Abstract
The coat proteins of different single-strand RNA phages use a common protein tertiary structural framework to recognize different RNA hairpins and thus offer a natural model for understanding the molecular basis of RNA-binding specificity. Here we describe the RNA structural requirements for binding to the coat protein of bacteriophage PP7, an RNA phage of Pseudomonas. Its recognition specificity differs substantially from those of the coat proteins of its previously characterized relatives such as the coliphages MS2 and Qβ. Using designed variants of the wild-type RNA, and selection of binding-competent sequences from random RNA sequence libraries (i.e. SELEX) we find that tight binding to PP7 coat protein is favored by the existence of an 8 bp hairpin with a bulged purine on its 5′ side separated by 4 bp from a 6 nt loop having the sequence Pu-U-A-G/U-G-Pu. However, another structural class possessing only some of these features is capable of binding almost as tightly.
INTRODUCTION
The coat proteins of the single-stranded RNA bacteriophages are translational repressors of viral replicase. They execute this function by specifically binding an RNA hairpin that encompasses the replicase start codon. Although this interaction is apparently conserved among all the RNA bacteriophages studied so far, the sequences of the coat proteins and the RNA structures they recognize have diverged during evolution. All the coat proteins possess a similar fold and bind RNA on the surface of a large β-sheet. Thus, they provide a model system for understanding how a single β-sheet structural framework can be adapted by mutation for the binding of diverse RNA structures. Figure 1 depicts the RNA-binding targets of the coat proteins of three different RNA phages, MS2, Qβ and PP7. The RNA sequence and structural requirements for recognition by MS2 coat protein have been determined in great detail, both by the analysis of a large series of individually constructed variants (1), and by selection in vitro of tight-binding RNAs from huge random sequence populations (2,3). Similar, but more limited experiments were also conducted for Qβ (3–5). The necessary structural features of these two RNAs are illustrated in Figure 1. Although each of the operators can be characterized as a stem–loop with a bulged adenosine, the similarities seem to end there, since the required length of the base-paired region, the size and sequence of the loop, and even the importance of the bulged A differ markedly. Apart from our prior identification of the sequence shown here as the natural PP7 translational operator (6), nothing is known about the RNA structural requirements for recognition by PP7 coat protein. However, simple inspection of the RNAs in Figure 1 indicates that the translational operator of PP7 has structural features that make it unrecognizable to the coat proteins of the other phages, an expectation verified by experiment (6). Here we present our elucidation of the RNA structural requirements for specific binding by PP7 coat protein.
Figure 1.
The structures of the MS2, Qβ and PP7 translational operators. For MS2 and Qβ prior work identified elements of their structures needed for binding by their respective coat proteins as illustrated in the generalized structures also shown here. N, any nucleotide; N′, a complementary nucleotide; Pu, purine; Py, pyrimidine.
MATERIALS AND METHODS
Construction of designed PP7 operator RNA variants
Synthetic DNA oligonucleotides containing the wild-type and mutant PP7 operator sequences flanked by EcoRI and BamHI sites were cloned into pT7-1 (U.S. Biochemicals). Their sequences can be seen in Figure 3. Transcription in vitro of the BamHI-cleaved plasmids with T7 RNA polymerase in the presence of [32P]ATP resulted in the production of labeled RNAs suitable for use in filter-binding experiments.
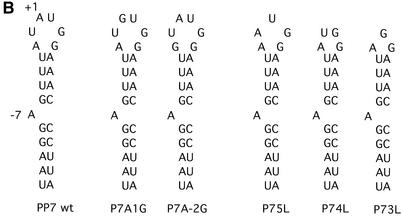
Figure 3.
The sequences and predicted secondary structures of the various designed PP7 translational operator variants used in this study. The bulge variants are shown in (A), the loop variants in (B) and the stem variants in (C).
Selection of PP7 coat protein-binding RNA aptamers
High-affinity RNA aptamers were isolated using the SELEX protocol described by Conrad et al. (7). The random sequence oligonucleotide template was 5′-GGGAGAATTCCGACC AGAAG—N30—TATGTCCGTCTACATGGATCCTCA-3′. Amplification of this template was achieved using the 5′ primer called 39.20 and the 3′ primer called 24.61 (7). A template was also produced that randomizes the loop sequence in an otherwise PP7-like structure. This template had the sequence 5′-GGAGAATTCCGACCAGAAGTAAGGAGT TTNNNNNNAAACCCTTAATAGAGGCAGATGATGGATCCAGT-3′. Its amplification was accomplished using 39.20 as the 5′ primer and a 3′ primer called 25.1 whose sequence was 5′-ACTGGATCCATCATCTGCCTCTAT-3′.
After selection, individual aptamers were cloned as EcoRI–BamHI fragments in pT7-1. The plasmids were then subjected to DNA sequence analysis and in vitro transcription to produce material for filter-binding analyses.
RNA-binding analysis
The PP7 coat protein was expressed in Escherichia coli and purified by methods described previously (6). 32P-labeled RNAs were produced by run-off transcription of plasmids cleaved with BamHI (8). Dissociation constants were determined in a protein-excess nitrocellulose filter-binding assay (9). RNA was present in these reactions at concentrations of 10 pM so that in the range of the various Kds, protein was always in excess. Binding curves were fitted to data using Kaleidagraph software (Abelbeck) and the equation F = P / (Kd + P), where F is the fraction of RNA bound, P is the protein concentration, and Kd is the dissociation constant. These binding assays were used to monitor the strength of the PP7 coat protein–RNA interaction during the course of the selection, and to determine the binding affinities of individual RNA variants.
RESULTS
Designed variants of PP7 RNA
Prior studies of the RNA operators of the bacteriophages MS2 and Qβ revealed that binding specificity is largely due to interaction with nucleotides found in single-stranded loops and bulges whose arrangement in space is due in large part to the pattern of base pairs (1) (Fig. 1). Therefore, we tested the importance of the identities of some nucleotides in the PP7 RNA operator loop and bulge so as to determine their importance for PP7 coat recognition. Variants were produced as synthetic oligonucleotide duplexes and cloned under the control of the T7 promoter in a plasmid. 32P-labeled RNAs were produced by transcription in vitro and their affinities for PP7 coat protein were determined in a protein-excess nitrocellulose filter-binding assay (9). The predicted structures of the designed variants are illustrated in Figure 3.
Note that a large number of binding curves were produced during this work. For simplicity we present only a few typical ones in Figure 2. These particular examples are for wild-type PP7 RNA and for three aptamers whose isolation is described later.
Figure 2.
Representative examples of binding curves. Shown here are the protein-excess nitrocellulose filter-binding curves for wild-type PP7 operator RNA and aptamers A, E and F (see Figs 4 and 5), which were selected for this illustration because they represent a range of binding behaviors (Kds from 0.7 to 25 nM).
The role of the bulged A
Wild-type PP7 operator RNA binds PP7 coat protein with Kd = 1 nM. Deletion of the bulge in variant P7NB (Fig. 3A) resulted in a nearly complete loss of binding activity. Replacement of the bulge with U (in P7UB) and C (in P7CB) elevated the Kd substantially to 63 and 38 nM, respectively (Table 1). A G-bulge variant was not included in this initial series due to its propensity [predicted by mfold (10)] for formation of an alternative secondary structure. To surmount this problem, we changed the three GC base pairs of the PP7 RNA duplex to CG base pairs, thereby allowing the production of a G-bulge variant with a well defined PP7-like secondary structure (P7GB-CG, Fig. 3A). The design of this variant assumes that the identities of nucleotides we altered within the base-paired regions are not important determinants of binding, an assumption that was born out by subsequent experiments (see later). The G-bulge variant was bound only slightly less well than wild-type (Kd = 2.5 nM).
Table 1. The dissociation constants of the designed PP7 operator variants depicted in Figure 3 for PP7 coat protein.
| RNA | Kd (nM) |
|---|---|
| PP7WT | 1 |
| P7NB | >1000 |
| P7UB | 63 |
| P7CB | 38 |
| P7GB-CG | 2.5 |
| P7A1G | 63 |
| P7A2G | 2 |
| P75L | 75 |
| P74L | 75 |
| P73L | >1000 |
| P7-WT | 1 |
| P7dl1 | 3.3 |
| P7mis2 | 6.7 |
| P7mis3 | 4.4 |
| P7short | 15 |
The importance of loop sequence and size
Since A residues in the loops of other operators have proven to play important roles in coat protein binding, we also tested the importance of the identities of the loop A nucleotides by substituting A +1 or A –2 with G residues in the P7A1G and P7A2G mutants (Fig. 3B). The replacement of A +1 with G increased the Kd to 63 nM, but the substitution of G for A –2 was well tolerated, elevating the Kd only 2-fold (Table 1). More extensive experiments, reported below, further elaborate the roles played by loop nucleotides and show that A or G, but not C or U, is tolerated at A –2.
Progressive deletion of nucleotides A –1, A +1 and U +2 changes the loop size from 6 to 5 to 4 and finally 3. Each change results in a serious defect (Table 1). It is not possible, of course, to distinguish effects of loop size and sequence changes, but the importance of the loop sequence is further elaborated in SELEX experiments described later.
The role of stem length and integrity
To test the role of the base-paired portions of the PP7 operator hairpin we created several variants that disrupt or delete base pairs in order to alter stem length (Fig. 3C). The variant called P7dl1 deletes the UA base pair at the top of the stem, thus moving the bulge closer to the loop. P7mis2 and P7mis3 introduce mismatches to progressively shorten the stem from the bottom. The results of filter-binding experiments indicate that each of these changes causes a relatively modest loss of coat protein-binding affinity ranging from 3.3- to 6.7-fold (Table 1). Combining the deletion of the UA at the top of the stem with mismatches in the three base pairs at the bottom of the stem still allows significant binding. The Kd of this variant (P7short) is elevated only 15-fold compared with wild type (Table 1). Thus, although the tightest binding requires a stem length and bulge placement similar to wild type, significant disruptions are reasonably well tolerated.
Selection in vitro of PP7 RNA loop variants that bind PP7 coat protein
The PP7 RNA operator contains a 6 nt loop having the sequence AUAUGG (Fig. 1). To identify the required features of this sequence we conducted a SELEX experiment. A pool of RNA sequences was created with the PP7 stem and bulged A intact, but with the loop sequence completely randomized. The initial pool had little or no affinity for PP7 coat protein (Kd was >1000 nM) but affinity steadily improved until after four rounds of selection the pool was bound with Kd = 1 nM. The selected RNAs were reverse transcribed, PCR amplified and cloned. Twenty-four clones were subjected to DNA sequence analysis, and among them we found 10 different sequences. Filter-binding experiments were conducted for each of these sequences and the results are shown in Table 2. The wild-type loop sequence, AUAUGG, was the most numerous of the sequences recovered, but several sequences with single, double, and even triple substitutions were also found. The wild-type sequence was the most tightly bound by coat protein but most of the selected variants had Kds no worse than ∼2-fold higher than wild type. Two positions, A +1 and G +3 were not substituted in any of the variants we found, and U –1 was substituted only once (variant L6 in Table 2). A –2 tolerates substitution with G, as we already knew from the designed variants described above, but apparently pyrimidines are not tolerated here. U +2 can be replaced by G, although we never isolated a variant with this change alone; it was always accompanied by the substitution of G +4 by A. The G +4 nucleotide seems relatively tolerant of A substitutions, but sequences with the single replacement of G +4 with C or U were the worst binders in this set. Taking together all the sequences shown in Table 2 we define the consensus sequence 5′-Pu-Py-A-G/U-G-N-3′. However, if we consider only the sequences that give Kds within ∼2-fold of wild type we may define a more restricted sequence required for tightest binding: 5′-Pu-U-A-G/U-G-Pu-3′.
Table 2. The affinities for PP7 coat protein of PP7 RNA loop sequence variants isolated by SELEX from a random loop sequence library.
| Name | Loop sequence | No. of isolates | Kd (nM) |
|---|---|---|---|
| L1 | AUAUGG | 5 | 1.0 |
| L2 | AUAUGA | 4 | 2.3 |
| L3 | AUAUGC | 3 | 7.0 |
| L4 | AUAUGU | 1 | 13 |
| L5 | AUAGGA | 1 | 1.9 |
| L6 | ACAGGA | 1 | 5.0 |
| L7 | GUAUGA | 4 | 1.9 |
| L8 | GUAUGU | 3 | 1.5 |
| L9 | GUAGGA | 1 | 1.0 |
| L10 | GUAUGG | 1 | 2.0 |
| PP7 WT | AUAUGG | 1.0 |
Selection in vitro from an N30 random sequence library
We also conducted a SELEX experiment starting with a random sequence pool containing no deliberately introduced similarities to the natural binding target. We followed essentially the method of Conrad et al. (7) using a random RNA sequence pool spanning 30 nt positions (N30). A starting amount of 1 µg (∼2 × 1013 individual molecules) of the synthetic N30 random sequence template was converted to double-stranded DNA by five cycles of PCR and subsequently transcribed in vitro to produce an RNA library for selection for binding to PP7 coat protein. To monitor the progress of the selection, filter-binding measurements were conducted after every three rounds. A constant amount of PP7 coat protein (20 pmol) was used but the RNA:protein molar ratio was varied from 100:1 in the first to third rounds, to 10:1 in the fourth to sixth rounds, to 1:1 in the seventh to ninth rounds, and finally to 50:1 in the 10th to 12th rounds. RNA populations having increasing affinities for PP7 coat protein were obtained. By the 12th round the pool had a Kd of 4 nM compared with wild-type PP7 RNA with Kd = 1 nM. Sequences were recovered from the 12th round by RT–PCR and cloned into pT7-1 as described in Materials and Methods. Forty individual clones were subjected to DNA sequence analysis. Figures 4 and 5 depict the secondary structures predicted for the various aptamers and Table 3 gives their Kds for binding PP7 coat protein and the number of occurrences of each.
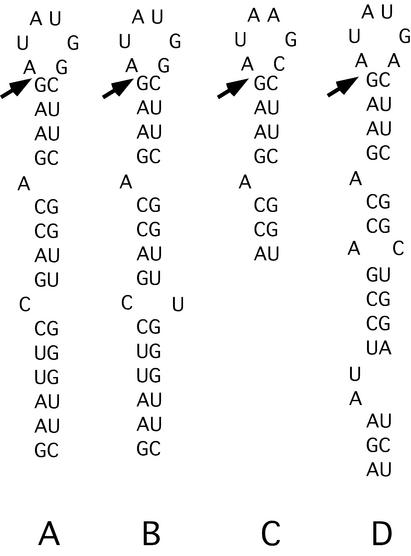
Figure 4.
Sequences and predicted secondary structures of so-called group I RNAs found by SELEX to tightly bind PP7 coat protein. The arrows indicate the junction of sequences derived from the common (to the left) and randomized (to the right) portions of the original template.
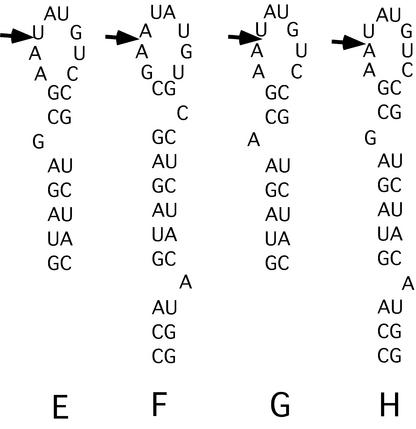
Figure 5.
Sequences and predicted secondary structures of group II RNAs found by SELEX to tightly bind PP7 coat protein. The arrows indicate the junction of sequences derived from the common (to the right) and randomized (to the left) portions of the original template.
Table 3. Affinities (in nM) for PP7 coat protein of aptamers selected for binding from the N30 random sequence library.
| RNA | Kd (nM) | No. of isolates |
|---|---|---|
| Aptamer A | 0.7 | 2 |
| Aptamer B | 0.8 | 3 |
| Aptamer C | 1.0 | 4 |
| Aptamer D | 1.5 | 3 |
| Aptamer E | 2.0 | 16 |
| Aptamer F | 25 | 1 |
| Aptamer G | 2.0 | 2 |
| Aptamer H | 2.0 | 2 |
| Others | 80–700 | 7 |
| PP7 WT | 1.0 |
Many of the aptamers bear a striking similarity to the natural PP7 translational operator. We call these group I aptamers (i.e. RNAs A, B, C and D in Fig. 4). The Kds of these PP7-like RNAs for PP7 coat protein range from 0.67 to 1.5 nM. These RNAs have secondary structures very like that of the true PP7 operator with a bulged A separated by 4 bp from the six-membered loop. Moreover, their loop sequences are identical (A and B) or highly similar (C and D) to the wild-type sequence, AUAUGG. Other aptamers (E, F, G and H in Fig. 5) have less similarity to the folded structure of the PP7 operator. We categorize these as group II RNAs. All have hairpins but the number of base pairs, the positions and identities of bulged nucleotides, and the sizes of the loops are different from group I RNAs. Notably, all of them have 8 nt loops that contain a perfect or near perfect fit to the wild-type loop sequence, emphasizing its importance for PP7 binding. Each of these aptamers also had adjacent stem–loops that we originally speculated might contribute to binding affinity, but subsequent experiments using synthetic versions that contain only the hairpins shown in Figure 4 demonstrated that the binding activities of these aptamers are due entirely to these hairpins. Moreover, synthetic versions of the neighboring hairpins had no detectable affinity for coat protein. Despite their divergence from the wild-type structure, the group II RNAs can bind PP7 coat protein tightly, the E, G and H aptamers all having Kds of 2 nM. F binds significantly less well, but still has Kd = 25 nM. All the group II aptamers, except F, have a bulged purine on the 5′ side of the stem, 2 bp below the loop.
It should be noted that the RNA aptamers isolated from the N30 population generally formed secondary structures that involved portions of the common sequences that flank the random sequence region. The sequences 5′ of the arrow in group I (Fig. 4), and sequences 3′ of the arrow in group II aptamers (Fig. 5) are derived from the primer sequences common to all the aptamers. Thus, the evolution of these sequences was partially constrained. Fortuitously, the common sequences 3′ of the random element shared certain features with the required loop sequence. Consequently, in the group II aptamers selection apparently found it easiest to find the required elements of operator RNA structure by incorporating that portion of the common sequence into the loop. Note also that the F aptamer (the most poorly bound of both the group I and II aptamers) contains mutations in the common sequence (just below the loop on the 3′ side) that cause it to deviate slightly from the others. We assume these mutations were due to low-level errors occurring during synthesis of the 3′ primer used for RT–PCR and were probably introduced into clone F in the amplification step after the last round of selection.
We also note the existence of seven RNAs (listed as ‘others’ in Table 3) whose predicted RNA secondary structures bore no obvious similarity to the PP7 RNA operator and that bound coat protein from 80- to 700-fold less tightly than wild-type (data not shown). We do not know whether these RNAs bind coat protein on its usual RNA-binding surface or bind it at some other site. In any case, we omit them from this analysis because their binding is comparatively weak.
DISCUSSION
Protein recognition of RNA typically takes advantage of RNAs capacity to fold into distinctive shapes (11). The exact nucleotide sequence of base-paired regions is often unimportant, but serves mainly to fold the molecule into a shape that properly positions single-stranded nucleotides so that base-specific interactions with protein can occur. Although important interactions with base pairs are sometimes seen, they are generally confined to regions near helical disruptions (bulges and helix ends) where the usually deep and narrow RNA major groove is rendered locally accessible.
Our results with the PP7 coat protein–operator RNA interaction are consistent with these general principles. We summarize in Figure 6 the important elements of the PP7 operator as defined by the designed variants (Fig. 3), by selection from a pool of randomized loop sequences (Table 2), and by the group I SELEX products (Fig. 4). While a base-paired stem is a required element of the translational operator, its sequence seems irrelevant. Thus, a number of tight-binding variants were isolated having little or no sequence similarity to wild-type in the base-paired regions. A bulged-A is apparently needed for tightest binding, although when pains were taken to avoid alternative RNA folding, a G here was almost as good. Replacement of the bulge with a pyrimidine resulted in greater decreases in binding affinity, and wholesale deletion of the bulge resulted in nearly complete loss of binding activity. For best binding the bulge should be placed on the 5′ side of the hairpin, at a distance of 4 bp from the loop, although a distance of 3 bp (or even 2 bp) was surprisingly well tolerated. The stem below the bulge should be 4 or 5 bp long for tightest binding, but shortening of this lower stem to 3 bp had a relatively modest effect.
Figure 6.
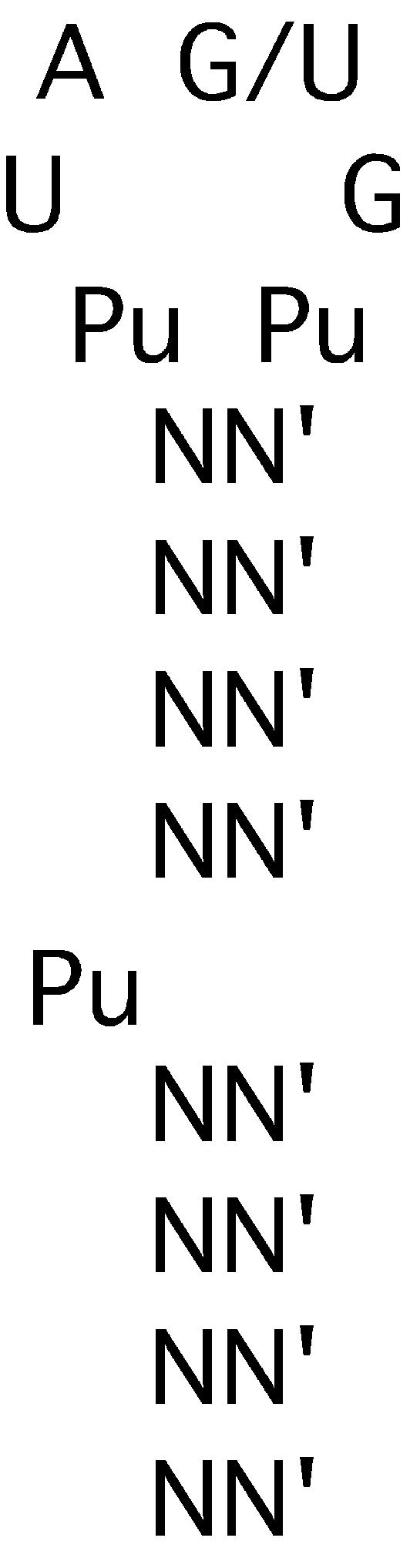
Summary of the required elements of the PP7 operator structure derived from the designed variants (Fig. 3) and the group I SELEX RNAs (Fig. 4).
The only requirement for a more or less definite sequence was found in the loop; tight-binding aptamers generally had loop sequences that were either exact mimics, or only slight variants of the wild-type loop. This presumably reflects the existence of specific amino acid–loop nucleotide contacts in the RNA–protein complex, but could also be the result of sequence-dependent RNA conformational requirements. In the case of the well-characterized MS2 RNA–protein complex (12,13) it is clear that the identities of some loop nucleotides (i.e. A –4 and U –5) are constrained by the requirements of their direct contacts with coat protein, while another (A –7) serves a mainly RNA-structural role. A similar combination of protein-contacting and RNA structure constraints probably operates here.
Although the group I aptamers conform to the picture shown in Figure 6, the existence of the group II RNAs makes it clear that other, related structures are also capable of tight interaction with coat protein. It is notable that each group II aptamer contains the required 6 nt sequence, but it is found within a larger loop of 8 nt. Besides having a larger loop, each of the tight-binding group II aptamers (E, G and H) differs from the group I RNAs in containing a bulged purine at a position 2 nt below the loop. Although we know from designed variants that deleting the bulge from wild-type RNA dramatically impairs binding (Kd > 1000 nM), aptamer F retains significant binding activity (Kd = 25 nM) without having any apparent properly placed bulge. The E, G and H aptamers bind with nearly wild-type affinities when they have a bulged A or G only 2 bp below the loop. Some of the other deviant features present in the group II RNAs may somehow compensate for these apparent defects. For example, there could be some importance to the fact that the group II clones have 8-nt loops with the required loop sequence starting 1 nt into the loop. Thus, even in the group II structures, the bulge is spaced 3 nt from the start of the required loop sequence. The fact that one of these three is unpaired may introduce added flexibility to maintain an appropriate spatial relationship of bulge and loop. Remember that moving the bulge 1 bp closer to the loop (i.e. 3 bp below the loop instead of 4 bp) in the P7dl1 variant had a modest effect, elevating Kd by only 3.3-fold.
These results are in a sense reminiscent of those obtained when SELEX was conducted with MS2 coat protein (2,3). RNA ligands were identified that possessed some of the features of the wild-type operator and retained significant binding activity, but they achieved the necessary contacts with coat protein by somewhat different means. For example, the F6 aptamer changed the length of the stem above the bulge from 2 to 3 bp and reduced the loop size from 4 to 3 nt. Designed variants showed that each of these changes by itself would be sufficient to greatly diminish binding (1), but when present together in the F6 aptamer the RNA was able to make contacts with coat protein highly similar to those observed in the wild-type complex (12–14). The spatial relationship between the bulged A and the loop is a crucial aspect of RNA recognition by MS2 coat protein, and it is retained in the F6 aptamer by means that were not obvious until the structure of its complex with coat protein was determined. Analogously, we suspect that the tight-binding type II aptamers of Figure 5 may make contacts with PP7 coat protein that are similar to those formed in the wild-type and group I complexes, even though the means by which they do this is not obvious in the absence of detailed structural information.
Although the various RNA phages have different coat protein and translational operator sequences, each virus seems to have preserved the interaction between them. The coat protein and its RNA ligand therefore represent a co-evolving pair in which mutations in one are compensated by changes in the other so as to preserve translational repressor and genome encapsidation functions. Moreover, the fact that the coat proteins of different RNA phages use homologous tertiary structures to bind diverse RNAs illustrates the adaptability of the β-sheet framework. We recently proposed a hypothetical model for the Qβ coat protein–RNA complex that rationalizes its RNA-binding specificity as a relatively straightforward variation on the MS2 model (5). Despite the overall divergence of their amino acid sequences and of their RNA targets, a close relationship between the MS2 and Qβ coat proteins is indicated by conservation of nearly all the important RNA-binding site amino acids and by the fact that single amino acid substitutions can confer to each protein the ability to bind the other’s RNA (15,16). PP7 diverges from MS2 more dramatically, however, and extensive efforts to confer to MS2 coat protein the RNA-binding specificity of PP7 have so far failed (F. Lim and D. S. Peabody, unpublished observations). The divergence between the MS2 and PP7 coat protein RNA-binding sites—they differ in 10 out of 15 potential RNA-binding site amino acids on the β-sheet—is reflected in correspondingly large differences in their respective RNA targets. Although both RNAs can be generally characterized as stem–loops containing a bulged adenosine, it is the size and sequence of the loop, and the differing placement of the bulge that must account for differences in binding specificity. We hope eventually to understand the interactions responsible for these specificity differences and to understand how the coat protein β-sheet can be adapted for the binding of divergent RNA structures.
Acknowledgments
ACKNOWLEDGEMENT
This work was supported by a grant from the NIH.
REFERENCES
- 1.Romaniuk P.J., Lowary,P.T., Wu,H.-N., Stormo,G. and Uhlenbeck,O.C. (1987) RNA binding site of R17 coat protein. Biochemistry, 26, 1563–1568. [DOI] [PubMed] [Google Scholar]
- 2.Schneider D., Tuerk,C. and Gold,L. (1992) Selection of high affinity RNA ligands to the bacteriophage R17 coat protein. J. Mol. Biol., 228, 862–869. [DOI] [PubMed] [Google Scholar]
- 3.Hirao I., Spingola,M., Peabody,D.S. and Ellington,A.D. (1999) The limits of specificity: an experimental analysis with RNA aptamers to MS2 coat protein variants. Mol. Divers., 4, 75–89. [DOI] [PubMed] [Google Scholar]
- 4.Whitherell G.W. and Uhlenbeck,O.C. (1989) Specific RNA binding by Qβ coat protein. Biochemistry, 28, 71–76. [DOI] [PubMed] [Google Scholar]
- 5.Spingola M., Lim,F. and Peabody,D.S. (2002) Recognition of diverse RNAs by a single protein structural framework. Arch. Biochem. Biophys., 405, 122–129. [DOI] [PubMed] [Google Scholar]
- 6.Lim F., Downey,T.P. and Peabody,D.S. (2001) RNA binding by PP7 coat protein. J. Biol. Chem., 276, 22507–22513. [DOI] [PubMed] [Google Scholar]
- 7.Conrad R.C., Giver,L., Tian,Y. and Ellington,A.D. (1996) In vitro selection of nucleic acid aptamers that bind proteins. Methods Enzymol., 267, 336–367. [DOI] [PubMed] [Google Scholar]
- 8.Draper D.E., White,S.A. and Kean,J.M. (1988) Preparation of specific ribosomal RNA fragments. Methods Enzymol., 164, 221–237. [DOI] [PubMed] [Google Scholar]
- 9.Carey J., Cameron,V., de Haseth,P.L. and Uhlenbeck,O.C. (1983) Sequence-specific interaction of R17 coat protein with its ribonucleic acid binding site. Biochemistry, 22, 2601–2610. [DOI] [PubMed] [Google Scholar]
- 10.Jacobson A.B. and Zuker,M. (1993) Structural analysis by energy dot plot of a large mRNA. J. Mol. Biol., 233, 261–269. [DOI] [PubMed] [Google Scholar]
- 11.Draper D.E. (1999) Themes in RNA-protein recognition. J. Mol. Biol., 293, 255–270. [DOI] [PubMed] [Google Scholar]
- 12.Valegård K., Murray,J.B., Stockley,P.G., Stonehouse,N.J. and Liljas,L. (1994) Crystal structure of an RNA bacteriophage coat protein–operator complex. Nature, 371, 623–626. [DOI] [PubMed] [Google Scholar]
- 13.Valegård K., Murray,J.B., Stonehouse,N.J., van den Worm,S., Stockley,P.G. and Liljas,L. (1997) The three-dimensional structures of two complexes between recombinant MS2 capsids and RNA operator fragment reveal sequence-specific protein–RNA interactions. J. Mol. Biol., 270, 724–738. [DOI] [PubMed] [Google Scholar]
- 14.Convery M.A., Rowsell,S., Stonehouse,N.J., Ellington,A.D., Hirao,I., Murray,J.B., Peabody,D.S., Phillips,S.E.V. and Stockley,P.G. (1998) Crystal structure of an RNA aptamer–protein complex at 2.8 Å resolution. Nature Struct. Biol., 5, 133–139. [DOI] [PubMed] [Google Scholar]
- 15.Spingola M. and Peabody,D.S. (1997) MS2 coat protein mutants which bind Qbeta RNA. Nucleic Acids Res., 25, 2808–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim F., Spingola,M. and Peabody,D.S. (1996) The RNA-binding site of bacteriophage Qβ coat protein. J. Biol. Chem., 271, 31839–31845. [DOI] [PubMed] [Google Scholar]