Abstract
The patterns of synonymous codon usage, both within and among genomes, have been extensively studied over the past two decades. Despite the accumulating evidence that natural selection can shape codon usage, it has not been possible to link a particular pattern of codon usage to a specific external selective force. Here, we have analyzed the patterns of synonymous codon usage in 40 completely sequenced prokaryotic genomes. By combining the genes from several genomes (more than 80 000 genes in all) into a single dataset for this analysis, we were able to investigate variations in codon usage, both within and between genomes. The results show that synonymous codon usage is affected by two major factors: (i) the overall G+C content of the genome and (ii) growth at high temperature. This study focused on the relationship between synonymous codon usage and the ability to grow at high temperature. We have been able to eliminate both phylogenetic history and lateral gene transfer as possible explanations for the characteristic pattern of codon usage among the thermophiles. Thus, these results demonstrate a clear link between a particular pattern of codon usage and an external selective force.
INTRODUCTION
The 20 amino acids that commonly occur in proteins are encoded by 61 different codons. This redundancy in the genetic code means that several ‘synonymous’ codons may encode the same amino acid. Consequently, one might argue that mutational changes affecting these codons would not be subject to natural selection, since the encoded protein sequence would be unaffected by such changes. A large body of indirect molecular evidence has accumulated, however, against such a simple assumption. First, it has been shown that different genomes each have their own characteristic patterns of synonymous codon usage (1). Secondly, and more convincingly, it has been shown that within genomes, highly expressed genes have shifted their codon usage toward a more restricted set of ‘preferred’ synonymous codons than other, less highly expressed genes (2–8). In many cases, it has been shown that codon usage mirrors the distribution of tRNA abundances (2–4), indicating that the ‘preferred’ codons are those that tend to match the more abundant anticodons. This correlation between the abundance of codons and their matching anticodons suggests that relative tRNA abundance is the selective force that determines synonymous codon usage (2–4). Although the relative tRNA abundances may well be the short-term determinants of codon usage, it has been suggested that over the course of long-term evolutionary change the tRNA abundances themselves may also evolve to match the genomic patterns of codon and nucleotide frequencies (9). In other words, it is not clear if, in the long term, the codon usage pattern is selected to match the relative abundances of the isoaccepting tRNAs or vice versa. In any case, there is strong evidence for a co-adaptation of the relative frequencies of codons and their respective anticodons within a genome. Despite this evidence, however, we cannot explain why a particular codon–anticodon combination might have a selective advantage over alternative synonymous codon–anticodon pairs that are also perfectly matched. Thus, although we have ample indirect evidence that a particular pattern of synonymous codon usage has biological significance, it is not as clear why that particular pattern is favored by selection in a given genome. Despite the accumulation of data on non-random patterns of synonymous codon usage, both between and within genomes, it has been difficult to identify an external selective force acting on synonymous codon usage.
Here, we present evidence that the pattern of synonymous codon usage within thermophilic prokaryotes is different from that within the mesophilic prokaryotes and that this difference is the result of natural selection linked to thermophily. Moreover, we show that this phenomenon affects all of the genes within the genome, that the pattern cannot be explained by a simple accident of phylogenetic history and that it is not due to horizontal gene transfer between mesophiles and thermophiles. This result indicates that natural selection acting through external environmental factors can indeed shape the genomic pattern of synonymous codon usage.
MATERIALS AND METHODS
We analyzed the patterns of synonymous codon usage in a total of 40 completely sequenced bacterial genomes (listed in Table 1). This set of genomes includes 32 eubacteria and eight archaea. Although the majority of the eubacterial species are mesophiles and the majority of the archaea are thermophiles, the list does include two eubacterial thermophiles (Aquifex aeolicus and Thermotoga maritima) and one mesophilic archaeal species (Halobacterium sp.). These three genomes have enabled us to distinguish between the effects of environmental selection and phylogenetic history.
Table 1. Total genomic G+C contents and optimal growth temperatures for the 40 genomes analyzed in our study.
Note that the variation in G+C contents is only weakly correlated with phylogenetic relatedness. Although the majority of thermophilic organisms are Archaea, there are two exceptions (A.aeolicus and T.maritima). In addition, there is one mesophilic archaeal species in our dataset (Halobacterium sp.).
In our analysis, we combined the genes from all 40 genomes (a total of 83 985 coding sequences) and calculated the relative synonymous codon usage (10) for each gene. We used correspondence analysis (11) to characterize the patterns of codon usage among this large set of genes and to map this pattern onto the distribution of codons on which the pattern is based (Fig. 1A and B). Correspondence analysis was carried out using the program CodonW1.4.2 (J. Peden, 2000; http://www.molbiol.ox.ac.uk/cu/). This ‘transgenomic’ analysis allowed us to gain information on both the intra-genomic and inter-genomic patterns of codon usage simultaneously. Moreover, it allowed us to directly compare the magnitude of the within-genome and between-genome variations in codon usage.
Figure 1.
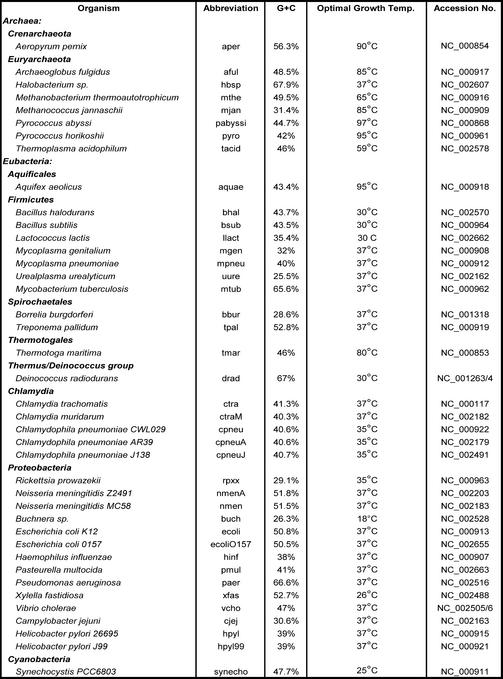
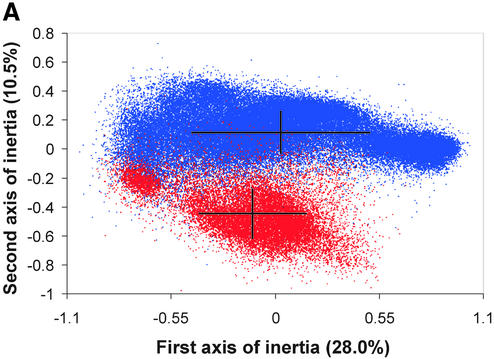
Correspondence analysis of the relative synonymous codon usage in 83 985 genes from 40 bacterial genomes (see Table 1). (A) Genes from thermophilic bacteria are shown in red while those from mesophilic bacteria are colored blue. For each of these two sub-groups, means and standard deviations are shown. Note that there is extensive overlap between thermophilic and mesophilic genes along the horizontal axis, whereas there is relatively little overlap between the two groups along the vertical axis. The difference between mesophiles and thermophiles along this latter axis is highly significant (P < 0.0001). (B) The distribution of synonymous codons along the first and second axes of the correspondence analysis. Here, we see that the GC-ending codons (shown in red) cluster to the right and the AT-ending codons (shown in green) cluster to the left.
RESULTS
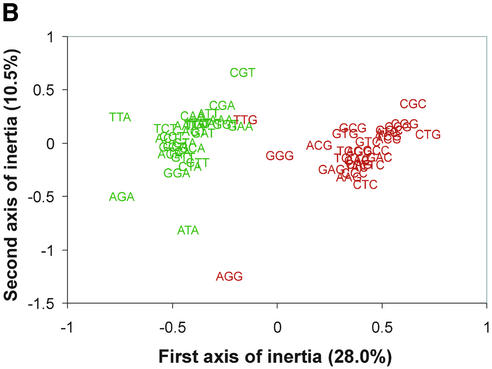
The genes from all 40 genomes were combined for the correspondence analysis of relative synonymous codon usage. Although all of the genes were combined, each gene could be identified in the output. Thus, we could, a posteriori, identify genes by genome or by type. For instance, in Figure 1, genes are identified based on whether they came from thermophilic or mesophilic species. Figure 1A shows the distribution of all of the genes on the first two axes of inertia of the correspondence analysis. Genes from thermophiles are shown in red, whereas those from mesophiles are shown in blue. Both mesophilic and thermophilic genes show a broad distribution along the horizontal axis (the first axis of inertia). It is clear from Figure 1A, however, that these groups of genes are significantly different with respect to their position along the vertical axis (the second axis of inertia). By looking at the corresponding distribution of codons (Fig. 1B), we see that the first axis of inertia is due to the separation of codons ending in A or T (shown in green) from codons ending in G or C (shown in red). Thus, the separation of genes along the horizontal axis is highly correlated with the overall G+C content of the genome to which they belong (Table 1). This can be seen more clearly in Figure 2, where we have grouped the genes by genome. Genes from GC-rich genomes, such as Mycobacterium tuberculosis and Pseudomonas aeruginosa cluster to the right of Figure 2, whereas genes from the AT-rich species, such as Methanococcus janaschii and Borellia burgdorferi, appear on the far left. Species with an intermediate G+C content, such as Escherichia coli and T.maritima, appear near the middle of the distribution. While variations in genomic G+C content explain most of the variation along the first axis of inertia, simple changes in nucleotide content do not explain the separation of the thermophilic and mesophilic genomes on the vertical axis. In Figure 2, it can be seen that all of the thermophilic genomes, including the two eubacterial thermophiles, are clearly separated along the second axis of inertia (vertical axis Fig. 2). Moreover, we can quantify this effect by comparing the position of each species on the second axis of inertia with its optimal growth temperature (see Table 1). The results of our regression analysis showed that this relationship is highly statistically significant (P << 0.00001). By examining the distribution of codons in Figure 1B, we can see that the major contributors to this pattern are the arginine (AGR and CGN) and isoleucine (ATH) codons, although many other codon groups also contribute to the separation between the thermophiles and the mesophiles (see Discussion below).
Figure 2.
Variation in codon usage within and between genomes. Genes shown are identified by genome and the means (±99.99% confidence intervals) for each genome are shown (the abbreviations for each organism are shown in Table 1). From this plot it is clear that the among-genome variation far exceeds the within-genome variation along both axes. Amongst the thermophiles we have circled the two eubacterial genomes (T.maritima and A.aeolicus). We have also circled the single mesophilic archaeal species (Halobacterium sp.).
Since the difference in synonymous codon usage between mesophiles and thermophiles is not due to a simple difference in the nucleotide content of the genomes, we investigated the possibility that it might be due to natural selection. To date, the best evidence for selection acting on codon usage among prokaryotes comes from the work of Ikemura and his colleagues, who demonstrated that highly expressed genes tend to have significantly different codon frequencies than other genes in the same genomes (2,3). Selection for optimal codon usage is not, however, the only evolutionary force acting on these genes. As stated by Gouy and Gautier (12), for each gene within the genome there is a balance between selection for optimal codons and other evolutionary forces such as mutation and genetic drift. These other forces are expected to affect all genes equally, whereas there is a predicted correlation between the strength of selection and the level of expression of each gene. Thus, although all genes are subject to some degree of selection, it is only among the most highly expressed genes that selection is strong enough to constitute the dominant evolutionary force (12). This, in turn, leads to a testable hypothesis: it has been proposed that if selection is the underlying cause of synonymous codon usage bias, then the bias should be more pronounced in the highly expressed genes than in the rest of the genome (13). To test this prediction, we compared the average codon usage of all genes within a genome with the average for the ribosomal protein genes from the same genome (Fig. 3). Among the thermophiles, the highly expressed ribosomal protein genes had a more extreme value on the second axis of inertia (the vertical axis) for all nine species. The same was true for a majority of the mesophilic genomes as well. These trends were statistically highly significant (P = 1 × 10–7 for the mesophiles and P = 2.6 × 10–3 for the thermophiles in paired t-tests). Essentially, the data show that the force responsible for the difference in codon usage between thermophiles and mesophiles acts more strongly upon highly transcribed genes than other genes within the genome.
Figure 3.
Evidence for selection on synonymous codon usage. Synonymous codon bias in highly expressed genes. Each arrow represents one genome, with the base of the arrow at the mean position for the whole genome and the arrowhead ending at the mean for the ribosomal genes within that genome. Note that the arrows tend to point away from the division between the thermophiles and the mesophiles; in other words, the ribosomal protein genes have a greater degree of synonymous codon bias than the genomes as a whole.
The fact that the difference in codon usage between thermophiles and mesophiles is more pronounced in the highly expressed genes provides strong evidence for selection. Nevertheless, we wanted to eliminate the possibility that such a pattern was simply due to the fact that most of the thermophiles studied are Archaea rather than Eubacteria. We can eliminate phylogenetic history as an explanation because the two eubacterial thermophiles (T.maritima and A.aeolicus) show a typically thermophilic pattern of codon usage in both their genomes as a whole (Fig. 2) and in their highly expressed genes (Fig. 3). Likewise, the mesophilic archaeal species, Halobacterium, shows a typically mesophilic pattern of codon usage. It should be noted that three ‘exceptional’ genomes represent more than 1000 individual genes in this analysis. Therefore, we can conclude that the separation in codon usage is between thermophiles and mesophiles, and not between eubacteria and archaea.
One might still argue that the common pattern of codon usage in eubacterial and archaeal thermophiles could be explained by horizontal gene transfer between the archaea and the thermophilic eubacteria (14–16). Indeed, codon usage has been used as an indicator of gene transfer between bacterial lineages (16,17) and there is evidence for such transfers between thermophiles (14,18). We addressed this question in two ways. First, we used the concatenated amino acid sequences of 10 ribosomal genes in both distance-based and maximum likelihood phylogenetic analyses (Fig. 4). We deliberately chose ribosomal proteins since they are included among the class of highly expressed genes that show the most pronounced differences between thermophiles and mesophiles in the patterns of synonymous codon usage (Fig. 3). The results of the phylogenetic test show very clearly that the Halobacterium ribosomal protein sequences, despite their mesophilic pattern of synonymous codon usage, group with the other archaea based on the amino acid sequences of the encoded proteins. Likewise, the two eubacterial thermophiles group with the Eubacteria. This is consistent with other recent reports based on whole genome analyses of sequence data (19,20). In other words, the synonymous codon usage patterns of these ribosomal protein genes identify them unambiguously as mesophiles or thermophiles, while the amino acid sequences encoded by these same genes group them into a conventional taxonomic arrangement of Eubacteria and Archaea. This provides convincing evidence that the nucleotides at the synonymous sites have undergone convergent evolutionary change and it shows that the nature of this change is directly related to thermophily or mesophily.
Figure 4.
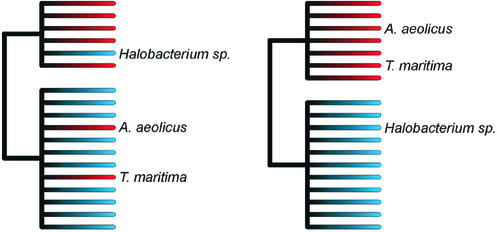
Summary of phylogenetic analyses based on the concatenated sequences of 10 ribosomal protein genes from each of the 40 genomes used in this study. Two possible phylogenetic groupings were compared. The grouping shown on the left side of the figure represents the accepted organismal tree. The tips of the branches are color coded (red for thermophiles and blue for mesophiles); it shows that both thermophiles and mesophiles are polyphyletic. The alternative hypothesis (shown on the right) describes the horizontal gene transfer hypothesis, whereby the thermophilic Eubacteria (A.aeolicus and T.maritima) would have acquired portions of their genomes, including the ribosomal protein genes, from thermophilic Archaea. In this case, the tree based on the ribosomal protein sequences would indicate that all thermophilic species would be monophyletic (again, as indicated by the color coding on the tips of the branches). Our results, using both distance-based and maximum likelihood methods of phylogenetic reconstruction (J. Felsenstein, PHYLIP v.3.6a2.1; http://evolution.genetics.washington.edu/phylip.html), greatly favor the first grouping (left) over the second (right), with 100% bootstrapping support.
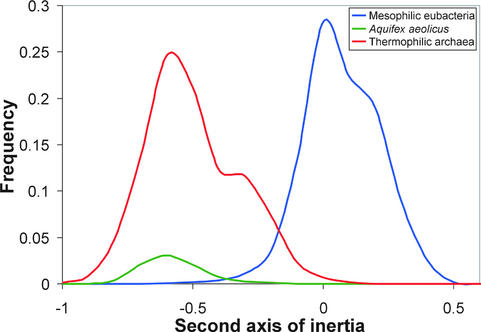
In addition to studying the ribosomal protein genes, we wished to do an independent and more general test of the possibility that horizontal gene transfer might be a significant factor in determining the codon usage of the thermophilic Eubacteria. For instance, we wondered if the genes within these genomes might show a bimodal distribution, reflecting the fact that a fraction of their genome had been derived from archaeal thermophiles by horizontal gene transfer. First, we plotted the frequency distributions for the values of the second axis of inertia for all genes from thermophiles and mesophiles (Fig. 5). This figure shows quite dramatically how separate both sets of genes are in terms of their codon usage. Our primary interest, however, was in looking for evidence of bimodality within a single genome, particularly within the genomes of the eubacterial thermophiles. The results show no trace of such bimodality. To illustrate this, we have plotted the distribution of genes for the A.aeolicus genome in Figure 5. As can be seen in Figure 5, the entire gene set for this eubacterial thermophile is unimodal and matches almost exactly with that of the entire set of non-AT-rich thermophile genes. This means that all of the genes within this genome have converged to a single pattern of codon usage regardless of their long-term evolutionary history.
Figure 5.
Frequency distributions of synonymous codon usage among thermophilic (red) and the mesophilic genes (blue) along the second axis of inertia. We have also plotted the A.aeolicus gene frequencies as a proportion of all thermophilic genes (shown in green). This genome, despite being eubacterial, shows a unimodal distribution that fits the distribution pattern of the other thermophilic genomes. The ‘shoulder’ in each curve is due to the AT-rich genomes in each category.
DISCUSSION
By combining the genes from all 40 genomes into a single data set, we were able to make a direct comparison between the intra-genomic and inter-genomic variations in codon usage. These results show very clearly that the inter-genomic differences can be very large relative to the variations between genes within a particular genome. This is illustrated in Figure 2, where we can see that the distribution of values for all genes within a genome is relatively tightly clustered around the mean of that genome. Examination of these results can also give us an insight into how rapidly codon usage patterns may change over the course of evolution. For instance, by exploiting the fact that these 40 species represent a wide range in divergence times, we can ask if codon usage is an evolutionarily conserved character. From Figure 2, it is clear that very closely related species, e.g. different species of Chlamydia, have similar patterns of codon usage. When we consider broader phylogenetic groupings such as the Proteobacteria, however, we see that this clustering of related taxa no longer holds. In fact, P.aeruginosa (Paer) and Buchnera sp. (Buch), both Proteobacteria, are found on opposite extremes of the scale for the first axis of inertia in Figure 2. We also see some dramatic cases of evolutionary convergence along the horizontal axis in Figure 2. For example, the codon usage pattern of the GC-rich archaeal species Halobacterium is very similar to that of the GC-rich gram-positive eubacterium M.tuberculosis and the gram-negative P.aeruginosa. This indicates that codon usage, while stable in the short term, is a labile character over the longer evolutionary term. It is particularly obvious that the codon usage of genes within a genome can ‘track’ the evolutionary changes in nucleotide content of the entire genome (compare Table 1 and Fig. 2). Given that codon usage is responsive to evolutionary changes in nucleotide composition, it is not surprising that it should also be responsive to other evolutionary pressures, such as the action of temperature-dependent selection.
Essentially, our results show that codon usage among these 40 genomes is determined by two major factors: nucleotide content and optimal growth temperature. Of these two factors, the G+C content of the genome explains more than 25% of the variation between genomes, whereas optimal growth temperature explains a further 10% of the variation. In this analysis, there are more than 50 axes in all, and the remaining variation is spread over a large number of the remaining axes. No other single axis explains even 5% of the variation in codon usage.
While it is clear that the second major factor affecting codon usage on a genome-wide scale is optimal growth temperature, it is not obvious what the nature of the selective force might be. At first glance, it seemed that the difference between thermophiles and mesophiles lay solely in their usage of arginine and isoleucine codons (Fig. 1B). However, when we recalculated the codon frequencies in the absence of these two codon groups, the difference between thermophiles and mesophiles remained. We also re-analyzed the data using 2-fold, 4-fold and 6-fold degenerate codons groups separately. In all cases, there was a difference between the genomes of the thermophiles and the mesophiles. This means that the effect is very pervasive. This pervasiveness, in turn, leads one to wonder if the selection is for some general property of the mRNAs that is particularly important under conditions of high temperature, rather than for specific codon–anticodon pairings. One possibility is that the process is driven by selection for increased mRNA stability at high temperature, rather than selection for translational efficiency. Increased mRNA stability would result in increased levels of translated protein per mRNA molecule. Thus mRNA stability could be subject to similar selection pressures as translational efficiency. Interestingly, both forms of selection would be more pronounced for highly expressed genes. It has been suggested that thermophilic genomes are purine rich (21) and such a purine preference could affect both mRNA stability and the frequency of synonymous codons within these genomes.
In summary, we have shown that the patterns of synonymous codon usage within a genome can change dramatically during the course of evolution. Our results show that the two major forces affecting the broad patterns of codon usage among prokaryote genomes are (i) the nucleotide composition of the genome and (ii) some form of natural selection linked to optimal growth temperature. It will be of interest to ask if those genomes that have changed their synonymous codon usage in response to these evolutionary forces have undergone a corresponding change in the relative abundances of isoaccepting tRNAs. A second question that merits further study is the biochemical basis of the selective advantage of certain codons under high temperature conditions and, in particular, if such selective forces are related to the selection on non-synonymous sites among thermophiles (22). The main conclusion that can be drawn from the results presented here is that synonymous codon usage patterns can be subject to natural selection and, specifically, that a particular environmental factor such as high temperature can underlie selection for a specific subset of codons in both eubacterial and archaeal lineages.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by a Research Grant from NSERC Canada (D.A.H.) and graduate scholarships from the University of Ottawa (D.J.L.) and NSERC (G.A.C.S.).
REFERENCES
- 1.Grantham R., Gautier,C. and Gouy,C. (1980) Codon frequencies in 119 individual genes confirm consistent choices of degenerate bases according to genome type. Nucleic Acids Res., 8, 1893–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ikemura T. (1981) Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J. Mol. Biol., 146, 1. [DOI] [PubMed] [Google Scholar]
- 3.Ikemura T. (1981) Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for synonymous codon choice that is optimal for the E. coli translational system. J. Mol. Biol., 151, 389–409. [DOI] [PubMed] [Google Scholar]
- 4.Ikemura T. (1982) Differences in synonymous codon choice patterns of yeast and correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes. Differences in synonymous codon choice patterns of yeast. J. Mol. Biol., 158, 573–597. [DOI] [PubMed] [Google Scholar]
- 5.Shields D.C. and Sharp,P.M. (1987) Synonymous codon usage in Bacillus subtilis reflects both translational selection and mutational biases. Nucleic Acids Res., 15, 8023–8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shields D.C., Sharp,P.M., Higgins,D.G. and Wright,F. (1988) “Silent” sites in Drosophila genes are not neutral: evidence of selection among synonymous codons. Mol. Biol. Evol., 5, 704–716. [DOI] [PubMed] [Google Scholar]
- 7.Stenico M., Lloyd,A.T. and Sharp,P.M. (1994) Codon usage in Caenorhabditis elegans: delineation of translational selection and mutational biases. Nucleic Acids Res., 22, 2437–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McInerney J.O. (1998) Replicational and transcriptional selection on codon usage in Borrelia burgdorferi. Proc. Natl Acad. Sci. USA, 95, 10698–10703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bulmer M. (1991) The selection-mutation-drift theory of synonymous codon usage. Genetics, 129, 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharp P.M. and Li,W.-H. (1987) The selection-mutation-drift theory of synonymous codon usage. Nucleic Acids Res., 15, 1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greenacre M.J. (1984) Theory and Applications of Correspondence Analysis. Academic Press, London, UK.
- 12.Gouy M. and Gautier,C. (1982) Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res., 10, 7055–7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xia X. (1998) How optimized is the translational machinery in Escherichia coli, Salmonella typhimurium and Saccharomyces cerevisiae? Genetics, 149, 37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aravind L., Tatusov,R.L., Wolf,Y.I., Walker,D.R. and Koonin,E.V. (1998) Evidence for massive gene exchange between archaeal and bacterial hyperthermophiles. Trends Genet., 14, 442–444. [DOI] [PubMed] [Google Scholar]
- 15.Nelson K.E., Clayton,R.A., Gill,S.R., Gwinn,M.L., Dodson,R.J., Haft,D.H., Hickey,E.K., Peterson,J.D., Nelson,W.C., Ketchum,K.A. et al. (1999) Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature, 399, 323–329. [DOI] [PubMed] [Google Scholar]
- 16.Kanaya S., Kinouchi,M., Abe,T., Kudo,Y., Yamada,Y., Nishi,T., Mori,H. and Ikemura,T. (2001) Analysis of codon usage diversity of bacterial genes with a self-organizing map (SOM): characterization of horizontally transferred genes with emphasis on the E. coli O157 genome. Gene, 276, 89–99. [DOI] [PubMed] [Google Scholar]
- 17.Wang H.C., Badger,J., Kearney,P. and Li,M. (2001) Analysis of codon usage patterns of bacterial genomes using the self-organizing map. Mol. Biol. Evol., 18, 792–800. [DOI] [PubMed] [Google Scholar]
- 18.Ochman H., Lawrence,J.G. and Groisman,E.A. (2000) Lateral gene transfer and the nature of bacterial innovation. Nature, 405, 299–304. [DOI] [PubMed] [Google Scholar]
- 19.Clarke G.D., Beiko,R.G., Ragan,M.A. and Charlebois,R.L. (2002) Inferring genome trees by using a filter to eliminate phylogenetically discordant sequences and a distance matrix based on mean normalized BLASTP scores. J. Bacteriol., 184, 2072–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.House C.H. and Fitz-Gibbon,S.T. (2002) Using homolog groups to create a whole-genomic tree of free-living organisms: an update. J. Mol. Evol., 54, 539–547. [DOI] [PubMed] [Google Scholar]
- 21.Lao P.J. and Forsdyke,D.R. (2000) Thermophilic bacteria strictly obey Szybalski’s transcription direction rule and politely purine-load RNAs with both adenine and guanine. Genome Res., 10, 228–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kreil D.P. and Ouzounis,C.A. (2001) Identification of thermophilic species by the amino acid compositions deduced from their genomes. Nucleic Acids Res., 29, 1608–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]