Abstract
Profile hidden Markov models (HMMs) are amongst the most successful procedures for detecting remote homology between proteins. There are two popular profile HMM programs, HMMER and SAM. Little is known about their performance relative to each other and to the recently improved version of PSI-BLAST. Here we compare the two programs to each other and to non-HMM methods, to determine their relative performance and the features that are important for their success. The quality of the multiple sequence alignments used to build models was the most important factor affecting the overall performance of profile HMMs. The SAM T99 procedure is needed to produce high quality alignments automatically, and the lack of an equivalent component in HMMER makes it less complete as a package. Using the default options and parameters as would be expected of an inexpert user, it was found that from identical alignments SAM consistently produces better models than HMMER and that the relative performance of the model-scoring components varies. On average, HMMER was found to be between one and three times faster than SAM when searching databases larger than 2000 sequences, SAM being faster on smaller ones. Both methods were shown to have effective low complexity and repeat sequence masking using their null models, and the accuracy of their E-values was comparable. It was found that the SAM T99 iterative database search procedure performs better than the most recent version of PSI-BLAST, but that scoring of PSI-BLAST profiles is more than 30 times faster than scoring of SAM models.
INTRODUCTION
Protein sequence homology detection is a central tool in genomics. The ability to infer a relationship between two proteins of known amino acid sequence using computers and without laboratory experimentation gives valuable information about many of the large and rapidly growing number of protein sequences.
Many related proteins have similar sequences, making their relationship easy to detect, but equally as many have diverged to a point where their structural and functional similarity is hard to detect from purely sequence-based data. Homology detection methods vary in their ability to detect some of these more distant relationships. It has been shown by Park et al. (1) that profile-based methods, which consider profiles of protein families, perform much better than pairwise methods, which consider individual protein sequences, and that of the profile-based methods hidden Markov models (HMMs) (2,3) perform best. A more recent study by Lindahl and Elofsson (4) confirmed the relative performance of pairwise and profile methods and showed, further, that at the family and superfamily levels profile methods are also superior to threading methods as represented by THREADER (5).
Currently there are two popular profile HMM software packages: HMMER (6) and SAM (7,8). Little is known about their relative performance or the features that are important for their success. The work described here focuses on these issues. Because of the large number of parameters affecting the performance of profile HMMs the approach taken was to use the default settings wherever possible, making the results representative of what an informed but inexpert user might achieve. It should be noted, however, that an expert user may be able to obtain an improved performance with either method by fine tuning the parameters and conditions (9). Great care was taken to compare the methods without bias.
Our results differ from those of other studies (4,10) that have used profile HMMs. This is discussed in detail in a separate section towards the end of this paper.
The general organisation of this paper is as follows. First we introduce the profile HMM procedure and the two packages, HMMER and SAM. Then we describe the tests we chose to measure their performance and explain the technical aspects of the comparison. Next we discuss the results and put the performance of profile HMMs into a broader context of other sequence-based methods, including PSI-BLAST. Finally, we compare our results to those of the previous studies, as mentioned above.
THE PROFILE HMM PROCEDURE
The operation of pairwise sequence comparison methods (e.g. simple BLAST, http://www.ncbi.nlm.nih.gov/BLAST, http://blast.wustl.edu) essentially consists of a single step: the program takes two sequences and calculates a score for their optimal alignment; this score may then be used to decide whether the two sequences are related. The use of profile HMMs in homology detection is more complicated because of the need to first construct the profile HMM. The procedure consists of three steps. (i) A multiple sequence alignment is made of known members of a given protein family. The quality of the alignment and the number and diversity of the sequences it contains are crucial for the eventual success of the whole procedure. (ii) A profile HMM of the family is built from the multiple sequence alignment. The model-building program uses information derived from the alignment together with its prior knowledge of the general nature of proteins. (iii) Finally, a model-scoring program is used to assign a score with respect to the model to any sequence of interest; the better the score, the higher the chance that the query sequence is a member (homologue) of the protein family represented by the model. In this way each sequence in a database can be scored to find the members of the family present in the database.
This assessment compares the performance of the model-building (step 2) and model-scoring (step 3) programs in the two packages. The performance is measured by the ability to detect members of a protein family in sequence databases given a multiple sequence alignment for that family. A variety of alignments were used in this work, some of which might be expected to be more suitable for one package, some for the other, and some about which there were no expectations. A detailed explanation of our tests is provided later in the paper.
THE TWO PROFILE HMM PACKAGES
In this section we briefly introduce the two profile HMM packages assessed in this work. The technical issues associated with their comparison are discussed later.
HMMER
Developed chiefly by Sean Eddy, the HMMER package (6; http://hmmer.wustl.edu) is freely available under the GNU General Public License and includes the necessary model-building and model-scoring programs relevant to homology detection. In addition, the package contains a program that calibrates a model by scoring it against a set of random sequences and fitting an extreme value distribution to the resultant raw scores; the parameters of this distribution are then used to calculate accurate E-values for sequences of interest. All HMMER models used in this study were calibrated in this way.
In this comparison version 2.2g of the package was used, which was released in August 2001.
SAM
Developed by the bioinformatics group at the University of California, Santa Cruz, the SAM package (7,8; http://www.cse.ucsc.edu/research/compbio/sam.html) is not open source, but it is free for academic use and the authors retain no rights over the models produced with the software. The package contains the necessary model-building and model-scoring programs as well as several scripts for running them. In particular, the fw0.7 script recommended in the documentation was used for all SAM model building; for model scoring the relevant program was used directly. Unlike HMMER, the package does not include a model-calibration program. The SAM model-scoring program calculates E-values directly using a theoretical function that takes as its argument the difference between raw scores of the query sequence and its reverse.
Perhaps the most important component of the SAM package is the target99 script (8) commonly known as T99, which automatically generates a multiple sequence alignment suitable for model building. The script takes as its input a single seed sequence or an initial alignment and iteratively searches a sequence database in a manner similar to PSI-BLAST (11). T99 is not under direct assessment here due to the lack of an equivalent component in the HMMER package.
Version 3.2 of SAM was used, released in July 2000.
REMOTE HOMOLOGY DETECTION TESTS
We subjected the profile HMM methods to two tests, which were to some extent complementary in their nature. We first give an overview of the tests, highlighting the key points and contrasting the differences, and then describe them in technical detail. The two tests were as follows. (i) A test against nrdb90 (12), a non-redundant database of all known protein sequences. The assessment was restricted to just two protein families (globins and cupredoxins) because of the need to classify all hits by hand, but in return we were able to explore a wide variety of multiple sequence alignments from which to build the HMMs, ranging from fully automated to expert curated and from purely sequence-based to structural. Because only two protein families were investigated, the large size of the database (in particular the fact that it contained many homologues of the two families) was crucial for statistical significance of the results. (ii) An all-against-all match using a database of approximately 3000 proteins of known structure. For each member of the database an alignment of homologous proteins was created using fully automated methods with single sequence inputs, and models created from these alignments were scored against the original database. Evolutionary relationships of all proteins in the test set were provided by the SCOP (13) database, allowing for automatic classification of the results. Although limited to a small number of alignment methods, this test was very comprehensive, as representatives from every protein family of known structure were included in the database.
We also briefly investigated the low complexity masking capabilities of the two methods and their computing time requirements. The protocols for these are described together with our findings in the Results section.
Next we describe the two remote homology detection tests in detail.
Test 1: The globin and cupredoxin families
A variety of models representing the globin and cupredoxin families, as defined by the SCOP (13) family ‘Globins’ and the superfamily ‘Cupredoxins’, were searched against the nrdb90 database (12). All hits were then classified as either true, false or uncertain based on the following criteria. (i) Database annotation. Globins and cupredoxins were chosen for this test because they are well known families with reliable annotations in the databases. (ii) Pairwise comparisons with well annotated homologues. For poorly annotated sequences with highly significant pairwise matches to well annotated homologues the annotations from the well annotated homologues were used as above. (iii) Structural understanding of the two families. Members of our group have carried out detailed structural and key residue analyses of the two families (14; J.Gough, unpublished results) and this knowledge was used in a number of cases. (iv) All hits about which clear-cut decisions could not be reached were classified as uncertain. The classification is available on our website (http://stash.mrc-lmb.cam.ac.uk/HMMER-SAM/).
The models used in this test were built from a large number of alignments listed below. Each alignment was used with both packages and the results were compared; the T99 alignments were either re-formatted or re-aligned beforehand (see the subsection on input alignments later in the paper).
The manual alignments used in test 1 were: (i) a revised version of the alignment (15) of selected members of the globin family based on a detailed structural analysis (denoted G-STR); (ii) the full PFAM globin alignment (16) (denoted G-PFAM); (iii) an alignment of the cupredoxin family (J.Gough, unpublished results) similar to Bashford et al. (15) (denoted C-STR). Here G stands for globins and C for cupredoxins, STR for structural and PFAM for a PFAM alignment. Each of these alignments was also used as input for the SAM T99 producedure; the resultant T99 alignments are called G-STR-T99, G-PFAM-T99 and C-STR-T99.
In addition, a single representative member of each family (1rse for globins and 1plc for cupredoxins) was used as a seed for T99; the resulting T99 alignments are denoted G-1S-T99 or C-1S-T99, where 1S stands for single sequence input. Finally, the same two sequences were used in a procedure comprised of a WU-BLAST (http://blast.wustl.edu, version 2.0a19) search of nrdb90, followed by a ClustalW alignment of hits with a P-value score better than 1 × 10–5; these are denoted G-1S-BL&CLW and C-1S-BL&CLW.
Test 2: the SCOP all-against-all
The SCOP database (13; http://scop.mrc-lmb.cam.ac.uk/scop/index.html) provides an accurate and reliable classification of all proteins of known structure based on structural, functional and sequence evidence. As many of the relationships between proteins that are clear at the structural and functional levels are difficult to detect at the sequence level it represents a hard test set for assessment of sequence comparison methods. Because it covers all proteins of known structure it is also very comprehensive. Starting with Brenner et al. (17) it has indeed been used in a number of previous studies of this type (1,4). The sequence set filtered to 40% sequence identity (which ensures that only remote homologues are present and hence that the test is suitably difficult) is provided by the ASTRAL compedium (18). Version 1.50 of the database was used and the test set consisted of 2873 sequences.
Taking each sequence in the set as a seed, two automatic methods were used to create multiple sequence alignments: T99 (1S-T99) and a WU-BLAST search of nrdb90 followed by a ClustalW alignment (1S-BL&CLW). These were identical to the corresponding methods used in test 1, in particular T99 was iterated on nrdb90. Models generated from these alignments were then scored against all seed sequences. Hits from all models were pooled, sorted by E-value and classified as either true, false or uncertain according to their SCOP classification. Hits to the same superfamily as the model were classified as true, hits to the same fold as uncertain and hits to a different fold as false. Two significant exceptions to this general scheme were that: cross-hits between the Rossmann and Rossmann-like folds (3.2, 3.3 and 3.4 in the 1.50 classification) were considered uncertain, and hits to the seed sequence from which the particular model was built were ignored altogether as too easy. Using these criteria there was a total of 36 612 possible true and 8 173 744 false pairwise relationships.
TECHNICAL ASPECTS OF THE COMPARISON
There are two technical aspects of the comparison between the two packages that are important for understanding the results: our model conversion procedure and a difference in the nature of multiple sequence alignments used for model building. A further aspect, the search mode, is needed for reproducibility of our results. This section is devoted to an explanation of these issues.
Input alignments: an important difference
The SAM model-building program distinguishes between two types of alignment columns: the aligned residues, marked by upper case letters or - characters for deletions, and unaligned insertions, with lower case letters or . characters (see Table 1 for illustration). The program strictly follows this convention and the number of aligned upper case columns in the multiple sequence alignment is therefore always equal to the number of segments in the final model. In contrast, in a HMMER-style alignment all columns are supposed to be aligned, though the HMMER model-building program treats the most divergent regions as insertions. With the important exception of alignments produced by the SAM T99 procedure, none of the alignments used in this assessment differentiated between aligned and unaligned regions.
Table 1. A comparison of alignment conventions.
| HMMER alignment convention | |
| 1aac | ---EAALKGPMMKKEQAY--SLTFTE----AG-TYDYHCTP--H--PFMR |
| 1bqk | ---DGAEA.FKSKINENY--KVTFTA----PG.VYGVKCTP--HYGMGMV |
| 2cbp | ---STCNTPAGAK---VY--TSGRDQIKLPKGQSY-FICNFPGHCQSGMK |
| 1nwp | VIAHTKVIGAGEK--DSV--TFDVSKLA--AGEKYGFFCSFPGHI-SMMK |
| 1rcy | -GTGFSPVPKDGK--FGYTDFTWHPT----AG-TYYYVCQIPGHAATGMF |
| SAM alignment convention | |
| 1aac | .--EAALKGPMMKKEQAY..SLTFTE....AG.TYDYHCTP..H..PFMR |
| 1bqk | .--DGAEA-FKSKINENY..KVTFTA....PG.VYGVKCTP..HygMGMV |
| 2cbp | .--STCNTPAGAK---VY..TSGRDQiklpKGqSY-FICNFpgHcqSGMK |
| 1nwp | vIAHTKVIGAGEK--DSV..TFDVSKla..AGeKYGFFCSFpgHi.SMMK |
| 1rcy | .GTGFSPVPKDGK--FGYtdFTWHPT....AG.TYYYVCQIpgHaaTGMF |
Note that SAM distinguishes between two types of columns: aligned (upper case residue or - for deletions) and unaligned (lower case residues and .). A SAM-style alignment therefore contains more information than a HMMER-style one.
When dealing with T99 alignments, which follow the SAM convention, our principal objective was to deny SAM any information not available to HMMER. This was achieved in one of two ways: by simply converting all letters to upper case and . characters to - (these alignments are called T99-UC) or by completely realigning the sequences with ClustalW (19), a popular multiple sequence aligment program (T99-CLW). (To counteract poisoning due to inserted domains we removed all insertions longer than 30 residues prior to realignment with ClustalW.) The situation was therefore entirely analogous to that for other alignments.
We also used the intact T99 alignments with SAM so that the effects, if any, of the above conversions could be seen (the alignments are called simply T99). Finally, it is possible to pass the SAM aligned–unaligned distinction to the HMMER model-building program in a non-default way, via the ‘--hand’ option in combination with the SELEX format. We wrote a program (a2m2selex.pl) to facilitate the conversion, available on our website (http://stash.mrc-lmb.cam.ac.uk/HMMER-SAM/). This method was only used in test 2, and T99 alignments used in this way are called T99HAND.
Conversion between HMMER and SAM models
In order to be able to compare the model-building and model-scoring components of the two packages separately, we wrote a program (convert.pl) to convert between HMMER and SAM model files, available from our website (http://stash.mrc-lmb.cam.ac.uk/HMMER-SAM/). Each alignment used in this assessment therefore gave rise to four sets of results: two using the default procedures for each package (denoted HH for HMMER and SS for SAM) and two where models built by one package were converted using our program and scored by the other package (denoted HS for HMMER-built models converted to the SAM format and scored by SAM, and vice versa for SH). In this way models produced by each program were scored independently by both model-scoring programs and, conversely, both model-scoring programs were assessed on essentially the same models.
Due to a difference in model topologies there was a small loss of information when converting from SAM models to HMMER ones, but no information was lost the other way; in particular, using the script to convert from the HMMER format to the SAM format and back again resulted in a file identical to the initial one. See our website for further information.
The local/local search mode was used
There are two popular ways of using profile HMMs: by forcing a global alignment to the model and a local one to the query sequence (the domain or global/local mode) or by using the local mode for both. (By global alignment we mean the mode whereby a match is forced to each segment of the model or every residue of the query sequence, as opposed to the local mode where only the best scoring region is considered.)
Which approach is better in a real application is debatable and dependent on the problem at hand. In our experience the local/local mode is the one better suited for genome annotations because it is more robust with respect to inserted domains and gene prediction errors. In test 2 each sequence in the test set is hand curated to represent a single complete domain in the corresponding protein structure. As a result, the global/local mode performs significantly better on this test, but this is not representative of real sequence databases.
In order to compare both methods under realistic conditions, we used the local/local mode for both methods in all of our tests.
PROFILE HMM RESULTS
Model building
In all the cases we investigated SAM models performed better than HMMER models when both were scored with the same program, i.e. SAM models converted to the HMMER format (SH) were better than native HMMER models built from the same alignment (HH), and HMMER models converted to the SAM format (HS) were worse than native SAM models (SS) (see Table 2A and Fig. 1 for illustration).
Table 2a. The numbers of true homologues found in tests 1 and 2, at 1% error rate, for a selected number of methods, concentrating on comparisons of model building and model scoring.
| Alignment | Model building and scoring method | |||
|---|---|---|---|---|
| HH | HS | SH | SS | |
| (i) | ||||
| C-STR | 117 | 113 | 129 | 116 |
| G-PFAM | 557 | 590 | 560 | 593 |
| G-PFAM-T99-CLW | 554 | 549 | 560 | 554 |
| (ii) | ||||
| 1S-BL&CLW | 5536 | 5247 | 5929 | 5965 |
| 1S-T99-CLW | 6620 | 7205 | 7559 | 8128 |
| 1S-T99, 1S-T99HAND |
7414 |
7813 |
8283 |
8784 |
| (iii) | Number of nrdb90 iterations | |||
| |
0 |
3 |
30 |
|
| PSI-BLAST | 4330 | 7897 | 7462 | |
(i) Results for test 1; (ii) for test 2; (iii) results of test 2 applied to PSI-BLAST (the case with zero nrdb90 iterations being pairwise NCBI BLAST). G stands for globins, C for cupredoxins, 1S for a single-sequence seed; PFAM indicates a PFAM alignment, STR a structural alignment, G-1S is the sequence of 1rse; T99 is the SAM T99 procedure, either default (T99), with all columns in the resultant alignment converted to upper case (T99-UC), or realigned with ClustalW (T99-CLW). Finally, HH and SS are the default procedures for HMMER and SAM, respectively, HS indicates a HMMER model converted to the SAM format and scored by SAM, and vice versa for SH. See text for further explanations.
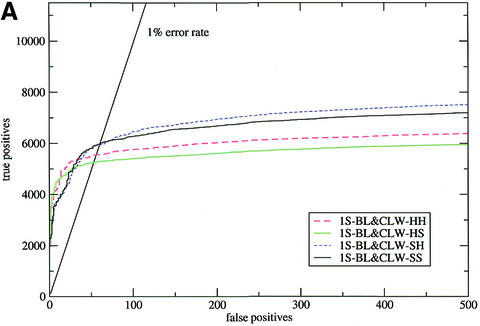
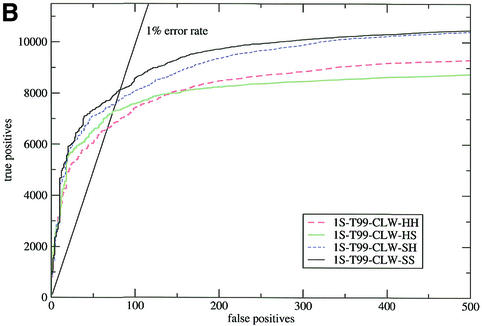
Figure 1.
Sensitivity plots for the SCOP all-against-all (test 2). The input alignments were: (A) the results of a WU-BLAST search of nrdb90 aligned with ClustalW; (B) T99 alignments realigned with ClustalW. In both figures, HH and SS are the default procedures for HMMER and SAM, respectively, HS indicates a HMMER model converted to the SAM format and scored by SAM, and vice versa for SH.
Furthermore, in all cases bar one this was true across all error rates. The single exception was the case of WU-BLAST followed by ClustalW (1S-BL&CLW in test 2; Fig. 1A) where HMMER-built models performed better at very low error rates (<0.5%). This behaviour was caused by a small number of ‘poisoned’ alignments, in which some of the sequences contained parts of domains from other superfamilies. When these alignments (<1% of the total) were removed from the test set SAM performed better across all error rates. HMMER model building was less susceptible to this type of error because the columns that contained poisoning sequences also tended to be poorly aligned and thus were averaged as insertions (see the earlier subsection on alignments).
Overall, this means that SAM consistently produces better models than HMMER, even though it is less robust with respect to poisoned alignments that do not follow its distinction between aligned and unaligned columns.
Model scoring
SAM scoring (HS and SS) performed better on high quality, diverse alignments; it did so overwhelmingly on both manual globins alignments (structural and PFAM, or G-STR and G-PFAM; data not shown for G-STR) and up to a certain error rate also on the T99 alignments in test 2 (1S-T99 and 1ST99HAND), but on our structural cupredoxin alignment (C-STR) HMMER scoring performed better. Similarly, while HMMER scoring was better for the hand curated PFAM alignments run through T99 and realigned with ClustalW (G-PFAM-T99-CLW), SAM scoring was better when the same procedure was applied to our hand curated structural alignment (G-STR-T99-CLW; data not shown) (see Table 2A and Fig. 1 for illustration).
As the above difficulties illustrate, we have not managed to extract any definitive rules governing the relative performance of the model-scoring programs, even though it was almost always the case that one of them performed better than the other on both sets of models.
Multiple sequence alignments
The previous two subsections show that SAM builds better models than HMMER and that the relative performance of the model-scoring components varies. However, often the most important factor affecting the overall performance was not the particular profile HMM program, but rather the input alignment used.
As can be seen in Table 2B, the best-performing alignment for globins was the PFAM alignment (G-PFAM), but the fully automated SAM-T99 procedure seeded from a single member of the family (G-1S-T99) with 433 sequences in the final alignment came second and performed better than the manual structural alignment consisting of only 12 sequences (G-STR). Running the PFAM alignment through the T99 procedure (G-PFAM-T99) worsened performance, though for the structural alignment this did lead to a marginal improvement at larger error rates (G-STR-T99). Realigning the T99 alignment with ClustalW resulted in a loss of performance in both cases (G-PFAM-T99-CLW and G-STR-T99-CLW) and this loss was greater than that from merely removing the aligned–unaligned distinction (G-PFAM-T99-UC and G-STR-T99-UC). We nevertheless chose ClustalW for test 2 as the difference between the HMMER and SAM models was smaller using it. The results for cupredoxins were similar. In test 2, using ClustalW to realign T99 alignments (1S-T99-CLW) again resulted in a ∼10% drop in performance.
Table 2b. The numbers of true homologues found in tests 1 and 2, at 1% error rate, for a selected number of methods, comparing suitability for model building of globin alignments in test 1.
| Method | Number of hits |
|---|---|
| G-PFAM-SS | 593 |
| G-1S-T99-SS | 582 |
| G-PFAM-T99-SS | 573 |
| G-STR-SS | 567 |
| G-STR-T99-SS | 567 |
| G-PFAM-T99-UC-SS | 565 |
| G-STR-T99-UC-SS | 561 |
| G-PFAM-T99-CLW-SS | 554 |
| G-STR-T99-CLW-SS | 548 |
G stands for globins, C for cupredoxins, 1S for a single-sequence seed; PFAM indicates a PFAM alignment, STR a structural alignment, G-1S is the sequence of 1rse; T99 is the SAM T99 procedure, either default (T99), with all columns in the resultant alignment converted to upper case (T99-UC) or re-aligned with ClustalW (T99-CLW). See text for further explanations.
To sum up, it is clear that a good alignment for use with profile HMM remote homology detection procedures needs to include a large number of diverse sequences, correctly aligned and with unalignable portions either excised or appropriately marked. The only way of producing such alignments is via an iterated database search, so the lack of a T99 equivalent in the HMMER package makes it less useful for an inexpert user.
E-value accuracy
The E-value is defined as the expected number of errors per query. In the context of test 2 this means that one would expect on average E false positives per model with an E-value score better than E to occur by chance. For the 2873 models used in test 2 one would therefore expect about 29 false positives with an E-value score better than 0.01 when results for all models are pooled together.
For pairwise methods with accurate E-value scoring schemes, e.g. NCBI BLAST, this is indeed what one gets (results not shown). Unfortunately matters are more complicated for profile methods, because of model poisoning already alluded to in the subsection on model building (also discussed in 9). Poisoned models systematically assign highly significant scores to sequences from poisoning superfamilies and thus greatly inflate false positive counts at low E-values. As a result, the customary plots of false positive counts against E-value (1,17) are dominated by poisoning and therefore say little about E-value accuracy for unpoisoned models.
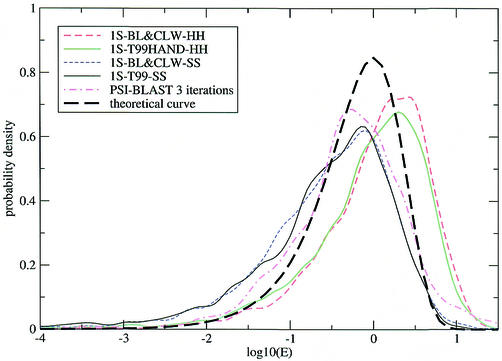
To circumvent this problem we plotted the distribution of E-values of the first false hit for each model (Fig. 2). It can be seen that the distributions for both profile HMM methods are in reasonably good agreement with the theoretical curve, although HMMER scoring is somewhat conservative and SAM scoring somewhat optimistic. HMMER E-values are more accurate in the crucial region around E = 0.01, presumably because the extreme value distribution function is being fitted there, whereas SAM E-values (based on a theoretical calculation) only become accurate in the high-E limit.
Figure 2.
Distribution of E-values E of first false positives in test 2. The probability density is with respect to the log10(E) x-axis. The experimental curves are smoothed (each model was added as a Gaussian of standard deviation 0.1 and area 2873–1), the theoretical curve is ln(10) E exp(–E). 1S-BL&CLW is the result of a WU-BLAST search of nrdb90 aligned with ClustalW, 1S-T99 the alignment produced by the T99 procedure; HH and SS are the default procedures for HMMER and SAM, respectively.
The numbers of models with first false positive below the 1 × 10–5 E-value level, indicative of the overall level of model poisoning for the particular alignment procedure, were 15–35 for WU-BLAST followed by ClustalW (1S-BL&CLW) and 100–120 for T99 (1S-T99-Sx, 1S-T99HAND-Hx and 1S-T99-CLW). All four combinations of model building and model scoring for the particular alignment procedure fitted within each range.
Low complexity masking
One aspect insufficiently covered by the comprehensive test 2 is the ability of the methods to deal with low complexity sequences, because of their absence from the PDB (14). As low complexity sequences are known to cause problems for sequence comparison methods, and because the HMMER ‘null2’ and the SAM ‘reverse null’ null models are very different, we chose 100 models from different SCOP folds at random and scored them against three databases of low complexity sequences with roughly 2000 sequences each. The databases were constructed as follows: (i) the program seg (20) was used to extract long sections of sequences from nrdb90; (ii) PDB sequences filtered to 30% identity were all reversed; (iii) PDB sequences filtered to 30% identity were all randomly shuffled.
WU-BLAST was also included in this test, to provide a comparison with pairwise methods. The results are summarized in Table 3; it is clear that unlike WU-BLAST, both profile HMM methods make very few hits to either database and therefore possess in their null models effective low complexity masking systems.
Table 3. Hits to low complexity databases.
| Database | Number of hits by each method | ||
|---|---|---|---|
| HMMER | SAM | WU-BLAST | |
| 1. seg | 0 | 0 | 33 |
| 2. reversed | 1 | 1 | 11 |
| 3. shuffled | 1 | 2 | 0 |
Computer time
To measure the speed of profile HMM programs, we recorded the times taken to build and calibrate 100 models and to score them against our test 2 dataset of 2873 sequences. It should be noted that each sequence in this dataset is a single protein domain and that the sequences are therefore significantly shorter than those in other databases. The test was performed on a computer with a single 1.3 GHz AMD Athlon processor and 768 MB of RAM running the Linux operating system (kernel version 2.4.2-2).
As shown in Table 4, model building is quick using both methods, except for HMMER model calibration, which is slow. Model scoring was on average 3.4 times faster using HMMER, but the additional need to calibrate all models means that HMMER is only faster for databases of more than approximately 2000 sequences; for smaller databases SAM is faster. This difference, although significant, will not affect the feasibility of large-scale experiments.
Table 4. Computer times per model against our test 2 dataset.
| Task | Average time (s) |
|---|---|
| HMMER model building | 0.4 |
| HMMER model calibration | 52.6 |
| SAM model building | 1.1 |
| HMMER model scoring | 17.1 |
| SAM model scoring | 58.9 |
| PSI-BLAST profile scoring | 1.6 |
COMPARISON TO OTHER METHODS
In order to see the performance of HMMER and SAM in the context of other sequence comparison methods, we re-ran test 2 on PSI-BLAST (11), the most popular iterative profile method, and WU-BLAST (http://sapiens.wustl.edu/blast), one of the best pairwise methods in Brenner et al. (17). Version 2.2.1 of PSI-BLAST was used, last updated in August 2001. This version included the improvements described in Schaefer et al. (21).
PSI-BLAST was first iterated on nrdb90 using each sequence in the test set as a seed, with a maximum of either three or 30 iterations. The resulting models were saved and run against our test set in a single iteration. In addition, we used one iteration of the PSI-BLAST binary (blastpgp) directly on our test set. This run is referred to as NCBI BLAST, since the underlying algorithm is a simple pairwise gapped BLAST.
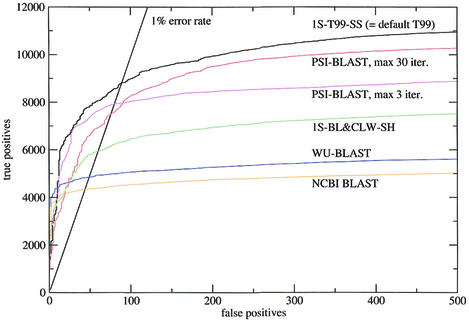
As shown in Figure 3, profile methods perform considerably better than pairwise methods (as represented by WU-BLAST and NCBI BLAST) and SAM-T99 is better than PSI-BLAST. The relative performance has remained approximately the same since the work of Park et al. (1), which was carried out on a smaller dataset, but the overall coverage has dropped: in our study, SAM T99 found 24% of the total number of possible true hits at the 1% error rate; in Park et al. (1) the corresponding figure for SAM-T98 was 34%.
Figure 3.
Sensitivity plots for the SCOP all-against-all. SCOP version 1.50 was used, filtered down to 2873 sequences of less than 40% sequence identity, with a total of 36 612 possible true pairwise relationships. See the text for further details.
Although SAM T99 detects ∼10% more true homologues than PSI-BLAST, profile HMM methods in general and SAM in particular are considerably slower (see Table 4): HMMER model scoring is 11 times, and SAM model scoring is 37 times slower than PSI-BLAST profile scoring. [For large databases PSI-BLAST appears to be even faster, by up to an order of magnitude, presumably due to indexing, which the profile HMM methods do not use. On the other hand, the SAM T99 procedure iterated on a large database does not actually search the entire database, but only a restricted set of WU-BLAST hits to the seed sequence (see 8 for details).]
E-values produced by PSI-BLAST appear to be more accurate than those of profile HMM methods (see Fig. 2).
DISCUSSION OF PREVIOUS ASSESSMENTS
There are two previous studies (4,10) that include some comparison of the two profile HMM packages. Lindahl and Elofsson (4) is a systematic benchmark, while Rehmsmeier and Vingron (10) use the two packages to demonstrate an improvement due to a novel approach.
Lindahl and Elofsson (4) found SAM to be better than HMMER at the superfamily level and vice versa at the family level. While these findings are in agreement with our results at the superfamily level, the authors used the global/global mode for HMMER and a local/local one for SAM (A.Elofsson, personal communication; see also our discussion earlier in the paper). It should be noted that the SCOP test strongly and artificially favours methods using the global alignment mode, because each sequence in the test set is hand curated to represent a single complete domain. Lindahl and Elofsson (4) also experienced difficulties running SAM and were often unable to produce error rates of <5%. These factors lead us to believe that the true performance of the SAM package at both family and superfamily levels is the same or better than that suggested (4), while the true HMMER performance is almost certainly worse.
Rehmsmeier and Vingron (10) found that HMMER performs better than SAM. They tested the two methods under substantially different conditions, which are less realistic for database searching: they considered the numbers of false positives for each model before the first true hit, regardless of E-value, and examined cases with up to 100 false positives. Under these circumstances we do not think a comparison between the two sets of results should be attempted.
In summary, we have reasons to believe that neither of the previous comparisons provides an accurate picture of the relative performance of HMMER and SAM.
CONCLUSIONS
We have examined the performance of two profile HMM packages for detection of remote protein homologues, HMMER and SAM. It is clear from our results that the most significant distinction between the two packages is the SAM T99 script. Not only do both packages require a multiple sequence alignment from which to build a model, but their performance is directly related to the quality of that alignment. The T99 script automatically produces high quality multiple alignments well suited for building HMMs, but so far there is no equivalent in the HMMER package.
In order to compare the model-building and model-scoring components of the two packages independently, we wrote a program to convert between the model formats used by the two packages. We found that SAM consistently produces better models than HMMER, i.e. the SAM model built from a given alignment and converted to the HMMER format using our script performs better than the native HMMER model built from the same alignment. The relative performance of the model-scoring components varies.
The E-value scores produced by both methods have similar reliability. If HMMER model calibration is included, then HMMER scoring is faster for searches of more than 2000 sequences, SAM being faster for smaller ones. This difference does not affect the feasibility of large-scale studies.
Comparing the performance of HMMs to other methods, the relative results on a SCOP all-against-all test (our test 2) are similar to those obtained by Park et al. (1), namely SAM T99 is somewhat better than PSI-BLAST and the pairwise methods perform poorly in comparison. We found PSI-BLAST scoring to be more than 10 times faster than HMMER scoring (excluding model calibration).
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Cyrus Chothia for many helpful discussions, and Bernard de Bono, Rajkumar Sasidharan, Cyrus Chothia and an anonymous referee for many helpful comments on the manuscript.
REFERENCES
- 1.Park J., Karplus,K., Barrett,C., Hughey,R., Haussler,D., Hubbard,T. and Chothia,C. (1998) Sequence comparisons using multiple sequences detect three times as many remote homologues as pairwise methods. J. Mol. Biol., 284, 1201–1210. [DOI] [PubMed] [Google Scholar]
- 2.Krogh A., Brown,M., Mian,S., Sjolander,K. and Haussler,D. (1994) Hidden Markov models in computational biology. J. Mol. Biol., 235, 1501–1531. [DOI] [PubMed] [Google Scholar]
- 3.Eddy S.R. (1995) Hidden Markov models. Curr. Opin. Struct. Biol., 6, 361–365. [DOI] [PubMed] [Google Scholar]
- 4.Lindahl E. and Elofsson,A. (2000) Identification of related proteins on family, superfamily and fold level. J. Mol. Biol., 295, 613–625. [DOI] [PubMed] [Google Scholar]
- 5.Jones D.T., Taylor,W.R. and Thornton,J.M. (1992) A new approach to protein fold recognition. Nature, 358, 86–89. [DOI] [PubMed] [Google Scholar]
- 6.Eddy S.R. (1998) Profile hidden Markov models. Bioinformatics, 14, 755–763. [DOI] [PubMed] [Google Scholar]
- 7.Hughey R. and Krogh,A. (1996) Hidden Markov models for sequence analysis. Extension and analysis of the basic method. Comput. Appl. Biosci., 12, 95–107. [DOI] [PubMed] [Google Scholar]
- 8.Karplus K., Barrett,C. and Hughey,R. (1999) Hidden Markov models for detecting remote protein homologies. Bioinformatics, 14, 846–856. [DOI] [PubMed] [Google Scholar]
- 9.Gough J., Karplus,K., Hughey,R. and Chothia,C. (2001) Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol., 313, 903–919. [DOI] [PubMed] [Google Scholar]
- 10.Rehmsmeier M. and Vingron,M. (2001) Phylogenetic information improves homology detection. Proteins, 45, 360–371. [DOI] [PubMed] [Google Scholar]
- 11.Altschul S.F., Madden,T.L., Schaeffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holm L. and Sander,C. (1998) Removing near-neighbour redundancy from large protein sequence collections. Bioinformatics, 14, 423–429. [DOI] [PubMed] [Google Scholar]
- 13.Murzin A.G., Brenner,S.E., Hubbard,T. and Chothia,C. (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol., 247, 536–540. [DOI] [PubMed] [Google Scholar]
- 14.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bashford D., Chothia,C. and Lesk,A.M. (1987) Determinants of a protein fold; unique features of the globin amino acid sequences. J. Mol. Biol., 196, 199–216. [DOI] [PubMed] [Google Scholar]
- 16.Bateman A., Birney,E., Durbin,R., Eddy,S.R., Howe,K.L. and Sonnhammer,E.L. (2000) The Pfam protein families database. Nucleic Acids Res., 28, 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brenner S.E., Chothia,C. and Hubbard,T. (1998) Assessing sequence comparison methods with reliable structurally identified distant evolutionary relationships. Proc. Natl Acad. Sci. USA, 95, 6073–6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brenner S.E., Koehl,P. and Levitt,M. (2000) The ASTRAL compendium for sequence and structure analysis. Nucleic Acids Res., 28, 254–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wootton J.C. and Federhen,S. (1993) Statistics of local complexity in amino acid sequences and sequence databases. Comput. Chem., 17, 149–163. [Google Scholar]
- 21.Schaefer A.A., Aravind,L., Madden,T.L., Shavirin,S., Spouge,J.L., Wolf,Y.I., Koonin,E.V. and Altschul,S.F. (2001) Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res., 29, 2994–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]