.R. (1999) Thermodynamic basis of the enhanced specificity of structured DNA probes. Proc. Natl Acad. Sci. USA, 96, 6171–6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vogelstein B. and Kinzler,K.W. (1999) Digital PCR. Proc. Natl Acad. Sci. USA, 96, 9236–9241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen W., Martinez,G. and Mulchandani,A. (2000) Molecular beacons: a real-time polymerase chain reaction assay for detecting Salmonella. Anal. Biochem., 280, 166–172. [DOI] [PubMed] [Google Scholar]
- 5.Fang X., Li,J.J. and Tan,W. (2000) Using molecular beacons to probe molecular interactions between lactate dehydrogenase and single-stranded DNA. Anal. Chem., 72, 3280–3285. [DOI] [PubMed] [Google Scholar]
- 6.Li J.J., Geyer,R. and Tan,W. (2000) Using molecular beacons as a sensitive fluorescence assay for enzymatic cleavage of single-stranded DNA. Nucleic Acids Res., 28, e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marras S.A., Kramer,F.R. and Tyagi,S. (1999) Multiplex detection of single-nucleotide variations using molecular beacons. Genet. Anal., 14, 151–156. [DOI] [PubMed] [Google Scholar]
- 8.de Baar M.P., Timmermans,E.C., Bakker,M., de Rooij,E., van Gemen,B. and Goudsmit,J. (2001) One-tube real-time isothermal amplification assay to identify and distinguish human immunodeficiency virus type 1 subtypes A, B and C and circulating recombinant forms AE and AG. J. Clin. Microbiol., 39, 1895–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsuo T. (1998) In situ visualization of messenger RNA for basic fibroblast growth factor in living cells. Biochim. Biophys. Acta, 1379, 178–184. [DOI] [PubMed] [Google Scholar]
- 10.Sokol D.L., Zhang,X., Lu,P. and Gewirtz,A.M. (1998) Real time detection of DNA.RNA hybridization in living cells. Proc. Natl Acad. Sci. USA, 95, 11538–11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molenaar C., Marras,S.A., Slats,J.C., Truffert,J.C., Lemaitre,M., Raap,A.K., Dirks,R.W. and Tanke,H.J. (2001) Linear 2′ O-methyl RNA probes for the visualization of RNA in living cells. Nucleic Acids Res., 29, e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuhn H., Demidov,V.V., Coull,J.M., Fiandaca,M.J., Gildea,B.D. and Frank-Kamenetskii,M.D. (2002) Hybridization of DNA and PNA molecular beacons to single-stranded and double-stranded DNA targets. J. Am. Chem. Soc., 124, 1097–1103. [DOI] [PubMed] [Google Scholar]
- 13.Goddard N.L., Bonnet,G., Krichevsky,O. and Libchaber,A. (2000) Sequence dependent rigidity of single stranded DNA. Phys. Rev. Lett., 85, 2400–2403. [DOI] [PubMed] [Google Scholar]
- 14.Bonnet G., Krichevsky,O. and Libchaber,A. (1998) Kinetics of conformational fluctuations in DNA hairpin-loops. Proc. Natl Acad. Sci. USA, 95, 8602–8606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cardullo R.A., Agrawal,S., Flores,C., Zamecnik,P.C. and Wolf,D.E. (1998) Detection of nucleic acid hybridization by nonradiative fluorescence resonance energy transfer. Proc. Natl Acad. Sci. USA, 85, 8790–8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sei-Iida Y., Koshimoto,H., Kondo,S. and Tsuji,A. (2000) Real-time monitoring of in vitro transcriptional RNA synthesis using fluorescence resonance energy transfer. Nucleic Acids Res., 28, e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsuji A., Koshimoto,H., Sato,Y., Hirano,M., Sei-Iida,Y., Kondo,S. and Ishibashi,K. (2000) Direct observation of specific messenger RNA in a single living cell under a fluorescence microscope. Biophys. J., 78, 3260–3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsuji A., Sato,Y., Hirano,M., Suga,T., Koshimoto,H., Taguchi,T. and Ohsuka,S. (2001) Development of a time-resolved fluorometric method for observing hybridization in living cells using fluorescence resonance energy transfer. Biophys. J., 81, 501–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsourkas A. and Bao,G. (2001) Detecting mRNA transcripts using FRET-enhanced molecular beacons. In Proceedings of the 2001 Bioengineering Conference. ASME BED, New York, NY, Vol. 50, pp. 169–170.
- 20.Ratilainen T., Holmen,A., Tuite,E., Haaima,G., Christensen,L., Nielsen,P.E. and Norden,B. (1998) Hybridization of peptide nucleic acid. Biochemistry, 37, 12331–12342. [DOI] [PubMed] [Google Scholar]
- 21.Zuker M. (2000) Calculating nucleic acid secondary structure. Curr. Opin. Struct. Biol., 10, 303–310. [DOI] [PubMed] [Google Scholar]