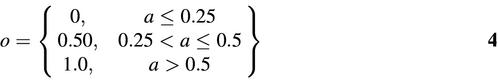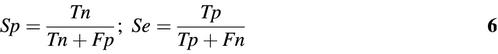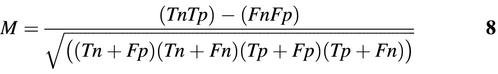Abstract
An mRNA transcript contains many potential antisense oligodeoxynucleotide target sites. Identifi cation of the most efficacious targets remains an important and challenging problem. Building on separate work that revealed a strong correlation between the inclusion of short sequence motifs and the activity level of an oligo, we have developed a predictive artificial neural network system for mapping tetranucleotide motif content to antisense oligo activity. Trained for high-specificity prediction, the system has been cross-validated against a database of 348 oligos from the literature and a larger proprietary database of 908 oligos. In cross- validation tests the system identified effective oligos (i.e. oligos capable of reducing target mRNA expression to <25% that of the control) with 53% accuracy, in contrast to the <10% success rates commonly reported for trial-and-error oligo selection, suggesting a possible 5-fold reduction in the in vivo screening required to find an active oligo. We have implemented a web interface to a trained neural network. Given an RNA transcript as input, the system identifies the most likely oligo targets and provides estimates of the probabilities that oligos targeted against these sites will be effective.
INTRODUCTION
The development of reliable in vivo gene inactivation strategies is an important scientific and therapeutic goal. Antisense oligodeoxynucleotide (ODN) technology allows the targeted reduction of mRNA expression through the in vivo application of short (10–20 nt) DNA molecules with a base sequence complementarity to a region of the transcript. Binding of an antisense molecule triggers cleavage and subsequent degradation of the transcript by mechanisms still under investigation (1). This technology provides a powerful tool for studying gene dynamics. It also shows promise for treatment, through direct control of gene expression, of diseases such as AIDS and cancer (2,3). Advances in chemistry have improved selectivity, stability and specificity of action of ODNs, resulting in several antisense drugs reaching human clinical trials (4). However, in spite of some notable successes, a number of problems associated with the use of ODNs have not yet been solved (5–7).
The designer of antisense oligos must choose between hundreds of potential sites along the RNA targeted for down-regulation. There is a great deal of variation in the efficacy of an oligo depending on the target site selected (8,9). Efficacy is usually measured by comparing the in vivo concentration of the target RNA (or protein product) in treated cells with the concentration in controls. In typical experiments, efficacy ranges from the complete knock-out of target RNAs (within the assay’s limits) to no apparent effect.
This variability presents a significant obstacle to the practical application of the technology. Expensive and time-consuming in vivo screenings are usually required to determine which of multiple ODNs is most effective. Several in vitro approaches have been developed to reduce time and cost, but because these methods do not perfectly mimic the cellular environment their ability to predict in vivo activity is limited (9–11).
Computational approaches to antisense efficacy prediction have been developed by several groups. Working from the hypothesis that ODN efficacy is determined by the structural and energetic favorability of oligo–RNA binding, calculations made for oligo, mRNA and hybrid duplex identify sites favoring oligo binding (12–14). It is difficult to assess the effectiveness of these methods. Each was tested with a different experimental dataset. One study utilized comparisons against in vitro binding assays only (14). Others were validated on limited datasets that make it difficult to derive statistically significant conclusions about their performance on previously untested data. None were cross-validated against a large database.
Though empirical evidence indicates that structural and energetic factors play an important role, these factors are not necessarily the sole moderators of antisense efficacy. Other considerations include biases in oligo delivery and the sequence specificity of RNase H. Tu et al. (15) observed that the single tetranucleotide motif TCCC, when present in an oligo, increases the likelihood of the oligo being effective from a background rate of <10% to ∼50%. Following Tu’s work, our previous efforts further explored the relationship between short textual motifs (primarily 3mers and 4mers) and antisense ODN effectiveness. An analysis of 349 oligos from the literature found several dozen other motifs correlated with in vivo antisense activity (16). While the mechanism behind this motif-related bias has not been explained, the clear correlation between motivic composition and efficacy has considerable potential as a predictive tool. The question is whether the local sequence of the antisense target region contains enough information to determine the activity of the oligo. Our hypothesis, motivated by the statistical studies referenced above, is that although there may be more than one mechanism at play, motif content by itself has predictive power for determining oligo efficacy.
In the present work we apply artificial neural networks (ANNs) to the challenge of motif-based prediction. This problem could be addressed using ANNs in either of two ways: as a classification task, in which the oligo sequence is identified as belonging to a class, e.g. active or inactive; or as a regression task, in which the sequence is mapped to a value within a function established by training, e.g. a quantitative measure of the activity of the oligo. While either approach to the problem could have been taken, we chose to view it as a regression task: networks were trained to map the motivic composition of an antisense oligo to a value quantifying its predicted in vivo activity.
MATERIALS AND METHODS
Important design issues for the problem have included data representation, network architecture, training method and learning parameters.
Data
The primary database used for this work consists of a set of oligos, collected from the literature, meeting the following criteria. (i) At least 10 oligos were assayed for in vivo effect on a given RNA target in mammalian species. (ii) Gene expression after oligo application was measured relative to untreated controls. (iii) Virus targets were excluded because assays are often performed by viral load, which can produce distinctly different results to gene expression assays. (iv) The experiments employed oligonucleotides with a phosphorothioate backbone. (v) The database currently contains entries for 348 oligos ranging from 10 to 22 nt in length. The oligos were compiled from 13 published experiments targeting 11 distinct mRNAs. Reported activity values range from 0 (complete knock-out) to 1 (no effect). One source reported oligos as inactive without providing a numeric value; the curators assigned activity values of 0.75 to these oligos. Available on the World Wide Web at http://antisense.genetics.utah.edu under ‘ODNBase’, the database is described in more detail in Giddings et al. (17).
The network architecture and design developed in experiments with ODNBase were cross-validated in experiments incorporating a larger, independent dataset provided by ISIS Pharmaceuticals (Carlsbad, CA). The database contains entries for 131 18-nt and 777 20-nt oligos tested against about 100 different transcripts. In the typical experiment, 80 DNA phosphorothioate oligonucleotides complementary to the target mRNA or pre-mRNA were synthesized and screened for antisense activity. The optimal tranfection agent (usually cationic lipid) and concentration of oligonucleotide were determined for each cell line. Total RNA was isolated from treated and untreated cells using RNAeasy (Qiagen) and target mRNA levels were measured using real time quantitative RT–PCR (TAQman; Perkin-Elmer). Each reported activity value is the average of duplicate measurements at a constant oligonucleotide dose and is expressed as percent untreated control.
The ISIS dataset differs from ODNBase in several ways. Oligo concentrations, determined as described above, were typically at least two orders of magnitude lower than those reported in the literature. The low dosage may account for the lower proportion of active oligos in the ISIS dataset: 4% compared with 16% in ODNBase. Also, the data were derived from experiments performed under homogenous conditions, with uniform protocols for agent and assay design, measurement, etc.; ODNBase was compiled from experiments performed under a variety of conditions.
Access to the ISIS database allowed us to assess the performance of the system on an independent dataset. Results from these cross-validation tests are presented to demonstrate the efficacy of the prediction method, but because we cannot provide them to other researchers for independent testing, these data were not used in the majority of experiments.
Input to network
Fundamentally, the input for the problem is a sequence string from the DNA alphabet and the output is a number relating to the activity of the corresponding oligo in vivo. An oligo of length n can be decomposed into (n – l + 1) overlapping motifs of length l. The four nucleotides (A, C, G, T) allow k = 4l possible motifs of this length. With these k motifs enumerated in some fashion (e.g. alphabetical order), oligo i can be represented by the number of occurrences (cij) in the oligo of each possible motif j. For a 20-nt oligo composed of 17 overlapping 4mers, most of the motif counts will be 0, with a few 1s and occasionally a larger number for a motif that occurs multiple times. Oligos typically range from 10 to 20 nt in length. To correct for different length oligos we can represent the proportion (pij) of oligo i that consists of motif j, rather than the direct motif count, by normalizing the counts by a factor Ni:
The proportion (pij) of oligo i that consists of motif j is calculated as:
The constant 10 scales the proportion values approximately to the range [0,1].
This mapping from sequence to activity can be represented as:
where ai is the empirically measured activity of oligo i. This representation ignores information about the positions of motifs within the oligo and thus is not a unique mapping between oligo and motif counts (since a single set of counts can be scrambled into multiple different oligos). Motifs were considered in a position independent manner because the prior publications correlating motif composition did not indicate the need for positional information. While they do not exclude the future use of positional information, our experiments demonstrate that a good model can be built without it.
Neural networks
For all experiments the Stuttgart Neural Network Simulator (SNNS) was used (18), http://www-ra.informatik.uni-tuebingen.de/SNNS. The system consists of a kernel, batch language and graphical interface. Initial experiments in network design were carried out with the graphical interface, using randomly chosen train and test data from the database (typically in a ratio of 90/10%, or 10-fold) to observe learning characteristics. After settling on a few architectures and ranges of training parameters, thorough cross-validation followed using the kernel, batch language and custom PERL scripts (available on request).
Various feed-forward network architectures were explored. These included fully connected networks mapping all 256 tetranucleotide motifs to input nodes, with one or two hidden layers containing between 4 and 20 nodes. The large size of these networks contributed to their inability to effectively generalize from the training set to test data. Two approaches to preventing over-fitting are available: to prune the system during or after training by removing nodes and weights that contribute little to the mapping, or to begin with a smaller network and fewer inputs. Pruning is recommended in situations where little is known about the relative importance of the various inputs. Optimal Brain Damage (19) and Optimal Brain Surgeon (20) are two popular pruning algorithms. Because previous studies had identified the correlation between particular motifs and activity, instead of pruning we opted to begin with a smaller network and limit the input to those motifs that are most highly correlated with activity. A χ2 test for significance (21) performed on the motifs for all oligos in the database was used to rank the motifs from most to least significant. The input set was thereby reduced to the 40 motifs exhibiting maximal statistical correlation to oligo activity (‘Chi-40’ networks). A table listing these 40 motifs is available as Supplementary Material.
The selection of the χ2 ranked tetranucleotide motifs is performed once for the whole ODNBase dataset. After beginning work using ODNBase we discovered that the compilation contained a duplicate entry. Because the duplicated oligo contains 10 motifs highly correlated with activity, removal of the duplicate changes the top-40 set considerably. To provide consistency with earlier experiments, we chose to continue using the original motif choices. Validation experiments training with the independent ISIS data used the same set of original motifs found most significant by χ2 selection on ODNBase; motifs were not re-selected, thus providing an independent cross-validation of these motif choices.
The motif length at which the correlation between motif content and activity is maximized remains an open question. There is likely a length l that optimizes the correlation. We performed some experiments exploring short motifs. At the length l = 1 (i.e. nucleotide composition) there was some bias present favoring C, but predictions based on this were weak. Limited tests with di- and tri-nucleotide motifs showed an improvement in prediction accuracy with each step up in length. At the transition from l = 3 to l = 4 the jump in input field size (from 64 to 256 nodes) predisposed the network to over-fitting. However, by reducing the input field for l = 4 using the χ2 pruning, the performance was again improved with this transition. Longer motifs have not yet been explored.
Each network has a single output unit corresponding to the activity of the oligo. Activity is usually quantified as the percentage of control (e.g. scrambled oligo) of the target RNA after oligo application, so values lie in the [0.0,1.0] interval with lower activities indicating greater efficacy. The output node can be trained with either the continuous-valued oligo activities or some function thereof. Experimental results led us to investigate the three-way threshold function given by:
where a is activity.
Training with threshold functions of this type improved generalization. Although the oligos in the public database were tested by different laboratories at various concentrations, high potency oligos may be more likely to display a measurable effect regardless of concentration, making their measurement more reliable. Thus, grouping oligos for training using equation 4 may facilitate the mapping from motif content to activity value.
Related to these observations, a log-scale function was developed to provide a non-threshold-based transformation that emphasizes the differences amongst high-activity oligos while de-emphasizing the differences between low-activity oligos by grouping the latter into a very narrow region. The function is:
where a is the empirical activity value and k is a scale constant for which we use the value of 100. Training with this function further improved generalization. This function was used in all experiments reported below.
It was also observed that a linear (identity) activation function on the output neuron improves performance (all other nodes use the standard logistic activation function). This held true for a variety of training conditions. This might be explained by the fact that the oligo activity values are already transformed with the non-linear functions of equations 4 or 5. These functions are better suited to the peculiarities of the data than the standard logistic function is, since the latter is symmetrical about the center and therefore accentuates differences in the central region of the curve, which is not the desired emphasis. The linear activation function on the output node has the side effect that the network can produce values outside the range [0,1], e.g. negative numbers. A negative number simply indicates that the network predicts an oligo will be highly effective, whereas a number greater than 1 is a prediction that the oligo is very ineffective.
While several supervised learning algorithms provided by SNNS were tested on the problem, all experiments reported herein were performed using the back-propagation (backprop) algorithm with a momentum term (22). Backprop performs connection weight adjustments in order to minimize the difference between training signal and network output. Weights are recursively adjusted as the model descends the gradient of a sum-of-squared-errors cost function calculated on each pass through the network. The rate of descent is controlled by the learning parameter η. The backprop momentum method utilizes two additional parameters: µ (momentum term) to reduce oscillation during learning, and c (flat spot elimination term), a constant value added to the derivative of the activation function to allow the network to avoid flat spots in the error space (23).
In all experiments, connection weights are initialized with random numbers in the range [–1,1]. The random weight initialization plays a large role in determining how effectively a particular network generalizes for the problem. Experiments described below were performed with either all or a 10 net subset (in the computationally intensive ‘–oligo’ process) of a common group of 50 randomly initialized nets. Patterns are presented to the networks in randomized order during each training cycle. Node activation values are calculated in topological order moving from the input to the output layer.
Interpretation of network output
Prediction performance is often measured in terms of specificity (Sp) and sensitivity (Se):
where Tn is true negative predictions, Fn is false negative predictions, Tp is true positive predictions and Fp is false positive predictions. A related quantity is the probability of a positive prediction being correct, given by
The Matthews calculation (24) generates a coefficient in the range [–1,1] quantifying the correlation between actual and predicted values:
A problem with these metrics is that they rely on the use of a threshold value that distinguishes between positive and negative cases in the prediction output. Sampling at only one threshold gives a limited perspective on performance, since across the space of possible thresholds there is natural variation due to noise. One way to overcome this limitation is to measure these threshold-dependent quantities at a variety of different thresholds to get a global view of their possible values. For example, along with the Matthews correlation coefficient at the threshold at which Sp, Sn, etc. are reported, we also report the maximum correlation observed over the range of threshold values (‘peak Matthews’). A more general means of dealing with the threshold dependency of specificity and sensitivity is ROC (receiver operating characteristic) analysis, which consists of sampling the values of Sp and Se at many different thresholds spanning the range from minimum to maximum model output (prediction values) and plotting them against each other (25). Because, for continuous models, there is generally an inverse relationship between specificity and sensitivity, the area under a ROC curve provides a concise measure of performance. The ROC curve for a perfect prediction model shows no tradeoff between specificity and sensitivity, so its area is 1.0. The opposite situation—a random prediction model—is a diagonal ROC curve from (0,1) to (1,0) with an area of 0.5. So the useful range of ROC curve areas is 0.5–1.0.
For the present task, ROC curves are sought which have their area distribution biased towards the high-specificity end of the curve. The goal is to find a few oligos that have a high likelihood of success for a given RNA. It is not a problem if there are many false negatives (low sensitivity), as long as a few targets are found likely to be active in vivo.
ROC analysis also requires a threshold, to classify the input data (i.e. the database of oligos) into categories of positive/negative (active/inactive). The choice of threshold has some effect on the ROC area, particularly if the threshold is an extreme value very near zero or one. To make useful measurements of accuracy the threshold is chosen so that the set of positives (or negatives) is not too small. For experiments herein, the value of 0.25 was used (i.e. reduction of RNA to 25% or less of control is considered an active oligo).
Another measure that has been used to report antisense prediction accuracy is the correlation coefficient (R) and significance value (P) (12,14), which indicate how well a set of predictions relate to experimental measurements. However, because it is difficult to translate an R-value into a useful measure of accuracy (e.g. the probability of a correct prediction), this approach was not emphasized in the present investigation.
Combining predictors
Several methods were used to combine the predictions of multiple networks and combine the predictions of network(s) with the results of other predictors. One approach is to average the outputs of several selected networks for a given oligo input. Another is logistic regression (http://m2.aol.com/johnp71/logistic.html) (26), which consists of transforming the linear regression of the data with a logistic function to generate a probability estimator. This was used to combine the outputs of a few predictors into an overall probability that an oligo will be active. It was applied in one instance to combine the predictions of several networks, and in another to combine a neural-network prediction with an estimate of the free-energy change associated with oligo–RNA duplex formation. The free energy change was calculated using the di-nucleotide energies given by Sugimoto et al. (27), without secondary structure or accessibility considerations.
Logistic regression can also be used to improve interpretation of the outputs from individual networks by mapping them into probability values. The process consists of performing cross-validation on the dataset and using the set of predicted activity values to calibrate the regression coefficient. The result is a function that maps from the network output for a given oligo to an estimator of the probability that the oligo will be active.
Cross-validation of network performance
Several cross-validation methods have been used to assess the system’s ability to generalize from training to test data. One is a ‘minus 10%’ or 10-fold system, where 10% of the database is randomly selected as the test set. Networks trained on the remaining 90% were then tested on the unseen 10%. This was used for initial network design experiments. Subsequently a ‘take one out’ approach has been used to more thoroughly assess performance of the chosen designs. Using PERL scripts, a single oligo is selected from the database for testing and the model is trained with the remainder. Following training, the model is tested for accuracy in predicting the activity of the single test oligo. The result for the test oligo is recorded and the procedure repeated, using the same training parameters, for each oligo in the database. For brevity, this process is hereafter referred to as ‘–oligo’ cross-validation.
One issue that has arisen regarding the use of the data in –oligo cross-validation is the substantial sequence overlap between oligos targeting the same transcript. There are two distinct reasons for this circumstance. The first is that experimenters tested oligos complimentary to overlapping regions of the target mRNA (‘oligo walking’). In the second case, identical (or nearly identical) oligos targeting the same mRNA were tested by separate laboratories. To eliminate any potential information leakage a regime was developed consisting of removing all oligos corresponding to a given RNA from the training database, training, and then testing the networks’ performance in predicting activity values of the excluded oligos. This process is repeated for each of the 11 unique RNAs against which oligos in the database were targeted. This process is called ‘minus-one-RNA’ cross-validation (–RNA). Although there is variation in the size of the split between the train and test sets using this scheme (test set sizes range from 11 to 59 oligos), in each round the oligos used for training versus testing are different, with each oligo present in a test set exactly once (i.e. when not present in the training set).
In a final set of experiments, networks were trained and tested respectively on each of two independent datasets, ODNBase and the ISIS database. This process was performed in both directions. We reason that because of oligo decomposition, information leakage between even highly similar oligos should be insignificant. For example, a single mismatch or gap in the central region of the alignment of two 20-nt sequences corresponds to a 25% difference in the constituent tetramers and a proportional difference in the input to the network. Smith–Waterman alignments (28) comparing each oligo in the ISIS data set to each oligo in ODNBase identified no identical oligos nor any cases where the full sequence of one oligo matched a continuous substring of another. Almost all aligned regions contained multiple mismatches. Most importantly, all paired oligos are dissimilar enough that they would be unlikely to bind the same RNA target. For example, in the highest similarity pairing a very short 10-nt oligo from the public dataset matches a 20-nt ISIS oligo in all 10 positions with a single gap. Because of the gap, only 4 of the 7 tetramers from the decomposition of the short oligo match tetramers from the longer oligo. The longer oligo shares only 4 of its 17 tetramers with the short oligo. Furthermore, the short oligo could only bind the same target as the larger by forming a bulge in the gap position, a conformation that is energetically unfavorable and therefore unlikely.
RESULTS AND DISCUSSION
Early in the project various tests were performed to investigate network architectures, training parameters, motif lengths and learning algorithms. The large space of options was quickly narrowed to a few ranges of values for these. This type of parameter space exploration can raise the concern that a large number of trials will eventually lead to a chance combination of parameters which work on that particular dataset but will not generalize to other data. However, in this work a multiplicity of parameter sets produced working predictors with only small variations in performance. Experiments with the data from ISIS Pharmaceuticals confirm that the generalization is not unique to the ODNBase data. Average results for groups of networks are presented as evidence that an effective mapping from motifs to activity is not an idiosyncrasy of a particular network but the result of an effective machine-learning system.
All experiments reported below were performed with a single group of 50 Chi-40 fully connected feed-forward networks (differing only in the random initialization of their connection weights) containing four units in the single hidden layer and a linear (identity) activation function on the output node (logistic function on all others). Networks were trained for 1000 cycles using back-propagation with learning parameters η = 0.1, µ = .0.5 and c = 0.1. The logarithmic (equation 5) function was used to transform oligo activity values in all experiments except A, which compared the use of the different transform functions. Table 1 lists the averages of various performance measures for the group of networks. Threshold-dependent performance measurements are reported for each experiment for a threshold at which all networks demonstrated a sensitivity ≥0.07 with specificity near maximum. To assess relative performance the networks in an identically trained group are ranked with respect to the various performance measures. The sum of these ordinals is used to identify networks that perform well both overall (as measured by the ROC area and the peak Matthews coefficient) and at the particular threshold used to classify network outputs. Table 2 lists results for some representative high-performance networks.
Table 1. Summary of performance of groups of networks differing only in random initialization of training weights.
| Experiment | No. of nets | Activity transformation | Cross validation | ROC range | Average ROC | Peak M range | Average peak M | Threshold | Average | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M | P+ | Fp | Tp | Sp | Se | |||||||||
| A1 | 50 | Logarithmic | ODN–RNA | 0.62–0.76 | 0.71 | 0.24–0.40 | 0.32 | 0.10 | 0.22 | 0.49 | 10.4 | 10.0 | 0.96 | 0.18 |
| A2 | 50 | Piecewise | ODN–RNA | 0.57–0.74 | 0.66 | 0.20–0.38 | 0.28 | 0.10 | 0.22 | 0.45 | 14.5 | 11.6 | 0.95 | 0.20 |
| B1 | 10 | Logarithmic | ODN–RNA | 0.73–0.76 | 0.74 | 0.30–0.39 | 0.35 | 0.10 | 0.21 | 0.48 | 10.5 | 9.7 | 0.96 | 0.17 |
| B2 | 10 | Logarithmic | ODN–oligo | 0.73–0.80 | 0.77 | 0.35–0.47 | 0.40 | 0.10 | 0.25 | 0.58 | 6.9 | 9.6 | 0.98 | 0.17 |
| D1 | 50 | Logarithmic | ISIS/ODN | 0.72–0.82 | 0.77 | 0.31–0.49 | 0.40 | 0.30 | 0.21 | 0.46 | 11.5 | 9.9 | 0.96 | 0.17 |
| D2 | 50 | Logarithmic | ODN/ISIS | 0.63–0.77 | 0.71 | 0.12–0.24 | 0.18 | 0.25 | 0.12 | 0.10 | 85.0 | 9.9 | 0.90 | 0.28 |
Reported values of P+, Fp, Tp, Sp and Se are averages at the listed threshold (an output value smaller than the threshold value indicates a prediction that the oligo is active).
Table 2. Results for representative high-performance networks, including the thresholds at which threshold-dependent values were measured.
| Experiment | Activity transformation | Cross-validation | ROC | Peak M | R | P | Threshold | M | P+ | Fp | Tp | Sp | Se |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 | Logarithmic | ODN–RNA | 0.74 | 0.39 | 0.30 | 1.0 E –8 | 0.10 | 0.27 | 0.55 | 10 | 12 | 0.97 | 0.21 |
| A2 | Piecewise | ODN–RNA | 0.73 | 0.35 | 0.34 | 4.4 E –7 | 0.10 | 0.27 | 0.52 | 12 | 13 | 0.96 | 0.23 |
| B1 | Logarithmic | ODN–RNA | 0.75 | 0.39 | 0.30 | 1.5 E –8 | 0.10 | 0.25 | 0.52 | 10 | 11 | 0.97 | 0.19 |
| B2 | Logarithmic | ODN–oligo | 0.77 | 0.40 | 0.37 | 2.1 E –12 | 0.10 | 0.29 | 0.67 | 5 | 10 | 0.98 | 0.18 |
| B2 + ΔG | N/A | N/A | 0.82 | 0.41 | 0.38 | 1.1 E –13 | 0.50 | 0.33 | 0.71 | 5 | 12 | 0.98 | 0.21 |
| D1 | Logarithmic | ISIS/ODN | 0.80 | 0.46 | 0.43 | 2.2 E –17 | 0.30 | 0.27 | 0.63 | 6 | 10 | 0.98 | 0.18 |
| D2 | Logarithmic | ODN/ISIS | 0.77 | 0.21 | 0.24 | 3.8 E –13 | 0.25 | 0.13 | 0.13 | 52 | 8 | 0.94 | 0.23 |
M = Matthews correlation coefficient; R = linear regression correlation coefficient; P = significance.
Effect of random initialization
To determine the effect of random initialization upon network performance an experiment was performed wherein the group of 50 networks was trained and tested using –RNA cross-validation against the 348 oligo database. The resulting ROC curves range from 0.62 to 0.76 in area. Though this is a wide spread of values, it is notable that all networks generate ROC curves with areas considerably greater than random prediction would produce. The predictions generate an average ROC area of 0.71 and an average peak Matthews correlation coefficient of 0.32. Using a threshold of 0.10 below which oligos are predicted as active the networks score an average P+ of 0.49, a significant improvement over a trial-and-error approach (Table 1, experiment A1). The best network in the group generates a ROC area of 0.74, peak Matthews correlation coefficient of 0.39, and probability score of 0.55 (Table 2, experiment A1).
In the above experiment networks were trained against oligo activity values transformed by the logarithmic function of equation 5. To compare the effectiveness of the two output-training functions the same group of networks was re-trained using activity values transformed by the piecewise function of equation 4. The resulting ROC curves range from 0.57 to 0.74 in area, with an average of 0.66. The average peak Matthews coefficient for these predictions is 0.28. Results for the best network from this experiment are presented in Table 2 (experiment A2).
All but seven nets generated a greater ROC area when trained with data transformed by the log function. The ROC curves for networks trained with log-transformed data also tend to have greater area in the high-specificity region. One explanation for the better performance is that the measured differences in activity between active oligos may be repeatable effects of motif content upon activity. If that is the case, the log function may work better because it not only retains but enhances the differences between the high activity oligos.
Comparison of cross-validation methods
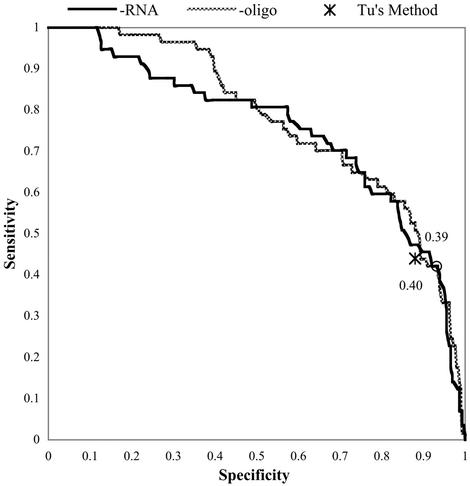
The two primary cross-validation methods were compared by re-training and testing the 10 networks that produced the highest ROC areas during –RNA cross-validation (experiment A1), now using –oligo cross-validation (Table 1, experiments B1 and B2). The average ROC area for the 10 networks using –oligo is 0.77, and using –RNA is 0.74. ROC curves for network number 3 of this experiment are illustrated in Figure 1, and the network’s performance measures are presented in Table 2, experiments B1 and B2. Though overall ROC area is reduced for –RNA cross-validation, in the critical high-specificity region, prediction ability is not significantly affected.
Figure 1.
ROC curves for a standard Chi-40 network tested using minus-one-oligo cross-validation and minus-one-RNA cross-validation. The circles indicate the points at which the networks scored their peak Matthews correlation coefficients (in this chart the circles are superimposed), the values of which are listed. Also shown for reference is the single point representing the sensitivity and specificity of Tu’s method (15) applied to predict the activities of oligos in the database.
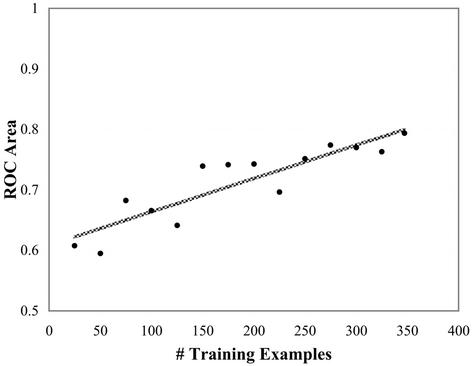
The slight reduction in accuracy due to the switch from –oligo to –RNA cross-validation prompts two possible explanations: (i) the elimination of some oligos eliminates information leakage (from oligo overlap) that was artificially inflating the performance measures; and (ii) the reduction in training set size used in –RNA cross-validation is decreasing accuracy. To assess the impact of the latter, an experiment was performed measuring the correlation between training set size and prediction accuracy. This was done in a manner similar to the –oligo cross-validation except that the training set size was varied from 25 oligos to the full database. In each of the 14 iterations each oligo is successively selected for testing and the training set of the proper size is randomly selected from the remaining database. Figure 2 shows the results of this procedure using the two networks that generated the highest ROC areas in –oligo cross-validation. The graph shows a strong correlation between prediction accuracy and the size of the training set (R = 0.89). The bumpiness of the curves is due to random selections of training sub-sets.
Figure 2.
Plot of the average of two experiments measuring ROC area versus cross-validation training set size for a Chi-40 network, and the regression line. R = 0.89, P = 1.57 × 10–5.
The average test set removed from the database in –RNA cross-validation is 32 oligos. With respect to the plot in Figure 2, this translates into a reduction of 0.012 in ROC area due to the loss of this many training examples when compared with the –oligo method. The average difference of 0.03 ROC curve area between the two primary cross-validation methods therefore appears to be a combined effect of the decrease in the number of training examples and some information leakage.
Results from combining predictors
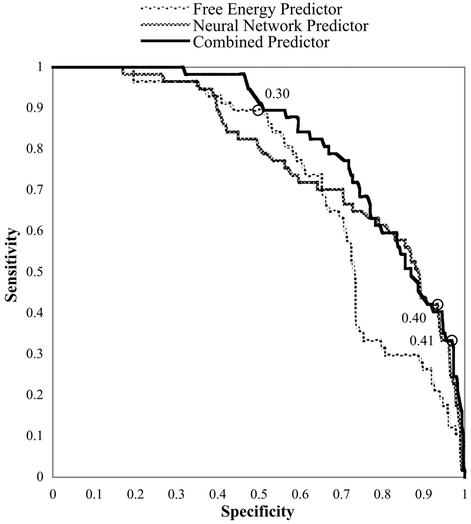
The combination of two or more predictors produced mixed results. The averaged prediction of multiple networks provided a result with an ROC area greater than that of the average network in the group, but generally there were one or more individual networks that made better predictions than the group averaging did. Better results were generated by combining a neural network output with the free energy (ΔG) calculation for binding between oligo and target using logistic regression (Table 2, experiment B2 shows results for the network alone, and experiment B2 + ΔG shows results for the combined predictor). The ROC curves for these experiments are shown in Figure 3. While the ΔG calculation by itself performs well in general, it does poorly in the high-specificity region. The combined prediction appears to benefit by the strengths of both independent predictors. It generates a ROC area of 0.82 and a peak Matthews correlation coefficient of 0.41, some of the best results obtained.
Figure 3.
ROC curves for the simple free-energy predictor, Chi-40 neural network predictor (–oligo cross-validation), and a logistic regression combining the two into a probability score. The circles indicate the points at which the networks scored their peak Matthews correlation coefficients, the values of which are listed.
Cross-validation using ISIS data
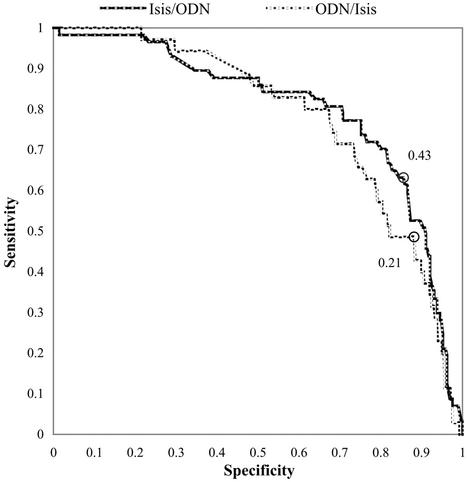
The group of 50 nets was used in a final set of experiments testing the ability of networks trained with one full data set to predict the efficacy of oligos in the other, independent database. Homogeneity in the experimental conditions under which the ISIS data were generated does not appear to impair networks trained with the ISIS dataset; they predict the activities of the oligos in ODNBase with ROC areas ranging from 0.72 to 0.82. The P+ scores range from 0.29 to 0.63, and the average peak Matthews correlation coefficient is 0.40 (Table 1, experiment D1). The best network in the group generates a peak Matthews correlation coefficient of 0.46 and a linear regression R-value of 0.43 with a significance of 10–17 (Table 2, experiment D1). Figure 4 shows the ROC curves for this network.
Figure 4.
ROC curves for a Chi-40 network trained/tested on each of the two independent datasets. The circles indicate the points at which the networks scored their highest Matthews correlation coefficients, the values of which are listed.
Networks trained with the ODN data also make useful predictions for the ISIS oligos. The best network produces a peak Matthews correlation coefficient of 0.21 and a linear regression R-value of 0.24 with a significance of 10–13 (Table 2, experiment D2). While ROC scores are strong, ranging from 0.63 to 0.77, the probability that an oligo predicted as active is actually active consistently falls near 1 in 10 (Table 1, experiment D2).
A possible explanation for the poorer performance testing on the ISIS data is that the large proportion of actives in ODNBase (0.16) is encouraging the system to set a low bar for activity. The ISIS dataset contains a relatively small percentage of actives: using a threshold of 0.25 to distinguish active oligos, only 4%, 35 of the 908, are active. The large proportion of positive predictions on a large dataset with few positives inclines the network to false positive predictions. Also, because the performance of a network depends on the size of the training set, networks developed with the public database would be expected to perform less well than those trained on the larger ISIS dataset. Nevertheless, using the predictions of the average network in the group one is still 2.6 times more likely to select an active oligo than by random selection from the ISIS dataset, again representing a considerable improvement over trial and error.
Practical implications
It is important to put into perspective what these results may mean to someone looking for an effective tool to find active oligos for an RNA target. We address this using a specific network in place on our web site whose cross-validation results are shown in Figure 1. With –oligo cross-validation, the ROC area is 0.77 (Table 2, experiment B2). At a threshold of 0.10 used to distinguish predictions as positive/negative, there are five false positive predictions and 10 true positive predictions, for a P+ of 0.67. For comparison, using the same database with Tu et. al’s method (TCCC selection), 29 true positives and 36 false positives predictions are produced for a P+ of 0.45.
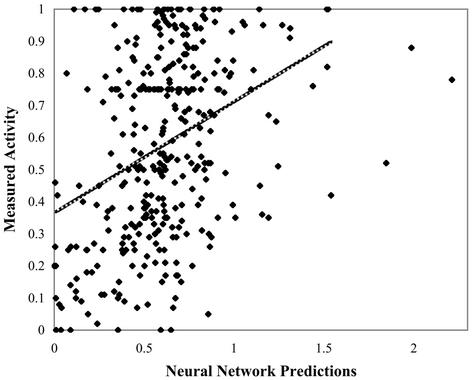
A linear regression analysis comparing the outputs from this network to empirical activity values produces a fit with an R-value of 0.37 and a significance of 10–12 (Fig. 5). The significance value indicates that it is quite unlikely these predictions were an accident of a particular experiment. The R-value falls between those reported above for cross-validation experiments incorporating the ISIS dataset.
Figure 5.
Linear regression of Chi-40 predicted versus actual oligo activities using –oligo cross-validation for the 348 oligo database. R = 0.37 with a significance of 2.1 × 10–12.
Note that the public database contains the bias that there are more positive examples than is expected in the general population of oligos to be tested. Estimates for the probability of finding active oligos by random selection on an mRNA vary, but it likely falls between 0.05 and 0.1. For comparison, using a threshold of 0.25 to distinguish active oligos, the proportion of positives in ODNBase is 0.16. A calculation was made to adjust for this discrepancy when cross-validating against the public data. Considering that the ratio of false positives to true positives will increase corresponding to the difference between the ratio of negative to positive oligos expected in nature compared with the database, a higher ratio of negatives (inactive oligos) in nature should lead to more mis-predictions. Correcting for this based on an estimated active oligo frequency of 0.10 in nature versus an active rate of 0.16 in the database, the above P+ values become 0.31 for Tu’s method and 0.53 for the neural network.
Web-based interface
We have created a web-based interface to the neural network predictors. It is available to non-commercial researchers free of charge with the execution of a license agreement (please contact the corresponding author for more information). The program scans across the input sequence, stepping from left to right one base at a time, with a default oligo size of 20 nt (user adjustable). At each step the network evaluates the complementary oligo. After all sites are evaluated the results are sorted from best to worst predicted oligo. The network score (lower better) is provided along with a probability estimator calculated by logistic regression. The probability value, based on the –oligo cross-validation data, gives a rough estimate of the probability that a given oligo will be active (opposite from the activity data, a higher number is better). Due to the cross-validation used this measure appears to provide somewhat low estimates of the probability of an oligo being active. An example of how the system would be used to predict effective targets against Hepatitis C is included in the Supplementary Material.
It is expected that an experimentalist wishing to find active sites on a target will enter an RNA sequence into the web site, select the top n oligos returned by the predictor and test them in the laboratory. The number n depends on resources, the need to find an extremely active oligo, and so on, but a reasonable number might be two to four. To illustrate, using a network with the same initial weights as the net available on the web to make –oligo cross-validated predictions for the 348 oligo database, the 20 oligos predicted most active by the network are shown in Table 3. Of the top 10 in Table 3, seven are in fact active (again with an activity threshold of 0.25). Of the 20 oligos in Table 3, 13 are active, with three near misses (empirical activity = 0.26). Even if the predictions are affected by the lower incidence of positive sites in nature than in our database, these results are good enough that the user is likely to find an effective oligo among the top two to four oligos predicted active by our system. Considering the network result from experiment B2 (Table 2) corrected for the background positive rate as above, with a P+ of 0.53, the savings on average should be at least 5-fold in the number of oligos that must be screened to find an active one. The reduction in effort should be greater if oligos are tested in order of predicted efficacy.
Table 3. The 20 highest ranked oligos according to ‘–oligo’ cross-validated predictions of a neural network, along with logistic regression scores and measured in vivo activity values.
| RNA | Oligo | In vivo activity | Network prediction | Regression probability | Correct? |
|---|---|---|---|---|---|
| MDR | TCCCCTTCAAGATCCATCCC | 0.20 | 0.00 | 0.59 | Y |
| c-raf | CTGATTTCCAAAATCCCATG | 0.26 | 0.01 | 0.59 | N |
| PKC-alpha | GTCAGCCATGGTCCCCCCCC | 0.46 | 0.01 | 0.59 | N |
| TNF | CTTCTTCCCTGTTCCCCTGGC | 0.20 | 0.01 | 0.58 | Y |
| VCAM-1.1 | TTTGTGTCCCACCTG | 0.10 | 0.01 | 0.58 | Y |
| ICAM-1.2 | TGCATCCCCCAGGCCACCAT | 0.00 | 0.01 | 0.58 | Y |
| IL-1 | GCCACCACAGCCTCTCCCTC | 0.42 | 0.02 | 0.57 | N |
| TNF | TGATCCACTCCCCCCTCCACT | 0.08 | 0.03 | 0.56 | Y |
| ICAM-1.3 | CCCCCACCACTTCCCCTCTC | 0.00 | 0.04 | 0.55 | Y |
| ICAM-1.1 | CCCCCACCACTTCCCCTCTCA | 0.07 | 0.04 | 0.55 | Y |
| ICAM-1.3 | GGGCGCGTGATCCTTATAGC | 0.80 | 0.07 | 0.52 | N |
| TNF | AAAGCTTTAAGTCCCCCGCCC | 0.25 | 0.08 | 0.51 | Y |
| MDR | CGGTCCCCTTCAAGATCCAT | 0.00 | 0.09 | 0.50 | Y |
| TNF | CCTATTCCCTTTCCTCCCAAA | 0.14 | 0.09 | 0.50 | Y |
| VCAM-1.1 | CCACCACTCATCTCG | 0.26 | 0.09 | 0.50 | N |
| TNF | AGGGAAGGAAGGAAGGAAGGG | 1.00 | 0.11 | 0.48 | N |
| TNF | TTCTTGCCCTCCCTCCCTACT | 0.12 | 0.12 | 0.47 | Y |
| TNF | CCTCTTTCCCTTACCCTCCTG | 0.10 | 0.12 | 0.47 | Y |
| TNF | GTTTCCCCTCCATCTCCCTCC | 0.26 | 0.12 | 0.46 | N |
| VCAM-1.1 | CGAGGCCACCACTC | 0.16 | 0.12 | 0.46 | Y |
Using a threshold for activity of 0.25, 70% of the top 10 predicted active, and 65% (with three near misses) of the top 20 predicted active, are actually active.
Conclusion
The good performance of these neural network predictors indicates the likelihood of a strong sequence-specific effect upon antisense oligodeoxynucleotide action. One possible explanation for the motif bias is RNase H sequence specificity. It is also possible that the oligo delivery process is biased towards particular motifs. Whatever the mechanism, we hope that these results combined with those of Tu et al. (15) and Matveeva et al. (16) are enough evidence of motif-based effects on oligo efficacy to motivate further exploration of this phenomenon.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by NIH Genome Scholar award HG00044 to M.C.G., DOE grant DE-FG03-99ER62732 to R.F.G., NIH Genome training grant 5T32HG00042, NIH grant 2R01GM48152, and a joint international research grant from the Japan Health Science Foundation (grant no. K-1005, PI, Y. Nakamura).
REFERENCES
- 1.Chiang M.Y., Chan,H., Zounes,M.A., Freier,S.M., Lima,W.F. and Bennett,C.F. (1991) Antisense oligonucleotides inhibit intercellular adhesion molecule 1 expression by two distinct mechanisms. J. Biol. Chem., 266, 18162–18171. [PubMed] [Google Scholar]
- 2.Geiger T., Muller,M., Monia,B.P. and Fabbro,D. (1997) Antitumor activity of a C-raf antisense oligonucleotide in combination with standard chemotherapeutic agents against various human tumors transplanted subcutaneously into nude mice. Clin. Cancer Res., 3, 1179–1185. [PubMed] [Google Scholar]
- 3.Jendis J., Strack,B. and Moelling,K. (1998) Inhibition of replication of drug-resistant HIV type 1 isolates by polypurine tract-specific oligodeoxynucleotide TFO A. AIDS Res. Hum. Retroviruses, 14, 999–1005. [DOI] [PubMed] [Google Scholar]
- 4.Gewirtz A.M. (1999) Myb targeted therapeutics for the treatment of human malignancies. Oncogene, 18, 3056–3062. [DOI] [PubMed] [Google Scholar]
- 5.Branch A.D. (1998) A good antisense molecule is hard to find. Trends Biochem. Sci., 23, 45–50. [DOI] [PubMed] [Google Scholar]
- 6.Stein C.A. (1999) Keeping the biotechnology of antisense in context. Nat. Biotechnol., 17, 209. [DOI] [PubMed] [Google Scholar]
- 7.Miraglia L., Watt,A.T., Graham,M.J. and Crooke,S.T. (2000) Variations in mRNA content have no effect on the potency of antisense oligonucleotides. Antisense Nucleic Acid Drug Dev., 10, 453–461. [DOI] [PubMed] [Google Scholar]
- 8.Bennett C.F., Condon,T.P., Grimm,S., Chan,H. and Chiang,M.Y. (1994) Inhibition of endothelial cell adhesion molecule expression with antisense oligonucleotides. J. Immunol., 152, 3530–3540. [PubMed] [Google Scholar]
- 9.Ho S.P., Britton,D.H., Stone,B.A., Behrens,D.L., Leffet,L.M., Hobbs,F.W., Miller,J.A. and Trainor,G.L. (1996) Potent antisense oligonucleotides to the human multidrug resistance-1 mRNA are rationally selected by mapping RNA-accessible sites with oligonucleotide libraries. Nucleic Acids Res., 24, 1901–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Southern E.M., Milner,N. and Mir,K.U. (1997) Discovering antisense reagents by hybridization of RNA to oligonucleotide arrays. Ciba Found. Symp., 209, 38–44. [DOI] [PubMed] [Google Scholar]
- 11.Matveeva O., Felden,B., Tsodikov,A., Johnston,J., Monia,B.P., Atkins,J.F., Gesteland,R.F. and Freier,S.M. (1998) Prediction of antisense oligonucleotide efficacy by in vitro methods. Nat. Biotechnol., 16, 1374–1375. [DOI] [PubMed] [Google Scholar]
- 12.Stull R.A., Taylor,L.A. and Szoka,F.C.,Jr (1992) Predicting antisense oligonucleotide inhibitory efficacy: a computational approach using histograms and thermodynamic indices. Nucleic Acids Res., 20, 3501–3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patzel V., Steidl,U., Kronenwett,R., Haas,R. and Sczakiel,G. (1999) A theoretical approach to select effective antisense oligodeoxyribonucleotides at high statistical probability. Nucleic Acids Res., 27, 4328–4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walton S.P., Stephanopoulos,G.N., Yarmush,M.L. and Roth,C.M. (1999) Prediction of antisense oligonucleotide binding affinity to a structured RNA target. Biotechnol. Bioeng., 65, 1–9. [PubMed] [Google Scholar]
- 15.Tu G.C., Cao,Q.N., Zhou,F. and Israel,Y. (1998) Tetranucleotide GGGA motif in primary RNA transcripts. Novel target site for antisense design. J. Biol. Chem., 273, 25125–25131. [DOI] [PubMed] [Google Scholar]
- 16.Matveeva O.V., Tsodikov,A.D., Giddings,M., Freier,S.M., Wyatt,J.R., Spiridonov,A.N., Shabalina,S.A., Gesteland,R.F. and Atkins,J.F. (2000) Identification of sequence motifs in oligonucleotides whose presence is correlated with antisense activity. Nucleic Acids Res., 28, 2862–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giddings M.C., Matveeva,O.V., Atkins,J.F. and Gesteland,R.F. (2000) ODNBase–a web database for antisense oligonucleotide effectiveness studies. Bioinformatics, 16, 843–844. [DOI] [PubMed] [Google Scholar]
- 18.Zell A., Mache,N., Hubner,R., Mamier,G., Vogt,M. and Herman,K.-U. (1993) SNNS (Stuttgart Neural Network Simulator). In Skrzypek,J. (ed.), Neural Network Simulation Environments. Kluwer, Boston.
- 19.Solla S., LeCun,Y. and Denker,J. (1990) Optimal Brain Damage. In Touretzky,D.S. (ed.), Advances in Neural Information Processing Systems 2. Morgan Kaufmann, San Mateo, pp. 598–605.
- 20.Stork D. and Hassibi,B. (1993) Second order derivatives for network pruning: Optimal Brain Surgeon. In Hinton,G.E. and Touretzky,D.S. (eds), Advances in Neural Information Processing Systems 5. Morgan Kaufmann, San Mateo, pp. 164–171.
- 21.Norman G.R. and Streiner,D.L. (1997) PDQ Statistics, 2nd Edn. Mosby, St Louis.
- 22.Rumelhart D.E., Hinton,G.E. and Williams,R.J. (1986) Learning internal representations by error propagation. In Rumelhart,D.E., McClelland,J.L. and Group,P.R. (eds), Parallel Distributed Processing. MIT Press, Cambridge, MA, Vol. 1, pp. 318–364.
- 23.Fahlman S.E. (1988) An Empirical Study of Learning Speed in Backpropagation Networks. Technical Report CMU-CS-88-162, Carnegie Melon University, Pittsburgh, PA.
- 24.Matthews B.W. (1975) Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim. Biophys. Acta, 405, 442–451. [DOI] [PubMed] [Google Scholar]
- 25.Hanley J.A. and McNeil,B.J. (1982) The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology, 143, 29–36. [DOI] [PubMed] [Google Scholar]
- 26.Hosmer D.J. and Lemeshow,S. (2000) Applied Logistic Regression, 2nd Edn. John Wiley and Sons, Chichester, UK.
- 27.Sugimoto N., Nakano,S., Katoh,M., Matsumura,A., Nakamuta,H., Ohmichi,T., Yoneyama,M. and Sasaki,M. (1995) Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry, 34, 11211–11216. [DOI] [PubMed] [Google Scholar]
- 28.Smith T.F. and Waterman,M.S. (1981) Identification of common molecular subsequences. J. Mol. Biol., 147, 195–197. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.