Abstract
The signal recognition particle (SRP) is a ribonucleoprotein complex involved in the recognition and targeting of nascent extracytoplasmic proteins in all three domains of life. In Archaea, SRP contains 7S RNA like its eukaryal counterpart, yet only includes two of the six protein subunits found in the eukaryal complex. To further our understanding of the archaeal SRP, 7S RNA, SRP19 and SRP54 of the halophilic archaeon Haloferax volcanii have been expressed and purified, and used to reconstitute the ternary SRP complex. The availability of SRP components from a haloarchaeon offers insight into the structure, assembly and function of this ribonucleoprotein complex at saturating salt conditions. While the amino acid sequences of H.volcanii SRP19 and SRP54 are modified presumably as an adaptation to their saline surroundings, the interactions between these halophilic SRP components and SRP RNA appear conserved, with the possibility of a few exceptions. Indeed, the H.volcanii SRP can assemble in the absence of high salt. As reported with other archaeal SRPs, the limited binding of H.volcanii SRP54 to SRP RNA is enhanced in the presence of SRP19. Finally, immunolocalization reveals that H.volcanii SRP54 is found in the cytosolic fraction, where it is associated with the ribosomal fraction of the cell.
INTRODUCTION
For proteins destined to reside outside the prokaryal cytoplasm or along the eukaryal secretory pathway, the process of translocating across the membrane bilayer begins with the recognition and correct targeting of such proteins to membrane-embedded translocation complexes. In all three domains of life, the processes of recognition and targeting rely on the signal recognition particle (SRP) pathway (1–3). In higher Eukarya, SRP consists of a 7S RNA onto which six proteins are attached (3–5). The RNA-bound SRP9/14 heterodimer serves to arrest protein translation upon interaction of the SRP54 subunit with the newly emerged signal sequence of a nascent polypeptide chain (6–8). SRP19 promotes the attachment of SRP54 to the SRP RNA (9), while the precise role of SRP68/72 remains to be defined. Interaction of SRP with the membrane is mediated by the SRP receptor, composed of the peripheral α-subunit and the integral β-subunit (10). In Bacteria such as Escherichia coli, the SRP pathway has been simplified to include a smaller 4.5S RNA molecule and only one polypeptide chain, the bacterial SRP54 homolog Ffh (reviewed in 1). In other bacterial species, such as Bacillus subtilis, a larger SRP RNA molecule and additional protein subunits may be present (11). The bacterial SRP receptor also represents a simpler version of its eukaryal counterpart, only comprising a single component, the FtsY protein. While many aspects of bacterial SRP-mediated protein targeting remain unclear, there is evidence that SRP is responsible for the delivery of a subset of plasma membrane proteins (12–14).
Genome sequencing, gene identification and numerous biochemical approaches have revealed the presence of SRP pathway components in the Archaea, the most recently described and least well understood form of life (reviewed in 15). In general, the archaeal SRP system incorporates selected aspects of the eukaryal and bacterial systems while also including archaeal-specific traits. Archaea contain an SRP RNA molecule bearing a striking overall similarity to the structure of its eukaryal counterpart and encode for two proteins also found in the eukaryal particle, i.e. SRP19 and SRP54. In contrast, the archaeal SRP receptor is reminiscent of its bacterial counterpart. Characteristic of the archaeal SRP RNA is the presence of helix 1, formed by residues near the termini of the RNA, and the lack of helix 7. Moreover, certain characteristic features of the SRP pathway components, such as the absence of regions of archaeal SRP19 or the SRP receptor, distinguish the archaeal SRP pathway from both the eukaryal and bacterial protein targeting systems, respectively (2). Insight into the assembly and structure of the archaeal signal recognition particle has come from recent reconstitution efforts using purified SRP components from Archaeoglobus fulgidus (16), Pyrococcus furiosus (17) and Methanococcus jannaschii (18,19). In this study, we report the expression and purification of SRP components from the halophilic archaeaon Haloferax volcanii, and their reconstitution into a ternary complex. Existing in molar concentrations of salt, haloarchaea like H.volcanii have modified their biochemistry to cope with the challenges of high salinity (20,21). As such, analysis of H.volcanii SRP provides insight into how halophilic ribonucleoprotein complexes assemble, how high salt levels modulate protein–RNA interactions, and how saline conditions might affect protein targeting.
MATERIALS AND METHODS
Materials
Haloferax volcanii DS2 was obtained from the American Type Culture Collection and grown aerobically at 40°C as previously described (22). Ampicillin, chloramphenicol, isopropyl-β-d-1-thiogalactopyranoside (IPTG) and kanamycin came from Sigma (St Louis, MO).
Synthesis of H.volcanii SRP RNA
The gene for H.volcanii SRP RNA (GenBank accession no. AF395888), including the T7 RNA polymerase promoter sequence, was assembled from 12 overlapping synthetic oligonucleotides (40–60 nt in length) as described previously (23). The termini were designed to be compatible with EcoRI and BamHI restriction sites for insertion into phRΔ35 (23) to obtain pHvSR. For synthesis of H.volcanii SRP RNA by run-off transcription, pHvSR was cleaved at a unique StyI site to yield a transcript of 310 nt, four residues longer than the predicted 3′-end. The linearized DNA was incubated with T7 RNA polymerase using previously described conditions (16). The RNA was recovered by extraction with phenol/chloroform followed by ethanol precipitation and dissolved in water. RNA concentration and integrity were determined by gel electrophoresis using a standard curve generated from known amounts of E.coli 5S ribosomal RNA.
Purification of H.volcanii SRP19
The gene encoding H.volcanii SRP19 was identified in a BLAST search using the sequence of SRP19 from Halobacterium sp. NRC-1 (GenBank accession no. NP280216) against the partially completed H.volcanii genome (http://wit-scranton.mbi.scranton.edu/Haloferax/genes_DNA. fasta). The H.volcanii SRP19 gene was synthesized de novo using previously described methods (23) from a set of 10 overlapping oligonucleotides (each 48–60 nt long), designed to favor frequently used E.coli codons. The cloned gene (GenBank accession no. AY138586), termed pET-Hv19, included NdeI and HindIII staggered ends for insertion into the bacterial expression vector pET23d (Novagen). Ligation mixtures were used to transform competent E.coli DH5α cells and transformants containing the desired plasmid clones were identified by restriction mapping and subsequently confirmed by sequencing. For expression of H.volcanii SRP19, competent E.coli BL21(DE3) pLysS cells were transformed with the pET-Hv19 DNA and subjected to a selection on Luria–Bertani (LB) agar plates containing ampicillin (200 µg/ml) and chloramphenicol (34 µg/ml) at 37°C overnight. Colonies were transferred to four cultures of 400 ml each and incubated in a shaker at 37°C to an OD600 of 0.3–0.4, at which time IPTG was added to a final concentration of 1 mM. After induction for 2 h, cells were harvested by centrifugation and resuspended in 14 ml of lysis buffer (50 mM NaCl, 5 mM DTT, 1 mM EDTA, 10% glycerol, 50 mM Na phosphate buffer, pH 8.0). The cells were frozen in liquid N2 and stored at –70°C.
All procedures related to purification of H.volcanii SRP19 were performed at 4°C. The suspended cells were thawed on ice and sonicated using a Sonic Dismembrator Model 300 (Fisher Scientific) seven times for 15 s at a setting of 35%, with 15 s intervals between each pulse, followed by a centrifugation (Sorvall SS34 rotor, 27 000 g, 20 min). SDS–PAGE of aliquots indicated that the vast majority of H.volcanii SRP19 was present in the pellet. Accordingly, the pellet was solubilized in 20 ml of lysis buffer to which solid urea was slowly added until a final urea concentration of 5 M was reached. The sample was again sonicated as above, dialyzed against lysis buffer containing 10 mM NaCl to remove the urea and spun at 40 000 r.p.m. for 2 h. The resulting supernatant (containing the majority of H.volcanii SRP19) was loaded onto a Q-Sepharose column (24 ml bed volume), pre-equilibrated in a buffer containing 10 mM NaCl, 1 mM EDTA, 1 mM DTT, 8% glycerol, 50 mM Na phosphate buffer, pH 8, and attached to an FPLC system at a flow rate of 0.5 ml/min. Elution was achieved by applying a 10 mM to 1 M NaCl gradient. SRP19 eluted at ∼450 mM NaCl. Fractions containing substantial amounts of H.volcanii SRP19 (as determined by SDS–PAGE) were pooled, concentrated to 6.5 µg/µl protein using a Centricon YM-3 concentrating device (Amicon) and stored in 50% glycerol at –20°C.
Purification of H.volcanii SRP54
To express sufficient quantities of H.volcanii SRP54, E.coli BL21 cells were transformed with plasmid pET-Hv54, encoding for H.volcanii SRP54 (GenBank accession no. AF395887) bearing a His6 tag at its C-terminus. Using oligonucleotides designed to introduce NdeI and XhoI staggered ends for insertion into the bacterial expression vector pET24b (Novagen), the SRP54-encoding gene was cloned from H.volcanii genomic DNA, prepared as previously described (24). Ligation mixtures were used to transform competent E.coli BL21 cells, with correct plasmids selected by restriction mapping and subsequent sequencing. The transformed bacterial cells were grown at 37°C in LB broth in the presence of 50 µg/ml kanamycin and induced at OD600 = 0.3 with 0.5 mM IPTG for 3 h. Cells were then harvested and disrupted by sonication (three times for 30 s with 30 s intervals between each pulse, 35% output, Misonix XL2020 ultrasonicator). Soluble proteins were separated from membrane proteins by ultracentrifugation (Sorvall Discovery M120 ultracentrifuge, S120ATS rotor, 73 000 r.p.m., 10 min, 4°C) and applied to Ni–NTA resin (Qiagen), previously equilibrated with 20 mM imidazole, 150 mM NaCl, 50 mM Tris–HCl pH 7.9. Following a 1 h incubation at 4°C, unbound proteins were removed by washing with the equilibration buffer. Specifically bound proteins were then eluted by addition of 500 mM imidazole, 150 mM NaCl, 50 mM Tris–HCl pH 7.9. Following SDS–PAGE, the identity of the eluted protein was confirmed as H.volcanii SRP54 by N-terminal amino acid sequencing.
Assembly of H.volcanii SRP
Haloferax volcanii SRP complexes were formed by combining 40 µg SRP RNA, 16 µg SRP19 and 12 µg SRP54 in 300 mM KOAc, 5 mM MgCl2, 1 mM DTT, 50 mM Tris–HCl, pH 7.9, in a volume of 400 µl. The amounts employed correspond to a 4–5-fold excess of protein over RNA so as to favor formation of the complex. Samples were incubated at 37°C for 10 min and then loaded onto the top of 40 to 10% sucrose gradients prepared in 50 mM MgSO4, 5 mM DTT, 50 mM Tris–HCl, pH 7.2, or 50 mM Tris–HCl, pH 7.2, containing either 1 M KCl or 1 M NaCl. The gradients were subjected to centrifugation in a Beckman NVT65 rotor (55 000 r.p.m., 4.5 h, 4°C). Eighteen fractions of ∼650 µl (24 drops) each were collected and analyzed by SDS–PAGE using a 600 µl aliquot of TCA-precipitated sample (for protein analysis) or a 13 µl aliquot for electrophoresis on 2% agarose gels (for RNA analysis). The amount of Coomassie blue or ethidium bromide stained material in each fraction aliquot was determined by densitometric scanning of the gels.
Immunoblotting
Immunoblotting was performed on nitrocellulose membranes (0.45 µm; Schleicher & Schuell, Dassel, Germany) using polyclonal serum raised against H.volcanii SRP54, at concentrations of 1:1000–4000. Antibody binding was detected using goat anti-rabbit horseradish peroxidase-conjugated antibodies and enhanced chemiluminescence. Isolation of H.volcanii membranes was achieved by sonication (three times for 30 s with 30 s intervals between each pulse, 35% output, Misonix XL2020 ultrasonicator) and ultracentrifugation in a Sorvall Discovery M120 ultracentrifuge (S120ATS rotor, 73 000 r.p.m., 10 min, 4°C). Membranes were contained within the pellet fraction. Protein concentrations were determined using Bradford reagent (Bio-Rad, Hercules, CA), with BSA as the standard.
RESULTS
Purification and characterization of H.volcanii SRP RNA, SRP19 and SRP54
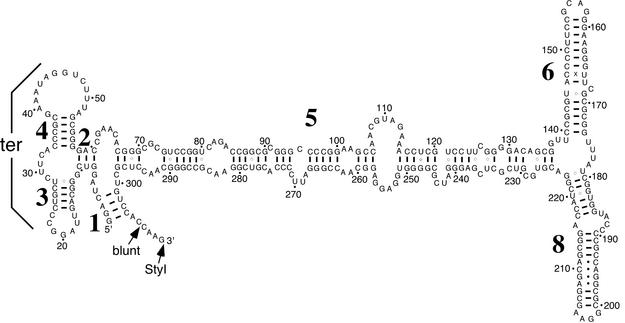
SRP RNA. Figure 1 shows the secondary structure of H.volcanii SRP RNA, with helix numbers indicated, as derived by comparative sequence analysis (25). Only those Watson–Crick and G-U base pairs that are supported by phylogenetic analysis are indicated. In addition, non-Watson–Crick pairs that are likely to exist in helices 6 and 8, as derived by comparison with the known structures of various homologous SRP RNAs (26), were included. As in other archaeal SRP RNAs (2,15,26,27), H.volcanii SRP RNA possesses a helix 1, absent in Eukarya, yet lacks helix 7, present in the eukaryal molecule. Furthermore, the tertiary pairing between the loops of helices 3 and 4 proposed to occur in other SRP RNAs (25,28) is also supported in the H.volcanii SRP RNA.
Figure 1.
Secondary structure of H.volcanii SRP RNA. Helices are numbered from 1 to 8 starting from the 5′-end. Numbering of residues is in increments of 10, as indicated by dots and numbers. Watson–Crick paired residues are connected by lines and G–U interactions by circles. A phylogenetically supported tertiary interaction (ter) between the loops of helices 3 and 4 is indicated.
The high degree of similarity between the secondary structures of H.volcanii SRP RNA and that of its relatives in higher eukaryotes raises the likelihood of the presence of binding sites for the six SRP proteins present in most Eukarya. However, consistent with what has been observed in other archaeal species, analysis of the H.volcanii genome revealed genes for SRP19 and SRP54, but no homologs for SRP9/14 or SRP68/72. The detection of only two H.volcanii SRP proteins agrees with the recent finding that purification of affinity-tagged SRP19 from transformed H.volcanii resulted in the co-capture of SRP RNA and SRP54 (29). However, the possible existence of additional H.volcanii SRP subunits, not readily identified through sequence comparisons, cannot be discounted.
To synthesize H.volcanii SRP RNA in vitro, the encoding gene was cloned under control of the T7 RNA polymerase promoter. For convenience, the RNA was modified to include a 4 nt extension of the predicted 3′-end. This modification is not expected to influence the binding of either SRP19 or SPR54, as the binding sites of these components and the termini-containing domain are located at opposite ends of the SRP RNA molecule. Electrophoresis of the in vitro transcribed RNA showed the molecule to be of the predicted size (not shown).
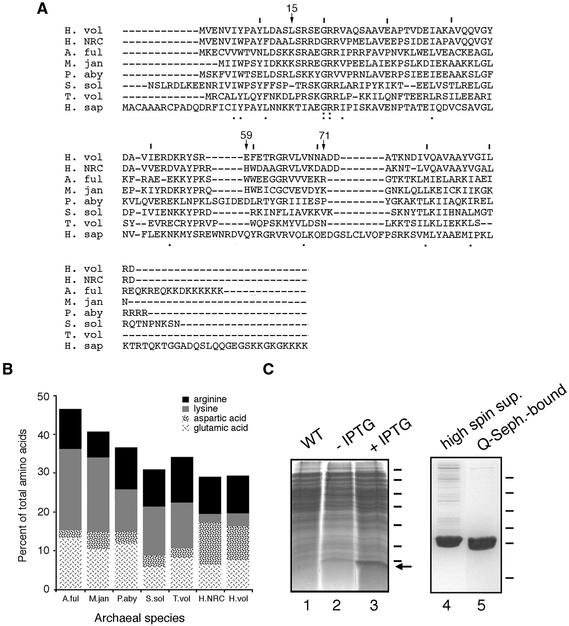
SRP19. By performing BLAST searches using the sequence of SRP19 from Halobacterium sp. NRC-1 (GenBank accession no. NP280216), the gene encoding H.volcanii SRP19 was identified in the partially completed H.volcanii genome. The gene encodes a polypeptide of 92 amino acids (10.153 kDa) sharing 68% identity with the Halobacterium sp. NRC-1 sequence and is also similar to other reported archaeal SRP19 genes (Fig. 2A). Because of its intimate association with RNA, SRP19 is typically a basic protein. Haloferax volcanii SRP19, however, has a predicted pKi value of 4.82. Closer examination of the H.volcanii SRP19 sequence revealed that the H.volcanii protein (like Halobacterium sp. NRC-1 SRP19) is enriched in aspartic acid residues, while containing fewer lysine residues than other archaeal SRP19 proteins (Fig. 2B). This raises the interesting question of how H.volcanii SRP19 is capable of binding to RNA despite its overall acidic character (discussed below).
Figure 2.
Purification of H.volcanii SRP19. (A) Alignment of H.volcanii (H. vol) SRP19 with SRP homologs from Halobacterium sp. NRC-1 (H. NRC), Archeoglobus fulgidus (A. ful), Methanococcus jannaschii (M. jan), Pyrococcus abyssi (P. aby), Sulfolobus solfataricus (S. sol), Thermoplasma volcanium (T. vol) and Homo sapiens (H. sap). Lines are placed above every tenth residue in the H.volcanii sequence, while H.volcanii SRP19 residues discussed in the text are denoted by an arrow and the residue number. Homology of the sequences is shown below each residue, with the colon depicting identity and the period depicting similarity. (B) The relative proportions of basic and acidic amino acid residues in archaeal SRP19 proteins are shown. The same species as addressed in (A) are shown. Arginine residues are depicted in black, lysine residues in grey, aspartic acid residues in a speckled pattern and glutamic acid residues in a scratch mark pattern. (C) Escherichia coli BL21(DE3) cells were transformed with pET-Hv19, encoding for H.volcanii SRP19, and induced with 1 mM IPTG for a period of 3 h. Lane 1, wild-type cells; lane 2, uninduced transformed cells; lane 3, induced transformed cells. Molecular weight markers are shown on the right and correspond to 116, 66, 45, 35, 25, 18.4 and 14.4 kDa, while the arrow depicts the position of SRP19. The supernatant of induced cells was applied to Q-Sepharose and eluted with a gradient of NaCl. Lane 4, high spin supernatant; lane 5, Q-Sepharose eluted protein. Molecular weight markers shown on the right correspond to 43, 29, 18.4, 14.4, 6.2 and 3 kDa.
To obtain purified H.volcanii SRP19 for biochemical analysis, the H.volcanii SRP19-encoding gene was used to transform E.coli BL21(DE3) pLysS cells. Upon induction with IPTG, overexpression of a 10 kDa species was detected (Fig. 2C, lane 3). Following cell lysis, differential centrifugation and dialysis, the soluble material was applied to Q-Sepharose (lane 4), resulting in a >95% purification of the protein (lane 5). Finally, as judged by its ability to interact with SRP RNA, the majority of the protein was active (see below).
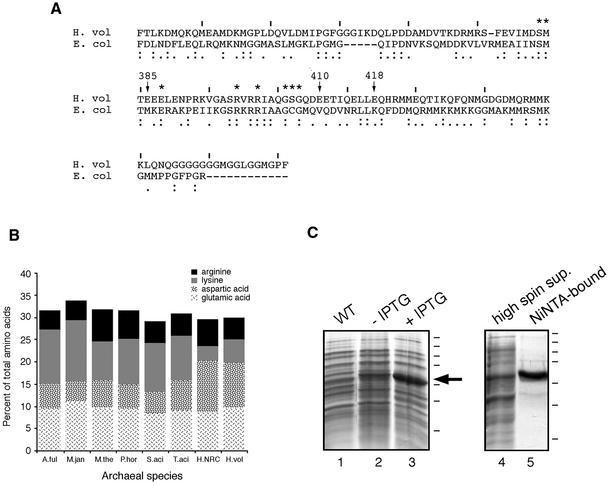
SRP54. The deduced amino acid sequence of H.volcanii SRP54 (GenBank accession no. AF395887) was compared with the sequences of other archaeal SRP54 homologs. The alignment of the H.volcanii SRP54 sequence with homologs in the SRP database (26) demonstrated that the H.volcanii SRP54 can be functionally divided into the NG domain, involved in guanidine nucleotide binding, and the M domain (Fig. 3A), which interacts with signal sequences and SRP RNA (30,31). As with H.volcanii SRP19, the haloarchaeal SRP54 protein is enriched in acidic residues (i.e. aspartic acid), as compared to its non-halophilic counterparts (Fig. 3B). The H.volcanii protein, moreover, lacks certain lysine residues present in non-haloarchaeal SRP54 proteins. A similar distribution of charged residues is also seen in the other haloarchaeal SRP54 sequence available, namely that of Halobacterium sp. NRC-1 (32).
Figure 3.
Purification of H.volcanii SRP54. (A) Alignment of the M domains of H.volcanii (H. vol) SRP54 and E.coli (E. col) Ffh. Haloferax volcanii SRP54 residues discussed in the text are denoted by an arrow and the residue number, while lines are placed above each tenth residue of the H.volcanii sequence. Asterisks denote those E.coli Ffh residues in contact with SRP RNA. Homology of the sequences is shown below each residue, with the colon depicting identity and the period depicting similarity. (B) The relative proportions of basic and acidic amino acid residues in archaeal SRP54 proteins are shown. A.ful, Archaeoglobus fulgidus; H.NRC, Halobacterium sp. NRC-1; H.vol, Haloferax volcanii; M.jan, Methanococcus jannaschii; M.the, Methanococcus thermoautotrophicum; P.hor, Pyrococcus horikoshii; S.aci, Sulfolobus acidocaldarius; T.aci, Thermoplasma acidophilum. Arginine residues are depicted in black, lysine residues in grey, aspartic acid residues in a speckled pattern and glutamic acid residues in a scratch mark pattern. (C) Escherichia coli BL21(DE3) cells were transformed with pET-Hv54, encoding for His6-tagged H.volcanii SRP54 and induced with 0.5 mM IPTG for a period of 3 h. The supernatant of induced cells was incubated with Ni–NTA resin and eluted with buffer containing 0.5 M imidazole. Lane 1, wild-type cells; lane 2, uninduced transformed cells; lane 3, induced transformed cells; lane 4, high spin supernatant; lane 5, Ni–NTA-bound proteins. Molecular weight markers are shown on the right and correspond to 250, 150, 100, 75, 50, 37 and 25 kDa, while the arrow depicts the position of SRP54.
Recent structural studies of the bacterial SRP54 homolog Ffh have identified residues in the SRP54 M domain implicated in SRP RNA binding (33). Sequence conservation and homology modeling (not shown) of the H.volcanii SRP54 protein indicate that the same amino acid residues are likely to be used for its interaction with the exceptionally well conserved SRP RNA helix 8, although at least one exception may exist (discussed below). Moreover, the increased acidic character of the haloarchaeal protein is not concentrated in the M domain, but rather is distributed throughout the protein. It thus appears that the enhanced negative charge of the protein is more related to the folding of a halophilic protein in highly saline surroundings rather than modulating H.volcanii SRP54 function (20,21). This is highlighted in the case of positions E410 and E418 of the H.volcanii protein, which replace valine and lysine residues at the corresponding positions in the E.coli Ffh M domain (33). In the bacterial protein, these residues lie on the protein surface.
For biochemical studies of H.volcanii SRP54, E.coli BL21 cells were transformed with plasmid pET-Hv54. Subsequent addition of IPTG induced the overexpression of a 50 kDa protein (Fig. 3C, lane 3), in good agreement with the predicted molecular weight of H.volcanii SRP54 (50.918 kDa). Following cell disruption, the soluble portion of the lysate, containing substantial amounts of the protein, was applied to a Ni–NTA affininity column. The H.volcanii SRP54 protein was eluted with 0.5 M imidazole (Fig. 3C, lane 5) and its identity was confirmed by N-terminal sequencing.
Reconstitution of H.volcanii SRP
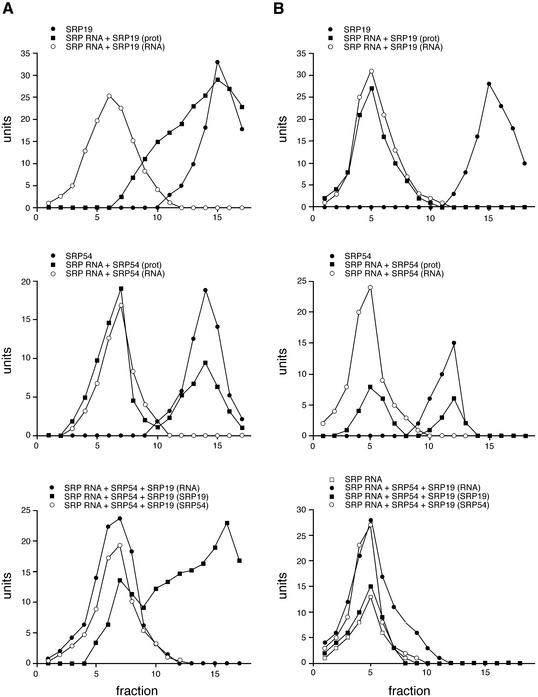
Initially, experiments were carried out to investigate the abilities of H.volcanii SRP19 and SRP54 to individually interact with SRP RNA. As shown in Figure 4A, reflecting the migration of SRP components in a sucrose gradient prepared in 1 M KCl, only limited, if any, interaction of SRP19 with SRP RNA was detected (upper panel). In contrast, a substantial portion of SRP54 co-migrated with SRP RNA on a sucrose density gradient (middle panel). When all three components were combined, substantial changes in the binding profile were detected (lower panel). In the presence of SRP19, essentially all of the SRP54 was co-localized to those fractions also containing SRP RNA. In addition, a significant amount of SRP19 could be detected in the SRP RNA–SRP54 complex. Essentially identical profiles were obtained when the gradients were prepared with 1 M NaCl (not shown). In control experiments using 5S ribosomal RNA from E.coli instead of SRP RNA, no co-migration of 5S rRNA, SRP54 and SRP19 could be detected, reflecting the specificity of the binding of the proteins to the H.volcanii SRP RNA (not shown).
Figure 4.
Reconstitution of H.volcanii SRP. Haloferax volcanii SRP19 and SRP54 were combined with SRP RNA either separately or together and the resulting complexes were examined by sucrose density gradient centrifugation. The levels of each component in the fractions collected from each gradient were determined densitometrically following electrophoresis and are presented as units. (A) Gradients prepared in 1 M KCl. (B) Gradients prepared in low salt. In both (A) and (B), the upper panels present the level of SRP19 in fractions collected from gradients containing SRP19 alone (filled circles), as well as the levels of SRP RNA (open circles) and SRP19 (filled squares) in fractions collected from gradients to which both components were added together. The middle panels in (A) and (B) present the levels of SRP54 in fractions collected from gradients containing SRP54 alone (filled circles), as well as the levels of SRP RNA (open circles) and SRP54 (filled squares) in fractions collected from gradients to which both components were added together. The lower panels in (A) and (B) present the levels of SRP RNA (filled circles), SRP19 (filled squares) and SRP54 (open circles) in fractions collected from high and low salt containing gradients containing all the components, respectively. In the lower panel of (B), the level of SRP RNA (open squares) in fractions collected from a low salt gradient to which SRP RNA alone was added is also shown.
The cytoplasm of halophilic archaea such as H.volcanii contains extremely high salt concentrations (34). Thus, the finding that substantial interaction between SRP RNA and the SRP protein components could be detected in sucrose density gradients prepared at the relatively low ionic strength of 300 mM potassium acetate was unexpected (Fig. 4B). Differences in the interactions between the various components in the two gradients were, however, detected. While SRP54 bound to SRP RNA when both high and low salt gradients were employed, less SRP54–RNA complex was obtained in the high salt containing gradients (compare middle panels, Fig. 4A and B). Moreover, SRP19 on its own bound efficiently to SRP RNA better in the low salt containing gradients (compare upper panels, Fig. 4A and B). Finally, when all three SRP components were combined under low salt conditions, the formation of a trimeric H.volcanii SRP complex could be demonstrated (Fig. 4B, lower panel).
Immunolocalization of SRP54
Antibodies against the bacterially expressed, polyhistidine-tagged version of H.volcanii SRP54 were raised in rabbits. As reflected in Figure 5A, the polyclonal serum was able to recognize the protein in induced bacterial cells (lane 2), as well as following Ni–NTA-based purification (lane 3). In the bacterial extracts, the antibodies also cross-reacted with an additional band of slightly higher molecular weight. This band likely corresponds to a native E.coli protein, since immunoblots of both wild-type and transformed cells, but not of the Ni–NTA-purified fraction, revealed staining of this additional band. SRP54 was also recognized by the antibodies in extracts of H.volcanii cells, although a number of bands of higher molecular weight were also detected, albeit more weakly (lane 5). In contrast, normal rabbit serum failed to recognize the 54 kDa band in either the transformed bacterial (lane 4) or H.volcanii cells (lane 6).
Figure 5.
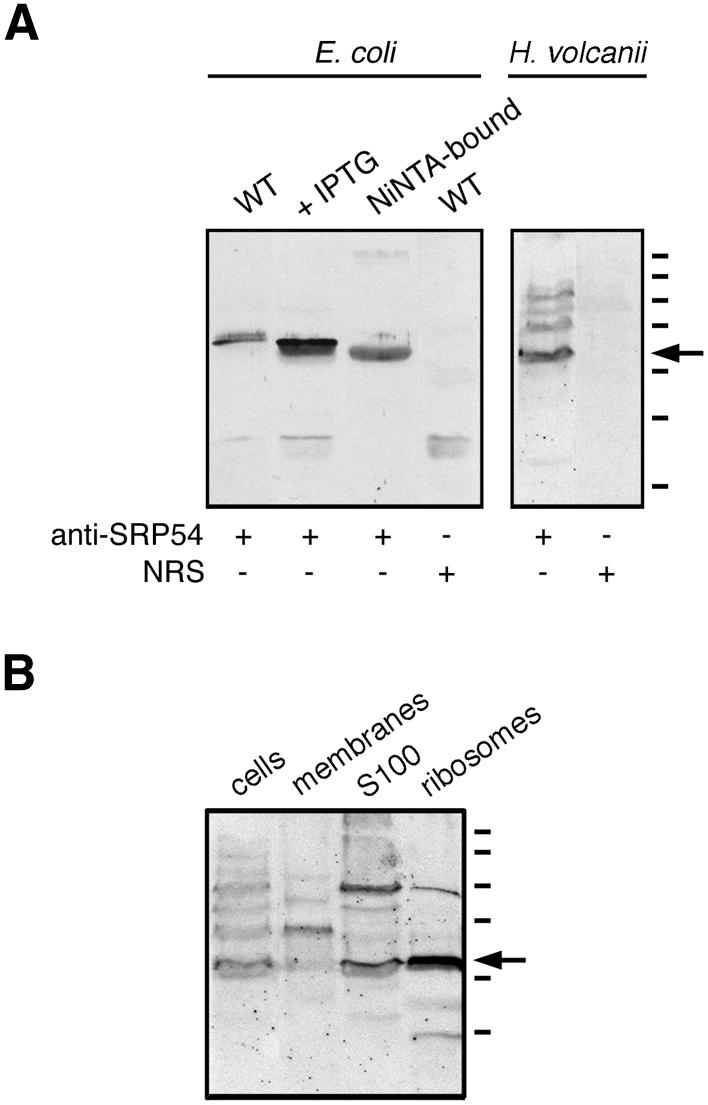
Immunodetection of H.volcanii SRP54. (A) Aliquots of wild-type E.coli cells, IPTG-induced E.coli cells transformed with plasmid pET-Hv54 encoding for H.volcanii SRP54, or Ni–NTA-purified H.volcanii SRP54 were probed with polyclonal antiserum raised against the H.volcanii SRP54. The specificity of antibody labeling of the bacterially expressed archaeal protein was confirmed by the failure of normal rabbit serum (NRS) to label such a band in E.coli cell extracts. Similarly, the polyclonal antibodies recognized SRP54 in an extract of H.volcanii. Again, NRS failed to label any such band. (B) Aliquots of H.volcanii cells, isolated membranes, the S100 cytoplasmic fraction and isolated ribosomes were probed with anti-H.volcanii SRP54 antibodies. The S100 and ribosomal fractions were prepared as described by Ban et al. (39). In both (A) and (B), the arrow depicts the position of the labeled 54 kDa protein recognized by the antibodies, while molecular weight markers shown on the right denote the 250, 150, 100, 75, 50, 37 and 25 kDa positions.
Next, the anti-H.volcanii SRP54 antibodies were used to determine the subcellular location of SRP54. Haloferax volcanii cells were grown to late exponential phase and intact cells, isolated membranes, a cytosolic fraction as well as ribosomes were examined by immunoblotting using the polyclonal preparation. As shown in Figure 5B, the antibodies labeled SRP54 in the cellular, cytosolic and ribosomal fractions of the H.volcanii cells.
DISCUSSION
With the recent purification, reconstitution and structural analyses of archaeal SRP from several species (16–19,29, 35,36), a clearer picture of this ribonucleoprotein complex in Archaea is beginning to emerge. In the current study, reconstitution of the SRP of the halophilic archaeon H.volcanii is presented. The availability of reconstituted H.volcanii SRP, as well as the recent purification of affinity-tagged SRP from H.volcanii cells (29), offers an opportunity to address the impact of high salt concentrations on SRP assembly, structure and function.
Despite the fact that both H.volcanii SRP19 and SRP54 are relatively enriched in aspartic acids and poorer in lysine residues, a comparison of SRP19– and SRP54–SRP RNA interactions suggests that the majority of these amino acid modifications do not play a direct role in H.volcanii SRP RNA–protein contacts. As such, haloarchaeal SRP19 and SRP54 appear to use the same mode of RNA binding despite their saline surroundings. However, as suggested by a comparison of H.volcanii SRP19 with the recently determined structures of complexes between human SRP RNA helix 6 and SRP19 (37) and between human or M.jannaschii SRP RNA and M.jannaschii SRP19 (18,19), some of the modifications found in the haloarchaeal SRP19 could affect the stability of the protein fold or indirectly modify the interaction of the protein with SRP RNA. In the crystal structure of the M.jannaschii SRP RNA–SRP19 complex (18,19), the contact between Leu12 and Gln56 appears to stabilize the protein fold. An interaction between Leu15 and Glu59 could fulfill a similar role in the H.volcanii protein, but would rely on amino acid residues bearing different properties. Hence, the contact between a basic and an uncharged polar residue employed in the methanogen would be replaced by a contact between an acidic and a non-polar residue, possibly bridged by salt, in the H.volcanii protein. Interestingly, in the solution structure of the A.fulgidus SRP19 protein (36), the equivalent Lys14 and Ser58 residues are not in contact, suggesting that RNA binding leads to a conformational change in the loop 3 region of the protein, resulting in an interaction between the two residues. The position occupied by Lys69 in M.jannaschii SRP19 may also be modified in the haloarchaeal protein. In the crystal structure of M.jannaschii SRP19 with the S domain of human SRP RNA (19), Lys69 is near one of the phosphate oxygens of G147, the first base of the helix 6 tetraloop involved in binding to SRP19. In H.volcanii SRP19 (as well as in the Halobacterium sp. NRC-1 protein), the lysine residue is replaced by an alanine. The presence of this less bulky residue might provide space for a sodium ion in the H.volcanii structure. However, in the structure between M.jannaschii SRP19 and M.jannaschii SRP RNA (18), this gap is filled by U164 due to an altered configuration of the helix 6 tetraloop and the side chain of Lys69 points away from the RNA.
Similarly, aspects of the interaction between H.volcanii SRP54 and SRP RNA may be unique to the haloarchaeon, as revealed by a comparison of the H.volcanii SRP54 M domain sequence with the structure of the E.coli SRP complex, the only SRP54–RNA complex for which structural information is available (33). Such an analysis shows that in the E.coli structure, M37 (numbering according to the E.coli structure, 1DUL.pdb) is separated from SRP RNA by only ∼6 Å. Thus, modification of this position to an acidic residue (E385) in the H.volcanii protein may carry implications for protein–RNA interactions. Interestingly, changes of the equivalent position in human SRP54 abolish RNA binding (C.Zwieb, unpublished observations).
In agreement with earlier reconstitution studies of archaeal SRP from A.fulgidus (16) and P.furiosus (17), a significant proportion of H.volcanii SRP54 could interact with SRP RNA in the absence of SRP19. In the presence of SRP19, however, all of the H.volcanii SRP54 present could be detected as part of the trimeric RNA–SRP19–SRP54 complex. However, whereas H.volcanii SRP54 readily bound to SRP RNA, the interaction of H.volcanii SRP19 with SRP RNA presented a somewhat more complicated picture. SRP19 bound to SRP RNA rather poorly in sucrose gradients prepared with high levels of salt, but binding was enhanced by the presence of SRP54. Weak binding of SRP19 to SRP RNA in high salt gradients may reflect impaired folding of the protein under the investigated conditions. The stimulation of SRP54 binding to SRP RNA in the presence of SRP19 in the high salt gradients may reflect a more active role for the RNA in the assembly and function of the H.volcanii SRP. In this context, it may be significant that examination of the genome of the thermoacidophilic archaeon Thermoplasma acidophilum failed to detect an SRP19 homolog (15,38), suggesting a possible dispensability for SRP19 in the archaeal ribonucleoprotein complex. Alternatively, the enhanced SRP54 binding may point to the importance of protein–protein contacts, as suggested by the structure of the M.jannaschii SRP S domain complex (18).
The isolation of functional haloarchaeal ribosomes has been a cornerstone for the study of this ribonucleoprotein complex (39). Such ribosomes, together with well characterized protein targeting components from halophilic archaea, will offer a powerful system for understanding the biogenesis of SRP-dependent proteins in Archaea. This study demonstrates that in exponentially growing cells, H.volcanii SRP54 associates with ribosomes, most likely as part of a fully assembled SRP. Thus, the present study represents an important step in the understanding of the haloarchaeal SRP. Continued study will provide insight into the molecular mechanisms that govern SRP-mediated protein targeting and how molecules recognize each other under high salt conditions. Moreover, together with functional haloarchaeal ribosomes (40,41), translocation-competent H.volcanii preprotein (V.Irihimovitch, Z.Konrad and J.Eichler, manuscript in preparation) and inverted membrane vesicles prepared from H.volcanii (42), the availability of the H.volcanii SRP will assist in future in vitro recreation of archaeal protein translocation.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank Gabriela Ring for gifts of the H.volcanii S100 fraction and ribosomes. C.Z. is supported by NIH grant GM-49034. J.E. is supported by the Israel Science Foundation (grant 291/99) and is the incumbent of the Murray Shusterman Career Development Chair in Microbiology.
REFERENCES
- 1.Herskovits A.A., Bochkareva,E.S. and Bibi,E. (2000) New prospects in studying the bacterial signal recognition particle pathway. Mol. Microbiol., 38, 927–939. [DOI] [PubMed] [Google Scholar]
- 2.Eichler J. and Moll,R. (2001) The signal recognition particle of archaea. Trends Microbiol., 9, 130–136. [DOI] [PubMed] [Google Scholar]
- 3.Keenan R.J., Freymann,D.M., Stroud,R.M. and Walter,P. (2001) The signal recognition particle. Annu. Rev. Biochem., 70, 755–775. [DOI] [PubMed] [Google Scholar]
- 4.Lütcke H. (1995) Signal recognition particle (SRP), a ubiquitous initiator of protein translocation. Eur. J. Biochem., 228, 531–550. [DOI] [PubMed] [Google Scholar]
- 5.Walter P. and Johnson,A.E. (1994) Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu. Rev. Cell Biol., 10, 87–119. [DOI] [PubMed] [Google Scholar]
- 6.Siegel V. and Walter,P. (1986) Removal of the Alu structural domain from signal recognition particle leaves its protein translocation activity intact. Nature, 320, 81–84. [DOI] [PubMed] [Google Scholar]
- 7.Thomas Y., Bui,N. and Strub,K. (1997) A truncation in the 14 kDa protein of the signal recognition particle leads to tertiary structure changes in the RNA and abolishes the elongation arrest activity of the particle. Nucleic Acids Res., 25, 1920–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mason N., Ciufo,L.F. and Brown,N. (2000) Elongation arrest is a physiologically important function of signal recognition particle. EMBO J., 15, 4164–4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gowda K., Clemons,W.M., Zwieb,C. and Black,S.D. (1999) Expression, purification, and crystallography of the conserved methionine-rich domain of human signal recognition particle 54 kDa protein. Protein Sci., 8, 1144–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Young J.C., Ursini,J., Legate,K.R., Miller,J.D., Walter,P. and Andrews,D.W. (1995) An amino-terminal domain containing hydrophobic and hydrophilic sequences binds the signal recognition particle receptor α subunit to the β subunit on the endoplasmic reticulum membrane. J. Biol. Chem., 270, 15650–15657. [DOI] [PubMed] [Google Scholar]
- 11.Nakamura K., Yahagi,S., Yamazaki,T. and Yamane,K. (1999) Bacillus subtilis histone-like protein, HBsu, is an integral component of a SRP-like particle that can bind the Alu domain of small cytoplasmic RNA. J. Biol. Chem., 274, 13569–13576. [DOI] [PubMed] [Google Scholar]
- 12.Seluanov A. and Bibi,E. (1997) FtsY, the prokaryotic signal recognition particle receptor homologue, is essential for biogenesis of membrane proteins. J. Biol. Chem., 272, 2053–2055. [DOI] [PubMed] [Google Scholar]
- 13.Ulbrandt N.D., Newitt,J.A. and Bernstein,H.D. (1997) The E. coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell, 88, 187–196. [DOI] [PubMed] [Google Scholar]
- 14.MacFarlane J. and Muller,M. (1995) The functional integration of a polytopic membrane protein of Escherichia coli is dependent on the bacterial signal-recognition particle. Eur. J. Biochem., 233, 766–771. [DOI] [PubMed] [Google Scholar]
- 15.Zwieb C. and Eichler,J. (2001) Getting on target: the archaeal signal recognition particle. Archaea, 1, 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhuiyan S.H., Gowda,K., Hotokezaka,H. and Zwieb,C. (2000) Assembly of archaeal signal recognition particle from recombinant components. Nucleic Acids Res., 15, 1365–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maeshima H., Okuno,E., Aimi,T., Morinaga,T. and Itoh,T. (2001) An archaeal protein homologous to mammalian SRP54 and bacterial Ffh recognizes a highly conserved region of SRP RNA. FEBS Lett., 507, 336–340. [DOI] [PubMed] [Google Scholar]
- 18.Hainzl T., Huang,S. and Sauer-Eriksson,A.E. (2002) Structure of the SRP19 RNA complex and implications for signal recognition particle assembly. Nature, 417, 767–771. [DOI] [PubMed] [Google Scholar]
- 19.Oubridge C., Kuglstatter,A., Jovine,L. and Nagai,K. (2002) Crystal structure of SRP19 in complex with the S domain of SRP RNA and its implication for the assembly of the signal recognition particle. Mol. Cell, 9, 1251–1261. [DOI] [PubMed] [Google Scholar]
- 20.Madern D., Ebel,C. and Zaccai,G. (2000) Halophilic adaptation of enzymes. Extremophiles, 4, 91–98. [DOI] [PubMed] [Google Scholar]
- 21.Madigan M.T. and Oren,A. (1999) Thermophilic and halophilic extremophiles. Curr. Opin. Microbiol., 2, 265–269. [DOI] [PubMed] [Google Scholar]
- 22.Mevarech M. and Werczberger,R. (1985) Genetic transfer in Halobacterium volcanii. J. Bacteriol., 162, 461–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zwieb C. (1991) Interaction of protein SRP19 with signal recognition particle RNA lacking individual RNA helices. Nucleic Acids Res., 19, 2955–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosenshine I., Zusman,T., Werczberger,R. and Mevarech,M. (1987) Amplification of specific DNA sequences correlates with resistance of the archaebacterium Halobacterium volcanii to the dihydrofolate reductase inhibitors trimethoprim and methotrexate. Mol. Gen. Genet., 208, 518–522. [Google Scholar]
- 25.Larsen N. and Zwieb,C. (1991) SRP-RNA sequence alignment and secondary structure. Nucleic Acids Res., 19, 209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorodkin J., Knudsen,B., Zwieb,C. and Samuelsson,T. (2001) SRPDB (Signal Recognition Particle Database). Nucleic Acids Res., 29, 169–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Althoff S., Selinger,D. and Wise,J.A. (1994) Molecular evolution of SRP cycle components: functional implications. Nucleic Acids Res., 22, 1933–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weichenrieder O., Wild,K., Strub,K. and Cusack,S. (2000) Structure and assembly of the Alu domain of the mammalian signal recognition particle. Nature, 408, 167–173. [DOI] [PubMed] [Google Scholar]
- 29.Rose W. and Pohlschroder,M. (2002) In vivo analysis of an essential archaeal signal recognition particle in its native host. J. Bacteriol., 184, 3260–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bernstein H.D., Poritz,M.A., Strub,K., Hoben,P.J., Brenner,S. and Walter,P. (1989) Model for signal sequence recognition from amino-acid sequence of 54K subunit of signal recognition particle. Nature, 340, 482–486. [DOI] [PubMed] [Google Scholar]
- 31.Römisch K., Webb,J., Herz,J., Prehn,S., Frank,R., Vingron,M. and Dobberstein,B. (1989) Homology of 54K protein of signal-recognition particle, docking protein and two E. coli proteins with putative GTP-binding domains. Nature, 340, 478–482. [DOI] [PubMed] [Google Scholar]
- 32.Ng W.V., Kennedy,S.P., Mahairas,G.G., Berquist,B., Pan,M., Shukla,H.D., Lasky,S.R., Baliga,N.S., Thorsson,V., Sbrogna,J. et al. (2000) Genome sequence of Halobacterium species NRC-1. Proc. Natl Acad. Sci. USA, 97, 12176–12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Batey R.T., Rambo,R.P., Lucast,L., Rha,B. and Doudna,J.A. (2000) Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science, 287, 1232–1239. [DOI] [PubMed] [Google Scholar]
- 34.Christian J.H.B. and Waltho,J.A. (1962) Solute concentrations within cells of halophilic and non-halophilic bacteria. Biochim. Biophys. Acta, 65, 506–508. [DOI] [PubMed] [Google Scholar]
- 35.Diener J.L. and Wilson,C. (2000) Role of SRP19 in assembly of the Archaeoglobus fulgidus signal recognition particle. Biochemistry, 39, 12862–12874. [DOI] [PubMed] [Google Scholar]
- 36.Pakhomova O.N., Deep,S., Huang,Q., Zwieb,C. and Hinck,A.P. (2002) Solution structure of protein SRP19 of Archaeoglobus fulgidus signal recognition particle. J. Mol. Biol., 317, 145–158. [DOI] [PubMed] [Google Scholar]
- 37.Wild K., Sinning,I. and Cusack,S. (2001) Crystal structure of an early protein-RNA assembly complex of the signal recognition particle. Science, 294, 598–601. [DOI] [PubMed] [Google Scholar]
- 38.Ruepp A., Graml,W., Santos-Martinez,M.L., Koretke,K.K., Volker,C., Mewes,H.W., Frishman,D., Stocker,S., Lupas,A.N. and Baumeister,W. (2000) The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature, 407, 508–513. [DOI] [PubMed] [Google Scholar]
- 39.Ban N., Nissen,P., Hansen,J., Moore,P.B. and Steitz,T.A. (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science, 289, 905–920. [DOI] [PubMed] [Google Scholar]
- 40.Sanchez M.E., Urena,D., Amils,R. and Londei,P. (1990) In vitro reassembly of active large ribosomal subunits of the halophilic archaebacterium Haloferax mediterranei. Biochemistry, 29, 9256–9261. [DOI] [PubMed] [Google Scholar]
- 41.Schmeing T.M., Seila,A.C., Hansen,J.L., Freeborn,B., Soukup,J.K., Scaringe,S.A., Strobel,S.A., Moore,P,B. and Steitz,T.A. (2002) A pre-translocational intermediate in protein synthesis observed in crystals of enzymatically active 50S subunits. Nature Struct. Biol., 9, 225–230. [DOI] [PubMed] [Google Scholar]
- 42.Ring G. and Eichler,J. (2001) Large-scale preparation and characterization of inverted membrane vesicles from the halophilic archaeon Haloferax volcanii. J. Membr. Biol., 183, 195–204. [DOI] [PubMed] [Google Scholar]