Abstract
A multicomponent complex of proteins and RNA is assembled on the newly synthesized pre-mRNA to form the spliceosome. This complex catalyzes a two-step transesterification reaction required to remove the introns and ligate the exons. To date, only six proteins have been found necessary for the second step of splicing in yeast, and their human homologs have been identified. We demonstrate that the addition of the selective chelator of zinc, 1,10-phenanthroline, to an in vitro mRNA splicing reaction causes a dose-dependent inhibition of the second step of splicing. This inhibition is accompanied by the accumulation of spliceosomes paused before completion of step two of the splicing reaction. The inhibition effect on the second step is due neither to snRNA degradation nor to direct binding to the mRNA, and is reversible by dialysis or add-back of zinc, but not of other divalent metals, at the beginning of the reaction. These findings suggest that the activity of a putative zinc-dependent metalloprotein(s) involved in the second step of splicing is affected. This study outlines a new method for specific reversible inhibition of the second step of splicing using external reagents, and suggests a possible role of divalent cations in the second step of mRNA splicing, most likely zinc.
INTRODUCTION
A multicomponent complex of proteins and RNA is assembled on the newly synthesized precursor messenger RNA (pre-mRNA) to form the spliceosome. This complex catalyzes a two-step transesterification reaction required to remove the introns and ligate the exons (1,2). During the first step of splicing, a 2′-hydroxyl of adenosine from the branch site attacks the phosphodiester bond at the 5′-splice site to form a 5′ exon and a 3′ exon-lariat intermediate. In the second step, the 3′-end hydroxyl group of the 5′ exon attacks the 3′-splice site, leading to exon ligation and the release of the intron lariat (3). Spliceosome formation proceeds via a coordinated assembly of complexes, E, A, B, C and I (4,5), or through a one-step assembly (6). Approximately 50 proteins are involved in the splicing reaction (6–9), some 15 of which are associated with purified complex C, in which splicing is arrested after the first step (10). To date, only six proteins have been found necessary for the second step of splicing in yeast (11–22), and their human homologs have been identified (23–29). Little is known, however, about the involvement of these and other factors in the second step.
It is estimated that ∼15% of the point mutations linked to human genetic diseases cause splicing defects (30); hence, understanding the elementary steps of the splicing machinery, spliceosome assembly, and the network of interactions between the splicing factors is a prerequisite to finding a cure for these diseases. Current methods to arrest splicing and spliceosome assembly include heat inactivation, ATP or magnesium depletion (31) and dephosphorylation of the nuclear extract (32); immunodepletion of the nuclear extract from the hPrp18 protein blocks step two of splicing and can be restored following re-addition of the protein (23); using mutations at the 5′-splice site or branch site sequence (33 and references therein) and at the 3′-splice site (10) to examine the rearrangement of splicing factors leading to the first or second step of splicing, respectively; studying the various U2 and U6 snRNP mutants shown to arrest splicing before the first or second step (23 and references therein; 34); and a bimolecular system that employs a pre-mRNA lacking the 3′-splice site to arrest splicing after the first step, proceeding to the second step after the addition of a second RNA molecule containing the 3′-splice site and second exon (35). These methods can stop splicing activity and freeze spliceosome assembly for analysis of the proteins and RNA components required for each step. None of them, however, involve a straightforward reversible method for inhibiting the second step of splicing using an external reagent.
The zinc-finger or -knuckle motif is a major structural motif involved in protein–nucleic acid and protein–protein interactions (36), and is present in the largest superfamily of transcription factors (37). Zinc ions coordinate this finger-like structure through bonds created with cysteine and histidine residues. Several spliceosomal proteins were found to possess the zinc-binding motif as well, including Slu7, SF3a60, SF3a66, SF1, ZNF265, 9G8, hLuc7p and U1-specific protein C (38–41). The zinc-finger domain, for example, is required for incorporation of SF3a60 and SF3a66 into the U2 snRNP (39), and was recently proposed to aid in the interaction of the SF1 protein with nucleotides located upstream of the branch point signal in the pre-mRNA (42). For some of these proteins the zinc-binding motif is essential for the splicing activity (39), while for others it exerts a modest effect (21). Since the RNA backbone carries a negative charge, it also carries a number of metal ions to neutralize the charge. The majority of these metal ions bind the RNA non-specifically, very weakly, and in rapid exchange (43). Little is known, however, about the requirement of zinc in the splicing reaction.
We demonstrate specific inhibition on the second step of mRNA splicing by the chelator of zinc, 1,10-phenanthroline (1,10-phe). The inhibition is accompanied by the accumulation of active spliceosomes paused before completion of step two of the splicing reaction. This effect on the second step is due neither to snRNA degradation nor to direct binding to the mRNA, and is reversible by dialysis or zinc add-back prior to the beginning of the reaction, suggesting that a putative zinc-dependent metalloprotein(s) is required for the second step of mRNA splicing. Two other chelators, neocuproine (NC) and bathocuproinedisulfonic acid (BC), exhibited a similar inhibiting effect on the second step of splicing, supporting the notion that a divalent cation is involved in the second step of splicing. This is the first indication that a putative zinc-dependent metalloprotein is involved in the second step of mRNA splicing. A new method for specific reversible inhibition of the second step of pre-mRNA splicing using external reagents is outlined, suggesting a possible role of zinc in the second step of splicing.
MATERIALS AND METHODS
Splicing substrates, reactions and spliceosome complex analysis
Standard splicing reactions (12.5 µl) contained 60% (v/v) HeLa nuclear extract, 2.4 mM MgCl2, 0.5 mM ATP, 20 mM creatine phosphate, 30 000 c.p.m. of mRNA precursor uniformly labeled with [α-32P]UTP (1 × 109 c.p.m./ng RNA), and the indicated concentration of 1,10-phe (Sigma), 1,7-phenanthroline (1,7-phe; Sigma), 2,9-dimethyl-1,10-phenanthroline (NC; Sigma) and 2,9-dimethyl-4,7-diphenyl-1,10-phenanthrolinedisulfonic acid (BC; Sigma). Following incubation at 30°C for 60 or 90 min, RNA was extracted by proteinase K treatment, phenol extraction and ethanol precipitation, and analyzed by polyacrylamide gel electrophoresis (PAGE) in 8% denaturing gels, essentially as described (44). Spliceosome complexes were analyzed in native 4% gels as described (45). The gels were run at 300 V for 6 h. The human β-globin gene (46) was recloned into pBluescript II SK+ plasmid (Stratagene), linearized by BamHI restriction enzyme digest, and transcribed from the T7 promoter. We previously described the adenovirus major late transcript plasmid (Adeno) (44,47). For the dialysis procedures, splicing reactions were inserted into Spectra/Por molecular porous membrane tubing (Spectrum Medical Industries; MWCO 12–14 000) and dialyzed against nuclear extract buffer (Buffer D) (48) at 500 times the volume of the reaction for 12 h at 4°C. 1,10-phe and 1,7-phe were prepared as 50 mM stock solutions in water. The pH was adjusted to 7.9 with NaOH, and the solutions were heated to 65°C to dissolve the reagents. Similar results were obtained when these reagents were prepared as 500 mM stock solutions in DMSO.
Northern blotting
Nuclear extract was incubated in in vitro splicing conditions for 60 min at 30°C, as mentioned above, with the indicated concentration of 1,10-phe and CuSO4. The RNA was extracted by proteinase K treatment, phenol extraction and ethanol precipitation, separated by electrophoresis in 10% polyacrylamide gel, and blotted onto a Hybond-XL nylon membrane (Amersham-Pharmacia). Hybridization was performed in a 50% formamide solution at 42°C with 32P-labeled complementary antisense riboprobes to U1, U2, U4, U5 and U6 snRNAs. After hybridization, blots were washed and exposed to X-ray film.
RESULTS
1,10-phe inhibits the second step of splicing
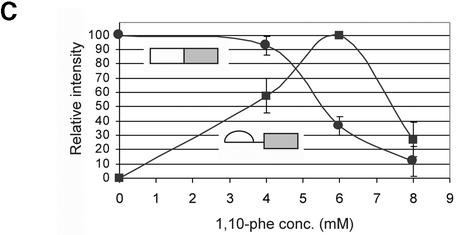
To examine the requirement for zinc in the mRNA splicing in vitro reaction, HeLa nuclear extract was incubated under in vitro splicing conditions with a radiolabeled adenovirus major late transcript (Adeno), and various concentrations of the potent zinc chelator 1,10-phe (see Fig. 1A for molecular structure). Following 90 min of incubation, the RNA was extracted and separated in 8% denaturing gel (Fig. 1B). At these conditions the splicing reaction reached the maximum level of mRNA accumulation after 60 min (Fig. 2A and data not shown). There was up to a 10-fold increase in the amount of the first-step splicing intermediates—3′ exon–intron lariat and 5′ exon intermediate—when 4 or 6 mM 1,10-phe was added to the splicing reaction. This resulted in at least a 2-fold decrease in products of the second step of mRNA splicing— intron lariat and mRNA—when 6 mM but not 4 mM 1,10-phe was added (Fig. 1B, compare lane 1 with lanes 2 and 3). Adding 8 mM 1,10-phe reduced the first step of splicing intermediate products (Fig. 1B, lane 4), and adding 10 mM or more 1,10-phe completely inhibited the splicing activity (data not shown). An average of three independent experiments shows an increase of 16% in total splicing products of reactions incubated with 6 mM 1,10-phe compared with reactions incubated without 1,10-phe (data not shown), suggesting that 4 and 6 mM 1,10-phe enhances splicing activity. Thus, 1,10-phe inhibited splicing in a dose-dependent manner, with the addition of 6 mM 1,10-phe leading to accumulation of intermediates of the first step of splicing and inhibition of the second step.
Figure 1.
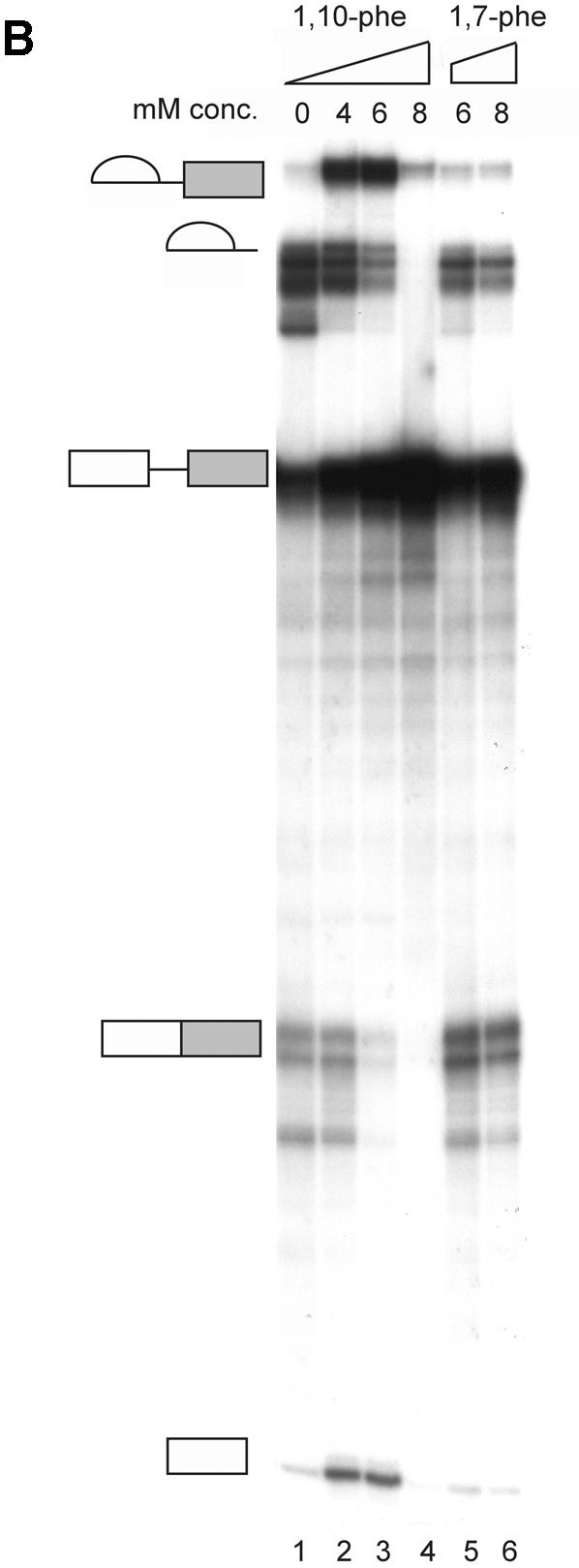
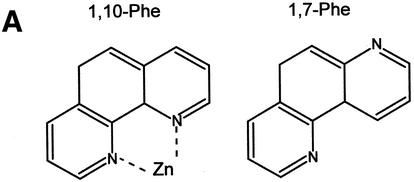
1,10-phe can efficiently inhibit the second step of mRNA splicing. (A) The molecular structure of 1,10-phe and 1,7-phe. Zinc is schematically added at its potential binding site. (B) A radiolabeled transcript of Adeno was incubated in an in vitro splicing reaction. The indicated concentration of the zinc chelating agent 1,10-phe or the non-chelating zinc agent 1,7-phe was added to each reaction and the reactions were incubated for 90 min at 30°C. RNA was extracted and separated by PAGE in 8% denaturing gel. RNA intermediates and products are schematically presented on the left; exons are represented by boxes and introns by lines. (C) The inhibition of the first and second step of splicing products—3′ exon-lariat (squares) and mRNA (circles)—as a function of the concentration of 1,10-phe in three different experiments. Minimum and maximum levels of the first and second steps of splicing products were normalized to a relative intensity of zero and 100, respectively.
Figure 2.
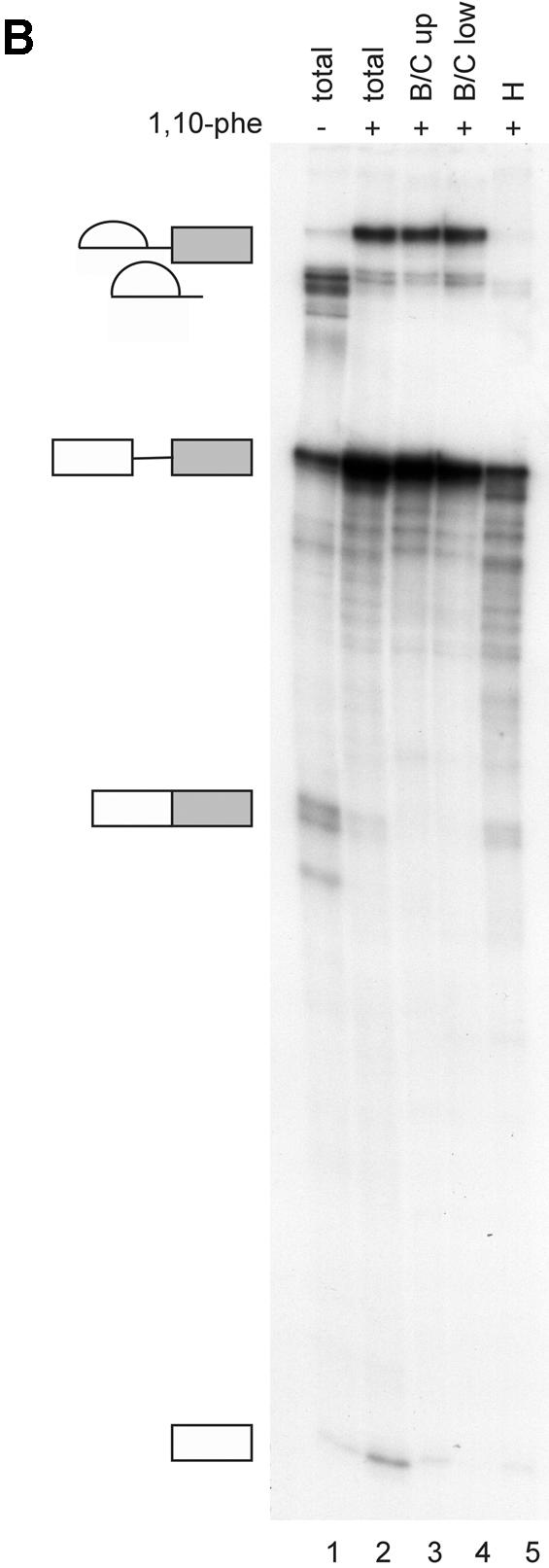
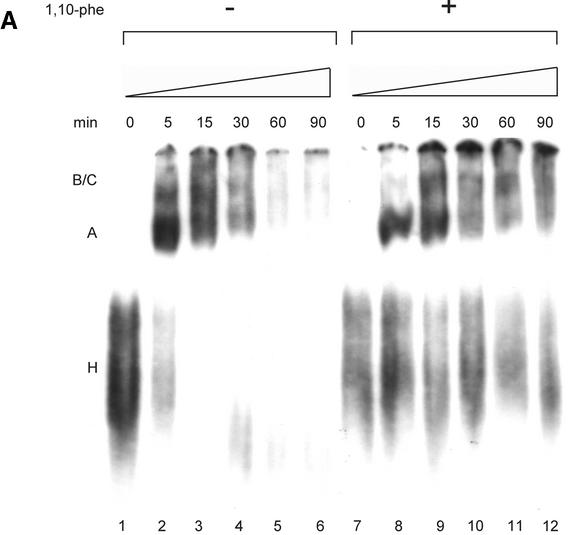
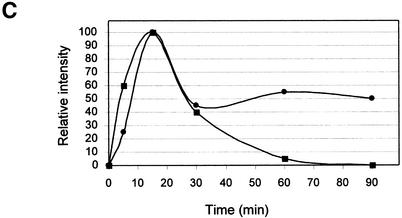
Analysis of spliceosomal complexes following the addition of 1,10-phe to the splicing reaction. (A) A radiolabeled Adeno transcript was incubated with nuclear extract under splicing conditions for the indicated duration at 30°C in the absence (–) or presence (+) of 6 mM 1,10-phe. Splicing complexes were fractionated in a 4% non-denaturing gel. (B) Analysis of the labeled mRNA contents in the B/C and H complexes. RNA from a splicing reaction incubated in the presence of 6 mM 1,10-phe for 60 min [(A), +1,10-phe; 60 min; B/C] was eluted from the gel, and the labeled RNA was extracted and separated in 8% denaturing gel. Complex B/C was divided into upper (B/C up) and lower (B/C low) parts according to the location in the gel. The RNA from complex B/C of the upper and lower parts was analyzed together with total RNA from a splicing reaction incubated in the absence (–) or presence (+) of 6 mM 1,10-phe. RNA intermediates and products are schematically represented to the left of the gel. (C) The relative intensity of the splicing complex B/C formed in the absence (squares) or presence (circles) of 1,10-phe as a function of time. Minimum and maximum intensities were normalized to zero and 100, respectively.
It is possible that 1,10-phe inhibits the splicing reaction non-specifically by intercalating into the RNA or the spliceosome. To test this assumption, a non-chelating structural isomer, 1,7-phe (see Fig. 1A for molecular structure), was added to an in vitro splicing assay; as shown in Figure 1B (lanes 5 and 6), 1,7-phe had no effect on splicing activity at concentrations of 6 and 8 mM. We concluded that the addition of 6 mM 1,10-phe to the splicing reaction efficiently and specifically inhibited the second step of splicing, and raising the concentration above 8 mM inhibited both steps of splicing. These findings suggest that the observed inhibition of splicing by 1,10-phe is due to its ability to act as a chelator of divalent cations, presumably zinc. Figure 1C illustrates the percentage of inhibition of the first and second steps of splicing as a function of the concentration of 1,10-phe in three independent experiments.
We determined that inhibition of the second step of splicing following addition of 1,10-phe is a general phenomenon by showing a similar pattern of second-step inhibition using human β-globin pre-mRNA (data not shown).
The effect of 1,10-phe on the spliceosome
To investigate the effect of 1,10-phe on spliceosome assembly, labeled Adeno was incubated under splicing conditions in the absence (–) or presence (+) of 6 mM 1,10-phe for different durations (Fig. 2A). In the absence of 1,10-phe (Fig. 2A, lanes 1–6), complex A was detected after 5 min of incubation and declined thereafter; this is similar to the data reported by Konarska and Sharp (45). Complex B/C appeared after 5 min, reached a peak at 15 min, and declined steadily thereafter. Thus, after 60 and 90 min of incubation both A and B/C complexes were barely detectable. When 6 mM 1,10-phe was added (Fig. 2A, lanes 7–12), complexes A and B/C appeared in the same time course as reactions incubated without 1,10-phe after 5 and 15 min, respectively. However, while the amount of complex A steadily decreased, complex B/C remained stable for at least 1 h of incubation. Moreover, complex H, which decreased in the reaction without 1,10-phe, remained stable after 60 min of incubation. Thus, inhibition of the second step of splicing with 6 mM 1,10-phe was accompanied by accumulation of complex B/C.
The RNA content within complex B/C of the second-step inhibited reaction was determined by two-step analysis. Labeled Adeno was incubated in a splicing reaction for 60 min with 6 mM 1,10-phe, and separated in a native gel. RNA was extracted from complex B/C and separated by electrophoresis in denaturing 8% PAGE (Fig. 2B). As expected, the splicing intermediates—the 3′ exon–intron lariat and 5′ exon intermediate—were detected only in complex B/C. It is noteworthy that 2-fold less 5′ exon intermediate was detected in complex B/C compared with the total RNA extracted from the unfractionated reaction, possibly due to a lower binding affinity of 5′ exon intermediate to these complexes. Complex H, detected in the initial stages without 1,10-phe and in all stages when incubated with 1,10-phe, was confirmed to have various degraded RNA and possibly mRNA products (Fig. 2B, lane 5) (49). Figure 2C illustrates the relative intensity of complex B/C in the absence and presence of 1,10-phe as a function of time. These results indicate that inhibition of the second step of splicing leads to accumulation of complex B/C containing splicing intermediates paused after completion of the first step of splicing, and that the second-step inhibition effect is not caused by degradation of the mRNA products.
The snRNAs are not degraded by 1,10-phe–copper complexes
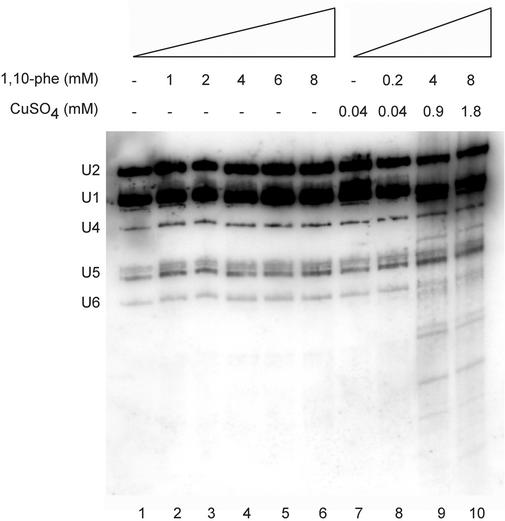
Several reports demonstrate that 1,10-phe can interact with copper to form a DNA/RNA chemical–nuclease complex (50,51) with the ability to specifically degrade RNA molecules at the single-stranded regions (51). We were concerned that degradation of one or several of the snRNAs led to the second-step inhibition effect in these experiments, although the optimal conditions for chemical–nuclease activity differed considerably from those in the splicing reaction with H2O2 and ascorbate (52), and no degradation of the pre-mRNA, splicing intermediates and products was detected when up to 10 mM 1,10-phe was added (see Fig. 1B and data not shown), or even in the purified spliceosomal complex (Fig. 2B). To clarify this we examined whether the snRNAs in the splicing reaction remained intact following incubation with 1,10-phe and copper. The splicing reaction was incubated with an increasing concentration of 1,10-phe, or both 1,10-phe and copper, at ratios required to degrade DNA (52); the RNA was purified and the snRNAs were examined by northern blotting. The addition of up to 8 mM 1,10-phe to the splicing reaction did not induce snRNA degradation (Fig. 3, lanes 1–6). However, when 1,10-phe and copper were added to the splicing reaction, snRNA degradation was detected at 0.9 and 1.8 mM copper (Fig. 3, lanes 9 and 10, respectively). These results indicate that the inhibition of the second step of mRNA splicing is not due to the formation of a DNA/RNA chemical–nuclease complex between 1,10-phe and copper, and the copper concentration in the nuclear extract is not sufficient to induce nuclease degradation.
Figure 3.
The snRNAs are not degraded by the addition of 1,10-phe. Nuclear extract was incubated in in vitro splicing conditions for 60 min at 30°C with the indicated concentration of 1,10-phe and CuSO4. RNA was extracted, separated by electrophoresis in a 10% polyacrylamide gel, and transferred to a nylon membrane. Following hybridization with U1, U2, U4, U5 and U6 complementary labeled antisense riboprobes, the membrane was washed and exposed to X-ray film. The snRNAs were identified according to size and are marked on the left.
NC and BC inhibit the second step of splicing
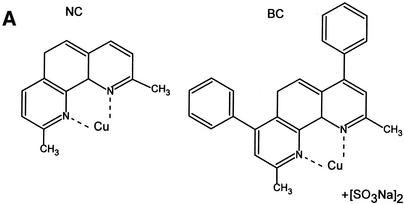
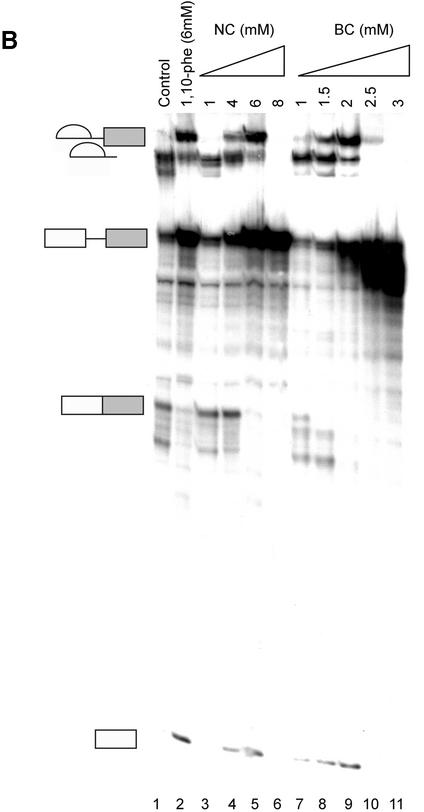
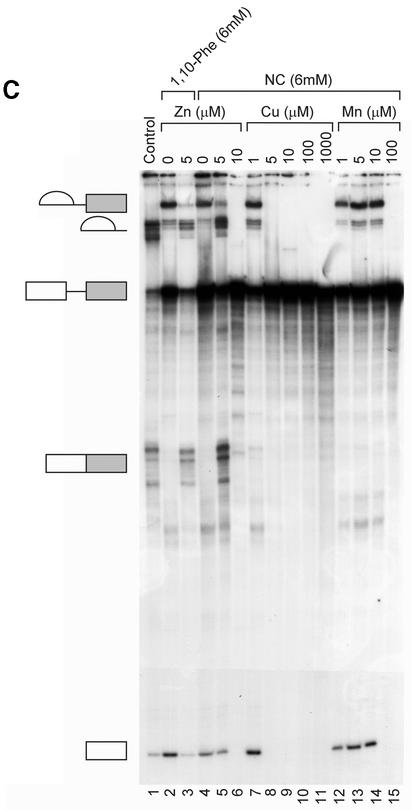
To extend our understanding of 1,10-phe’s effect on splicing, we analyzed dose-dependent splicing reactions in the presence of two copper-specific chelators, NC and BC, which share a high degree of structural similarity with 1,10-phe (see Fig. 4A for molecular structure). NC and BC are primarily copper chelators, with a 5.2 pKi of NC for copper compared with 4.1 for zinc (53), that form a complex with divalent cations similarly to 1,10-phe (52,54). The addition of NC to the splicing reaction led to a similar second-step inhibition pattern as that seen with 1,10-phe. The addition of 6 mM NC led to accumulation of first-step splicing products and inhibition of the second step, while 8 mM inhibited formation of all splicing products (Fig. 4B, lanes 5 and 6, respectively; also see Fig. 5C, lane 4). Interestingly, the addition of 1.5 mM BC caused accumulation of first-step splicing products and decreased second-step products (Fig. 4B, lane 8). Increasing BC concentration to 2 mM inhibits the second step by 70% and increases accumulation of first-step splicing products (Fig. 4B, lane 9). At 2.5 mM BC, the first splicing step was inhibited by 90% and the entire second step (Fig. 4B, lane 10). However, BC caused pre-mRNA degradation when 2.5 mM or more was added to the splicing reaction. We concluded that the addition of 6 mM NC and 2.5 mM BC to this nuclear extract inhibited the second step of splicing. Raising the NC concentration to 8 mM and the BC to 3 mM inhibited both steps of splicing. These results suggest that the inhibition is through chelation of a divalent cation, most likely copper.
Figure 4.
NC and BC can efficiently inhibit the second step of mRNA splicing. (A) The molecular structure of NC and BC. Copper is schematically added at its potential binding site. (B) A radiolabeled transcript of Adeno was incubated in an in vitro splicing reaction with the indicated concentration of NC and BC. RNA was extracted and separated in 8% denaturing gel. RNA intermediates and products are schematically presented on the left.
Figure 5.
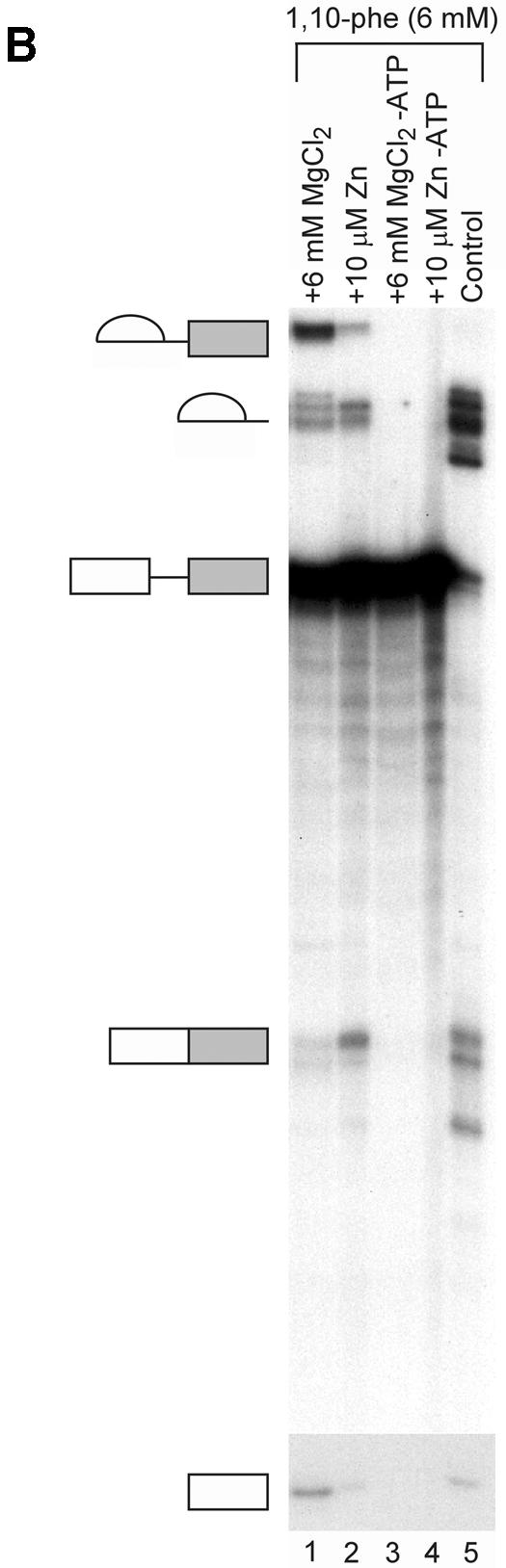
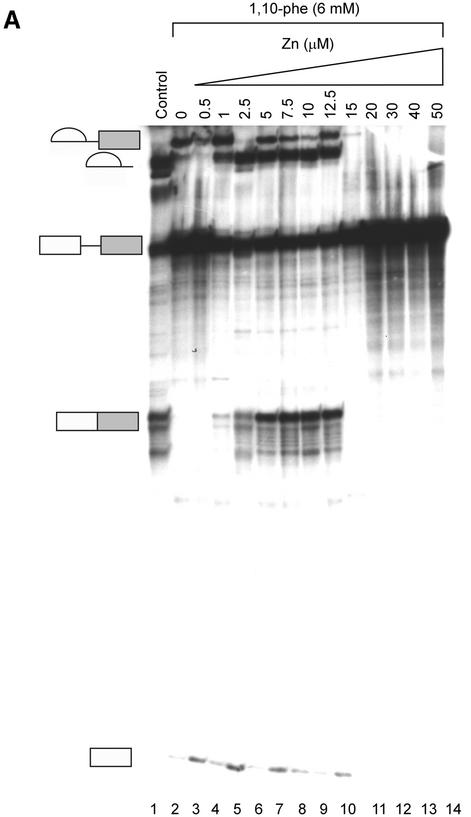
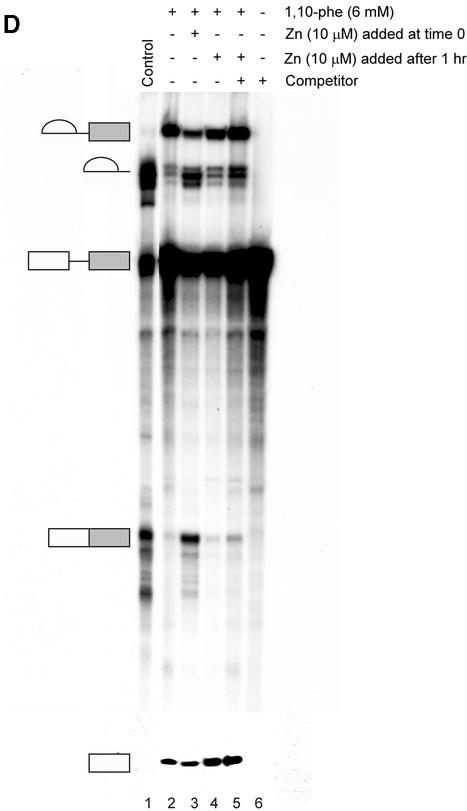
(Opposite and above) Relieving the inhibitory effect of 1,10-phe on the second step of splicing. (A) Zinc add-back effect on 1,10-phe-inhibited mRNA splicing. A radiolabeled Adeno transcript was incubated for 90 min at 30°C in nuclear extract under standard splicing conditions in the absence (lane 1) or presence (lanes 2–14) of 6 mM 1,10-phe and the indicated concentration of zinc (lanes 3–14). (B) The inhibition is specific to the zinc atoms and not to the magnesium atoms. Pre-incubated 1,10-phe with zinc (Zn) or magnesium (MgCl2) were added to the splicing reaction as indicated (lanes 1–4), and RNA was purified and separated in 8% denaturing gel. (C) Zinc, copper and manganese add-back effect on NC-inhibited mRNA splicing. A radiolabeled Adeno transcript was incubated in the splicing conditions mentioned above in the absence (lane 1) or presence (lanes 4–15) of 6 mM NC and the indicated concentration of zinc (Zn, lanes 2–6), copper (Cu, lanes 7–11), and manganese (Mn, lanes 12–15). RNA products were purified and separated in 8% denaturing gel. (D) The addition of zinc after completion of the first step of splicing cannot recover the second step. A radiolabeled Adeno transcript was incubated for 60 min at 30°C in nuclear extract under standard splicing conditions in the absence (lane 1) or presence (lanes 2–5) of 6 mM 1,10-phe. Zinc at 10 µM concentration was added at the beginning of the incubation (lane 3), or after 60 min of incubation (lanes 4 and 5), and all the reactions were incubated for another 60 min. A 250 times molar excess of non-radioactive Adeno RNA was added after 60 min of incubation (lane 5), or from the beginning of the incubation (lane 6). RNA was purified and separated in 8% denaturing gel.
The inhibition effect is specific to zinc and is reversible
Since all three chelators tested inhibited splicing, we undertook to determine which divalent cation was involved in the inhibition. An add-back experiment was performed to examine the specificity of inhibition and to test whether the inhibition is due to the chelation of zinc or copper. Labeled Adeno was incubated in a splicing reaction in the presence or absence of 6 mM 1,10-phe. RNA was extracted and separated in 8% denaturing gel. The addition of 6 mM 1,10-phe led to an accumulation of intermediates of the first step of splicing and a decrease in products of the second step (Fig. 5A, lanes 1 and 2, respectively). To determine whether the inhibitory effect on the second step of splicing was reversible, 6 mM 1,10-phe was added to the nuclear extract, followed by increasing concentrations of zinc, and then the labeled Adeno transcript (Fig. 5A, lanes 3–14). The addition of 1–5 µM zinc restored the second step of splicing in a dose-dependent manner (seen clearly in the mRNA product); the addition of 5–12.5 µM zinc completely restored the second step; the addition of 15 µM zinc presents a stepwise reduction in the splicing reaction (seen in this exposure in the 3′ exon-lariat intermediate); 20–50 µM zinc completely inhibited the splicing activity. This indicates that the inhibition effect was specific to the zinc, and similar to other add-back studies demonstrating that an excess of zinc inhibits the enzymatic activity (55). The addition of up to 10 µM zinc (without 1,10-phe) to the splicing reaction had no effect on splicing activity (data not shown). Remarkably, add-back of zinc to the splicing reaction to which 10 mM 1,10-phe (which completely inhibited the reaction) had been added, led to a complete non-reversible inhibition effect on the splicing activity (data not shown). This implied that a different zinc-binding protein(s) essential for the first step of splicing was brought into play when the concentration of 1,10-phe was increased from 6 to 10 mM in the splicing reaction. It is also possible that a reversible complex formed between a zinc-binding protein, zinc, and 1,10-phe at 6 mM 1,10-phe, while the chelation irreversibly removed the zinc from the metalloprotein(s) at 10 mM 1,10-phe, and the binding site became occupied by another divalent cation that affected the structure and/or function of the metalloprotein(s). The addition of a pre-mixed complex of 6 mM 1,10-phe and 6 mM MgCl2 to the splicing reaction led to the accumulation of intermediate products paused after completion of the first step of splicing (Fig. 5B, lane 1). These results point to the specificity of the inhibition to the zinc atoms and not to the magnesium atoms, which are also essential for the splicing activity (56). They also show that zinc is required for the second step of mRNA splicing, presumably within a putative zinc-dependent metalloprotein.
To further confirm that the chelation of zinc was responsible for the effect on the second step of splicing, we carried out a similar add-back experiment using NC, which showed a similar dose-dependent inhibition effect on the second step of splicing as 1,10-phe. This time the inhibited reaction was supplemented with various other metals. Again, an identical release of the second-step inhibition was seen when 5 µM zinc was added to a splicing reaction incubated with 6 mM 1,10-phe or NC (Fig. 5C, compare lanes 2 and 3 with lanes 4 and 5). The second-step splicing inhibition could not be released by 1 µM copper, and both steps of splicing were inhibited by 5 µM to 1 mM copper (Fig. 5C, lanes 7–11). The addition of 1–10 µM manganese could not release the second-step inhibition, and 100 µM manganese inhibited the splicing activity (Fig. 5C, lanes 12–15). This suggests that the inhibition effect is due to the chelation of zinc and not copper (see Discussion).
We next undertook to determine whether zinc add-back can restore the second step of splicing after completion of the first step. The addition of 6 mM 1,10-phe inhibited the second step of splicing and led to the accumulation of first-step intermediates, while addition of 6 mM 1,10-phe and 10 µM zinc, and then the labeled pre-mRNA, restored the second step of splicing (Fig. 5D, lanes 2 and 3, respectively). To examine the ability of zinc to restore second-step activity after step two had been inhibited and step one had been carried out, we incubated the splicing reaction with 6 mM 1,10-phe for 1 h, added 10 µM zinc and incubated for another hour (Fig. 5D, lane 4). This restored the second-step activity by only 5%, suggesting that the addition of zinc to the majority of the spliceosomes that paused after completion of the first step does not release the inhibition. However, the lower percentages of mRNA recovery might be due to zinc entering certain spliceosomes paused before the second step, thus restoring the second-step activity. It might also be because pre-mRNAs that were not assembled into the spliceosome during the first hour of incubation were assembled and spliced during the second hour of incubation, as recently demonstrated (6). To test these two possibilities, we added a 250 molar excess of unlabeled Adeno (used as a competitor) and 10 µM zinc after 1 h of incubation with 6 mM 1,10-phe, and incubated the reaction for another hour (Fig. 5D, lane 5). However, when labeled Adeno pre-mRNA was incubated with a 250 molar excess of unlabeled Adeno from the beginning of the incubation, it completely eliminated the splicing activity, indicating that an excess of unlabeled Adeno inhibited splicing activity on newly assembled spliceosomes, as previously seen (57). The comparable level of restoration of second-step splicing activity detected with or without the addition of unlabeled competitor suggests that zinc can penetrate a small fraction of the spliceosomes that paused after completion of the first step, reverse the inhibition, and allow these spliceosomes to complete the reaction (Fig. D, compare lanes 4 and 5, respectively). But, addition of zinc to the majority of the spliceosomes that paused after completion of the first step by 1,10-phe did not restore the second step. We concluded that the addition of zinc to 1,10-phe prior to the beginning of the reaction can reverse the inhibition effect, while the addition of zinc to assembled spliceosomes that completed the first step renders the zinc incapable of restoring the second-step activity, presumably because of an inability to enter the spliceosomes.
To follow up these observations, we examined whether the inhibition effect could be released by dialyzing the 1,10-phe from the splicing reaction. Labeled Adeno transcript was incubated under splicing conditions in the absence (Fig. 6, lanes 1 and 2), or presence (Fig. 6, lanes 3 and 4) of 6 mM 1,10-phe. Following a 90 min incubation at 30°C, one part of the reaction was removed, and dialyzed for 12 h against the nuclear extract buffer (Buffer D) (48) to remove the free 1,10-phe from the splicing reaction. The reactions were removed from the dialysis tube, a new set of splicing cofactors was added (ATP, MgCl2 and creatine phosphate), and the reactions were incubated for another 90 min. The addition of 6 mM 1,10-phe led to the accumulation of a splicing intermediate paused after completion of the first step of splicing (Fig. 6, lane 3). The dialysis process and the re-incubation of the splicing reaction restored the second step of splicing to the reaction inhibited by 6 mM 1,10-phe (Fig. 6, lane 4). Also, a 250 molar excess of unlabeled Adeno (used as a competitor) was added to the reaction inhibited by 6 mM 1,10-phe, which eliminated re-assembly of new spliceosomal complexes on the labeled Adeno following the dialysis (57). This restored the second step of splicing to the same level as splicing incubated without the competitor indicating that the dialysis removes the 1,10-phe from the spliceosomes paused before the second step, thus restoring to these spliceosomes the second-step activity (data not shown).
Figure 6.
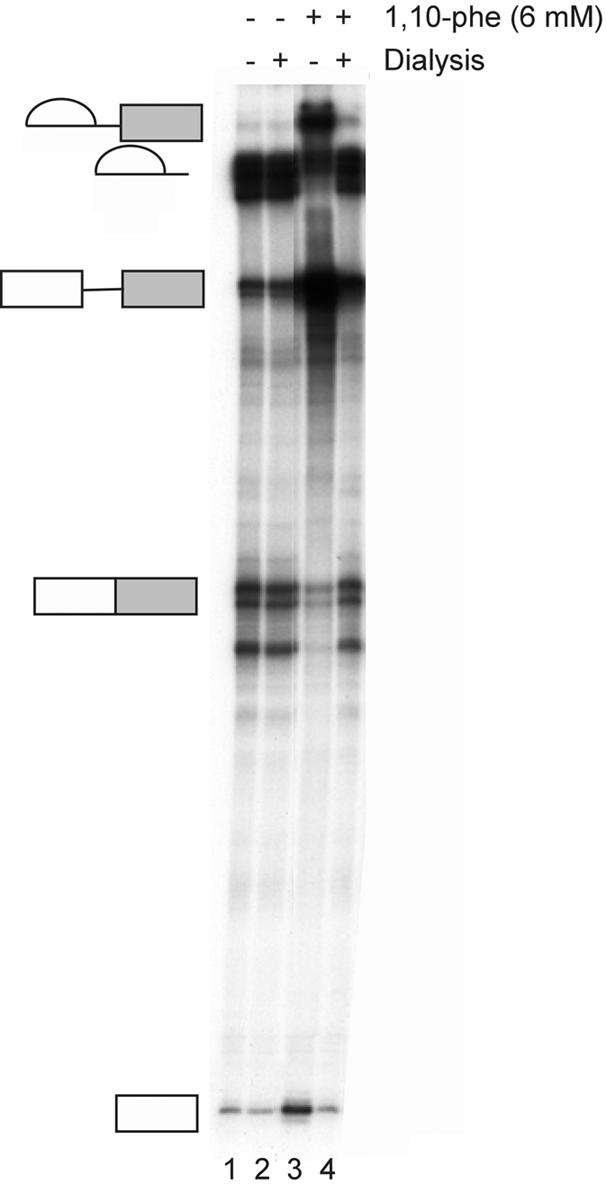
Removal of 1,10-phe restores the second step of splicing. A radiolabeled Adeno transcript was incubated in nuclear extract under standard splicing conditions in the absence (lanes 1 and 2) or presence (lanes 3 and 4) of 6 mM 1,10-phe. Following 90 min at 30°C, the reactions were dialyzed against the nuclear extract buffer (Buffer D) (48) for 12 h. ATP, creatine phosphate and MgCl2 were then added, followed by incubation for another 90 min at 30°C. RNA was purified and separated in 8% denaturing gel.
The restoration of splicing activity following inhibition by 6 mM 1,10-phe reduced the likelihood that free chelator, remaining in the reaction after the dialysis, inhibited the second step by interaction with the 3′ end of the first exon following the first step of splicing (the nucleophyl required to attack the 3′-splice site during the second step). The double-distilled water used in this experiment contained ∼10 p.p.b. zinc, which probably restored the required zinc to the nuclear extract treated with 1,10-phe (Shamryahu Blumberg, personal communication; 58). Most importantly, this indicated that the inhibition can be reversed by a 12 h dialysis, and served as further proof that the inhibition is due neither to steric interference at the RNA’s 5′-splice site nor to formation of a complex between the 3′ end of the first exon and 1,10-phe following the first step of splicing.
DISCUSSION
We have demonstrated the first known effect of zinc chelation on mRNA splicing: the addition of 6 mM of the zinc-specific chelator 1,10-phe inhibits the second step of splicing, and raising the concentration of 1,10-phe to 8 mM or higher blocks both steps of splicing. The inhibition leads to accumulation of spliceosomal complexes paused after completion of the first step of splicing. The add-back of zinc but not other divalent cations from the beginning of the reaction reverses the inhibition effect, and prolonged dialysis restores the second step of splicing, indicating the specificity of the inhibition to the chelation of the zinc. These results suggest that chelation of zinc with 1,10-phe either removes the zinc from the active site of a metalloprotein(s) involved in the second step of mRNA splicing, or leads to the formation of a complex with the zinc still attached to the protein, which blocks the function of the metalloprotein(s). The chelation of zinc might also interfere with the structural integrity of a metalloprotein(s) involved in the second step of mRNA splicing. A possible candidate for the chelating effect is Slu7, which has a zinc-knuckle motif near the N-terminal and is required for the second step of mRNA splicing (21; ref. 38 for the human homolog). However, we cannot rule out that the chelation affects other proteins that are essential for the second step of splicing, especially those in which the zinc-finger or -knuckle motif is involved in protein–nucleic acid or protein–protein interactions. We also note that mutations in the yeast Slu7 zinc-knuckle motif caused only a moderate decrease in yeast splicing (21) (see Note Added in Proof).
It is possible that 1,10-phe chelates the magnesium required for the second step of mRNA splicing (56). However, since the pKi of 1,10-phe for zinc is 6.36 and for magnesium is 1.2 (53), 1,10-phe will interact with magnesium only if the concentration of magnesium is >1 M. The magnesium concentration in the splicing reaction is 2.4 mM. The high-binding constant to magnesium, and the inability of magnesium to restore second step of mRNA splicing in the add-back assay (Fig. 5B), argue against the inhibition effect on the second step of mRNA splicing by magnesium and for zinc. Caution must be exercised in the use of affinity constant values, as they are for defined conditions, usually incubated with purified proteins rather than with extracts, and do not take into consideration the formation of ternary complexes between chelates and bimolecules (54).
We were surprised to find that NC and BC, which are commonly used as copper chelators (54,59), exerted a similar effect on splicing inhibition as the zinc chelator (Fig. 4B). One possible explanation is that NC and BC chelate zinc rather than copper in the nuclear extract. Another explanation is that NC and BC chelate copper from binding sites of other factors not involved in the second-step inhibition. The stripping of copper from proteins in the nuclear extract causes them to become potential competitors for the zinc metals in the reaction. As a result, a zinc-dependent metalloprotein involved in the second step of splicing might be depleted of its metal even if it does not involve a direct interaction with a chelator (Nigel Robinson, personal communication). This assumption remains to be tested, but could also account for the irreversibility of the inhibition at high concentrations (10 mM 1,10-phe). In other words, zinc is removed from a metalloprotein involved in the second step of splicing, other divalent cations are free to interact with the protein, and its activity is impaired. The zinc chelation assay can be used as a new method to specifically and efficiently inhibit the second step of mRNA splicing, which can subsequently be released by prolonged dialysis.
Support for the notion that the inhibition is not due to the formation of a 1,10-phe complex with the 3′ end of the first exon or its intercalation into the RNA comes from a number of our findings: the release of the second-step inhibition following dialysis (Fig. 6); the accumulation of a lariat intron in the reactions in which the second step was inhibited; and the inability of 1,7-phe and a strong cis-diol-forming reagent to inhibit splicing (3-aminophenylboronic acid; data not shown).
It is intriguing that the inhibitory effect on the splicing reaction commences at millimolar concentrations that are in great excess of the spliceosomal component’s concentration (60), assuming that the 1,10-phe distribution in the nuclear extract is homogenous. Nevertheless, this inhibition is rather specific since it is reversible upon zinc addition and is carried out within a range of concentrations similar to that used in many other studies: e.g. the dialysis against 5 mM 1,10-phe for 50 h necessary to remove zinc ion from T7 RNA polymerase (61), and the 5 mM 1,10-phe required to inhibit the mussel matrix metalloproteinases (62–65). Similarly, the half-life for the removal of zinc ion by 0.8 mM 1,10-phe is ∼30 h for Escherichia coli polymerase I (66). These reports support the relatively high concentration required to exert splicing inhibition reported here, especially at the moderately short intervals used in our experiments.
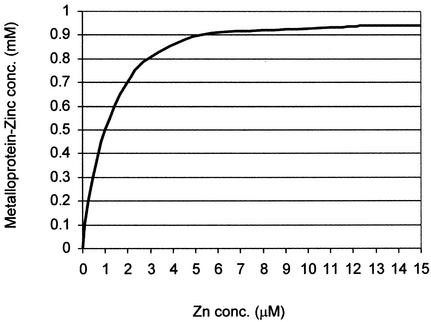
The observation that on the one hand a concentration of 6 mM 1,10-phe is required to inhibit the second step, but on the other that this block is rescued by the addition of 5 µM zinc, is intriguing. However, the reaction mixture is complex with a substantial number of low-affinity metal-binding sites. The chelating agent scavenges from these first. Only when these are depleted does it remove metal from higher affinity sites on the metalloprotein(s) involved in the second step. Conversely, the sites on the metalloprotein(s) will be the first to be repopulated when zinc is added back. Thus, a lot of chelator is needed to be added to inactivate the second step, but not much metal to restore activity. The Supplementary Material describes the calculations carried out to semi-quantify the concentration of zinc that binds the metalloprotein(s) with affinity similar to the metalloprotein(s) required for the second step of splicing. The solution to these calculations, presented in Figure 7, indicates that the affinity of zinc to the metalloprotein(s) is 10 000 times stronger than 1,10-phe, and that 5–15 µM zinc can restore 90–100% binding of the metalloprotein:zinc complex.
Figure 7.
The zinc concentration bound to metalloproteins in the nuclear extract. The graph presents the solution to the calculations in the Supplementary Material. It demonstrates that metalloproteins which have high affinity for zinc will require a high concentration of 1,10-phe to remove the zinc from the protein, but a relatively small amount of zinc to fully restore the function of the metalloprotein(s).
NOTE ADDED IN PROOF
While this manuscript was in press the following paper was published indicating that single mutations in Slu7’s zinc-knuckle had no effect on cellular growth in yeast (Shelley-Ann,J., Turner,W. and Schwer,B. (2002) RNA, 8, 1068–1077).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Dr Pinchuk Ilya for his assistance with the calculations in the Supplementary Material. This work was supported by an award from the Israel Academy of Science and in part by a grant from the Israel Cancer Association and the Leukemia Research Foundation to G.A. N.S. was partially supported by the Rieger Foundation scholarship.
REFERENCES
- 1.Konarska M.M. (1998) Recognition of the 5′ splice site by the spliceosome. Acta Biochim. Pol., 45, 869–881. [PubMed] [Google Scholar]
- 2.Nilsen T.W. (2002) The spliceosome: no assembly required? Mol. Cell, 9, 8–9. [DOI] [PubMed] [Google Scholar]
- 3.Burge M., Tuschi,T. and Sharp,P.A. (1999) Splicing of precursors to mRNA by the spliceosome. In Gesteland,R.F. and Atkins,J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 525–560.
- 4.Horowitz D.S. and Krainer,A.R. (1994) Mechanisms for selecting 5′ splice sites in mammalian pre-mRNA splicing. Trends Genet., 10, 100–106. [DOI] [PubMed] [Google Scholar]
- 5.Rosbash M. and Seraphin,B. (1991) Who’s on first? The U1 snRNP-5′ splice site interaction and splicing. Trends Biochem. Sci., 16, 187–190. [DOI] [PubMed] [Google Scholar]
- 6.Stevens S.W., Ryan,D.E., Ge,H.Y., Moore,R.E., Young,M.K., Lee,T.D. and Abelson,J. (2002) Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol. Cell, 9, 31–44. [DOI] [PubMed] [Google Scholar]
- 7.Collins C.A. and Guthrie,C. (2000) The question remains: is the spliceosome a ribozyme? Nature Struct. Biol., 7, 850–854. [DOI] [PubMed] [Google Scholar]
- 8.Das R., Zhou,Z. and Reed,R. (2000) Functional association of U2 snRNP with the ATP-independent spliceosomal complex E. Mol. Cell, 5, 779–787. [DOI] [PubMed] [Google Scholar]
- 9.Hong W., Bennett,M., Xiao,Y., Feld Kramer,R., Wang,C. and Reed,R. (1997) Association of U2 snRNP with the spliceosomal complex E. Nucleic Acids Res., 25, 354–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gozani O., Patton,J.G. and Reed,R. (1994) A novel set of spliceosome-associated proteins and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO J., 13, 3356–3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schneider S. and Schwer,B. (2001) Functional domains of the yeast splicing factor Prp22p. J. Biol. Chem., 276, 21184–21191. [DOI] [PubMed] [Google Scholar]
- 12.Horowitz D.S. and Abelson,J. (1993) A U5 small nuclear ribonucleoprotein particle protein involved only in the second step of pre-mRNA splicing in Saccharomyces cerevisiae. Mol. Cell. Biol., 13, 2959–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vijayraghavan U. and Abelson,J. (1990) PRP18, a protein required for the second reaction in pre-mRNA splicing. Mol. Cell. Biol., 10, 324–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Umen J.G. and Guthrie,C. (1995) The second catalytic step of pre-mRNA splicing. RNA, 1, 869–885. [PMC free article] [PubMed] [Google Scholar]
- 15.Teigelkamp S., Whittaker,E. and Beggs,J.D. (1995) Interaction of the yeast splicing factor PRP8 with substrate RNA during both steps of splicing. Nucleic Acids Res., 23, 320–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones M.H., Frank,D.N. and Guthrie,C. (1995) Characterization and functional ordering of Slu7p and Prp17p during the second step of pre-mRNA splicing in yeast. Proc. Natl Acad. Sci. USA, 92, 9687–9691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwer B. and Guthrie,C. (1991) PRP16 is an RNA-dependent ATPase that interacts transiently with the spliceosome. Nature, 349, 494–499. [DOI] [PubMed] [Google Scholar]
- 18.Schwer B. and Gross,C.H. (1998) Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J., 17, 2086–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collins C.A. and Guthrie,C. (1999) Allele-specific genetic interactions between Prp8 and RNA active site residues suggest a function for Prp8 at the catalytic core of the spliceosome. Genes Dev., 13, 1970–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siatecka M., Reyes,J.L. and Konarska,M.M. (1999) Functional interactions of Prp8 with both splice sites at the spliceosomal catalytic center. Genes Dev., 13, 1983–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frank D. and Guthrie,C. (1992) An essential splicing factor, SLU7, mediates 3′ splice site choice in yeast. Genes Dev., 6, 2112–2124. [DOI] [PubMed] [Google Scholar]
- 22.Vijayraghavan U., Company,M. and Abelson,J. (1989) Isolation and characterization of pre-mRNA splicing mutants of Saccharomyces cerevisiae. Genes Dev., 3, 1206–1216. [DOI] [PubMed] [Google Scholar]
- 23.Horowitz D.S. and Krainer,A.R. (1997) A human protein required for the second step of pre-mRNA splicing is functionally related to a yeast splicing factor. Genes Dev., 11, 139–151. [DOI] [PubMed] [Google Scholar]
- 24.Ono Y., Ohno,M. and Shimura,Y. (1994) Identification of a putative RNA helicase (HRH1), a human homolog of yeast Prp22. Mol. Cell. Biol., 14, 7611–7620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Z. and Reed,R. (1998) Human homologs of yeast prp16 and prp17 reveal conservation of the mechanism for catalytic step II of pre-mRNA splicing. EMBO J., 17, 2095–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ortlepp D., Laggerbauer,B., Mullner,S., Achsel,T., Kirschbaum,B. and Luhrmann,R. (1998) The mammalian homologue of Prp16p is overexpressed in a cell line tolerant to Leflunomide, a new immunoregulatory drug effective against rheumatoid arthritis. RNA, 4, 1007–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ben Yehuda S., Dix,I., Russell,C.S., Levy,S., Beggs,J.D. and Kupiec,M. (1998) Identification and functional analysis of hPRP17, the human homologue of the PRP17/CDC40 yeast gene involved in splicing and cell cycle control. RNA, 4, 1304–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindsey L.A. and Garcia-Blanco,M.A. (1998) Functional conservation of the human homolog of the yeast pre-mRNA splicing factor Prp17p. J. Biol. Chem., 273, 32771–32775. [DOI] [PubMed] [Google Scholar]
- 29.Luo H.R., Moreau,G.A., Levin,N. and Moore,M.J. (1999) The human Prp8 protein is a component of both U2- and U12-dependent spliceosomes. RNA, 5, 893–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blencowe B.J. (2000) Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases [published erratum appears in Trends Biochem. Sci. (2000) 25, 228]. Trends Biochem. Sci., 25, 106–110. [DOI] [PubMed] [Google Scholar]
- 31.Wassarman D.A. and Steitz,J.A. (1992) Interactions of small nuclear RNA’s with precursor messenger RNA during in vitro splicing. Science, 257, 1918–1925. [DOI] [PubMed] [Google Scholar]
- 32.Mermoud J.E., Cohen,P.T. and Lamond,A.I. (1994) Regulation of mammalian spliceosome assembly by a protein phosphorylation mechanism. EMBO J., 13, 5679–5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Champion-Arnaud P., Gozani,O., Palandjian,L. and Reed,R. (1995) Accumulation of a novel spliceosomal complex on pre-mRNAs containing branch site mutations. Mol. Cell. Biol., 15, 5750–5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fabrizio P. and Abelson,J. (1990) Two domains of yeast U6 small nuclear RNA required for both steps of nuclear precursor messenger RNA splicing. Science, 250, 404–409. [DOI] [PubMed] [Google Scholar]
- 35.Anderson K. and Moore,M.J. (1997) Bimolecular exon ligation by the human spliceosome. Science, 276, 1712–1716. [DOI] [PubMed] [Google Scholar]
- 36.Mackay J.P. and Crossley,M. (1998) Zinc fingers are sticking together. Trends Biochem. Sci., 23, 1–4. [DOI] [PubMed] [Google Scholar]
- 37.Klug A. (1999) Zinc finger peptides for the regulation of gene expression. J. Mol. Biol., 293, 215–218. [DOI] [PubMed] [Google Scholar]
- 38.Chua K. and Reed,R. (1999) Human step II splicing factor hSlu7 functions in restructuring the spliceosome between the catalytic steps of splicing. Genes Dev., 13, 841–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nesic D. and Kramer,A. (2001) Domains in human splicing factors SF3a60 and SF3a66 required for binding to SF3a120, assembly of the 17S U2 snRNP and prespliceosome formation. Mol. Cell. Biol., 21, 6406–6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mazroui R., Puoti,A. and Kramer,A. (1999) Splicing factor SF1 from Drosophila and Caenorhabditis: presence of an N-terminal RS domain and requirement for viability. RNA, 5, 1615–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adams D.J., van der Weyden,L., Mayeda,A., Stamm,S., Morris,B.J. and Rasko,J.E. (2001) ZNF265—a novel spliceosomal protein able to induce alternative splicing. J. Cell Biol., 154, 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Z., Luyten,I., Bottomley,M.J., Messias,A.C., Houngninou-Molango,S., Sprangers,R., Zanier,K., Kramer,A. and Sattler,M. (2001) Structural basis for recognition of the intron branch site RNA by splicing factor 1. Science, 294, 1098–1102. [DOI] [PubMed] [Google Scholar]
- 43.Feig A.L. and Uhlenbeck,O.C. (1999) The role of metal ions in RNA biochemistry. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 287–319.
- 44.Ast G. and Weiner,A.M. (1996) A U1/U4/U5 snRNP complex induced by a 2′-O-methyl-oligoribonucleotide complementary to U5 snRNA. Science, 272, 881–884. [DOI] [PubMed] [Google Scholar]
- 45.Konarska M.M. and Sharp,P.A. (1986) Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell, 46, 845–855. [DOI] [PubMed] [Google Scholar]
- 46.Krainer A.R., Maniatis,T., Ruskin,B. and Green,M.R. (1984) Normal and mutant human beta-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell, 36, 993–1005. [DOI] [PubMed] [Google Scholar]
- 47.Ast G. and Weiner,A.M. (1997) A novel U1/U5 interaction indicates proximity between U1 and U5 snRNAs during an early step of mRNA splicing. RNA, 3, 371–381. [PMC free article] [PubMed] [Google Scholar]
- 48.Dignam J.D., Lebovitz,R.M. and Roeder,R.G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res., 11, 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Konarska M.M. and Sharp,P.A. (1987) Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell, 49, 763–774. [DOI] [PubMed] [Google Scholar]
- 50.Sigman D.S., Graham,D.R., D’Aurora,V. and Stern,A.M. (1979) Oxygen-dependent cleavage of DNA by the 1,10-phenanthroline-cuprous complex. Inhibition of Escherichia coli DNA polymerase I. J. Biol. Chem., 254, 12269–12272. [PubMed] [Google Scholar]
- 51.Murakawa G.J., Chen,C.H., Kuwabara,M.D., Nierlich,D.P. and Sigman,D.S. (1989) Scission of RNA by the chemical nuclease of 1,10-phenanthroline-copper ion: preference for single-stranded loops. Nucleic Acids Res., 17, 5361–5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Burkitt M.J., Milne,L., Nicotera,P. and Orrenius,S. (1996) 1,10-Phenanthroline stimulates internucleosomal DNA fragmentation in isolated rat-liver nuclei by promoting the redox activity of endogenous copper ions. Biochem. J., 313, 163–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sillen L.G. and Martell,A.E. (1964) Stability Constants of Metal-Ion Complexes. The Chemical Society, Burlington House, London.
- 54.Zhu B.Z. and Chevion,M. (2000) Copper-mediated toxicity of 2,4,5-trichlorophenol: biphasic effect of the copper(I)-specific chelator neocuproine. Arch. Biochem. Biophys., 380, 267–273. [DOI] [PubMed] [Google Scholar]
- 55.Holland D.R., Hausrath,A.C., Juers,D. and Matthews,B.W. (1995) Structural analysis of zinc substitutions in the active site of thermolysin. Protein Sci., 4, 1955–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sontheimer E.J. (2001) The spliceosome shows its metal. Nature Struct. Biol., 8, 11–13. [DOI] [PubMed] [Google Scholar]
- 57.Yeakley J.M., Morfin,J.P., Rosenfeld,M.G. and Fu,X.D. (1996) A complex of nuclear proteins mediates SR protein binding to a purine-rich splicing enhancer. Proc. Natl Acad. Sci. USA, 93, 7582–7587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ganzi G.C. (1984) Preparation of high-purity laboratory water. Methods Enzymol., 104, 391–403. [DOI] [PubMed] [Google Scholar]
- 59.Kruszewski M., Iwanenko,T., Bouzyk,E. and Szumiel,I. (1999) Chelating of iron and copper alters properties of DNA in L5178Y cells, as revealed by the comet assay. Mutat. Res., 434, 53–60. [DOI] [PubMed] [Google Scholar]
- 60.Weinberg R.A. and Penman,S. (1968) Small molecular weight monodisperse nuclear RNA. J. Mol. Biol., 38, 289–304. [DOI] [PubMed] [Google Scholar]
- 61.Coleman J.E. (1974) The role of Zn(II) in transcription by T7 RNA polymerase. Biochem. Biophys. Res. Commun., 60, 641–648. [DOI] [PubMed] [Google Scholar]
- 62.Mannello F., Canesi,L., Gazzanelli,G. and Gallo,G. (2001) Biochemical properties of metalloproteinases from the hemolymph of the mussel Mytilus galloprovincialis Lam. Comp. Biochem. Physiol. B Biochem. Mol. Biol., 128, 507–515. [DOI] [PubMed] [Google Scholar]
- 63.Windle H.J. and Kelleher,D. (1997) Identification and characterization of a metalloprotease activity from Helicobacter pylori. Infect. Immun., 65, 3132–3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sevlever D., Mann,K.J. and Medof,M.E. (2001) Differential effect of 1,10-phenanthroline on mammalian, yeast and parasite glycosylphosphatidylinositol anchor synthesis. Biochem. Biophys. Res. Commun., 288, 1112–1118. [DOI] [PubMed] [Google Scholar]
- 65.Correa L.M., Cho,C., Myles,D.G. and Primakoff,P. (2000) A role for a TIMP-3-sensitive, Zn(2+)-dependent metalloprotease in mammalian gamete membrane fusion. Dev. Biol., 225, 124–134. [DOI] [PubMed] [Google Scholar]
- 66.Poiesz B.J., Seal,G. and Loeb,L.A. (1974) Reverse transcriptase: correlation of zinc content with activity. Proc. Natl Acad. Sci. USA, 71, 4892–4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.