Abstract
Insig-1 and Insig-2 are closely related proteins of the endoplasmic reticulum that play crucial roles in cholesterol homeostasis by inhibiting excessive cholesterol synthesis and uptake. In sterol-depleted cells Insig-1 is degraded at least 15 times more rapidly than Insig-2, owing to ubiquitination of Lys-156 and Lys-158 in Insig-1. In this study, we use domain-swapping methods to localize amino acid residues responsible for this differential degradation. In the case of Insig-2, Glu-214 stabilizes the protein by preventing ubiquitination. When Glu-214 is changed to alanine, Insig-2 becomes ubiquitinated, but it is still not degraded as rapidly as ubiquitinated Insig-1. The difference in the degradation rates is traced to two amino acids: Ser-149 in Insig-1 and Ser-106 in Insig-2. Ser-149, which lies NH2-terminal to the ubiquitination sites, accelerates the degradation of ubiquitinated Insig-1. Ser-106, which is COOH-terminal to the ubiquitination sites, retards the degradation of ubiquitinated Insig-2. The current studies indicate that the degradation of ubiquitinated Insigs is controlled by serine residues flanking the sites of ubiquitination.
Keywords: proteasome, ubiquitin
Sterol-induced binding of Insig proteins to two endoplasmic reticulum (ER) membrane proteins, SREBP cleavage activating protein (Scap) and 3-hydroxy-3-methylglutaryl-CoA reductase (HMG CoA reductase), is central to feedback inhibition of cholesterol synthesis in mammalian cells. Scap is an escort protein for SREBPs (1), which are membrane-bound transcription factors that activate transcription of all of the known genes encoding cholesterol biosynthetic enzymes (2). In sterol-depleted cells, Scap transports SREBPs from the ER to the Golgi apparatus where SREBPs undergo proteolytic activation. The accumulation of intracellular sterols triggers binding of Scap to Insigs, a reaction that leads to the ER retention of Scap and prevents delivery of its bound SREBPs to the Golgi for proteolytic release from membranes (3, 4).
HMG CoA reductase catalyzes the rate-limiting step in the synthesis of cholesterol (5). Sterols stimulate its binding to Insigs, which leads to the ubiquitination of reductase by an Insig-bound ubiquitin ligase, gp78 (6), and subsequent proteasomal degradation of reductase. Through this action, Insigs slow the rate of mevalonate production and reduce cholesterol synthesis. In combination, these Insig-mediated reactions (ER retention of Scap and accelerated degradation of HMG CoA reductase) prevent potentially toxic overaccumulation of cholesterol in cells.
Mammalian cells harbor two Insig proteins. Human Insig-1 is comprised of 277 aa (4), whereas human Insig-2 contains 225 aa (4). Both are deeply embedded in ER membranes by virtue of six membrane-spanning helices (7). The proteins exhibit an amino acid identity of 59% with the differences confined mostly to the short hydrophilic segments at the NH2 termini (4). Insig-1 and Insig-2 are functionally similar in that both cause the ER retention of the Scap/SREBP complex and the accelerated degradation of HMG CoA reductase (3, 4, 8, 9).
Despite the functional similarity between Insig-1 and Insig-2, the degradation of Insig-1 is much more rapid than that of Insig-2 in normal culture medium (10). When cells are incubated with sterols, Insig-1 is stabilized by binding to Scap, an event that blocks the ubiquitination and subsequent proteasomal degradation of Insig-1 (11). Insig-1 is degraded especially rapidly in mutant CHO cells deficient in Scap, regardless of the presence of sterols (11). In contrast, our unpublished data reveal that Insig-2 is relatively stable in the absence or presence of sterols or Scap.
In this study, we use domain-swapping methods to localize amino acid residues in Insig-1 and Insig-2 that determine the differences in their degradation rates. Our studies reveal that Ser-149 of Insig-1 is required for rapid degradation, whereas Ser-106 and Glu-214 of Insig-2 are responsible for its stability.
Results
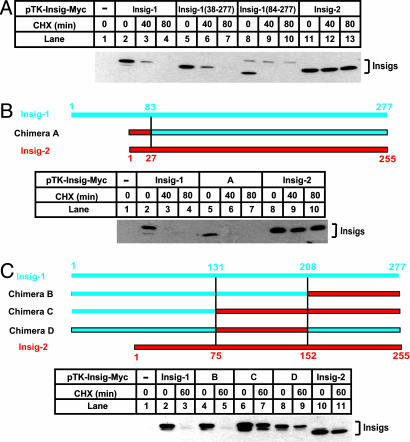
Fig. 1 shows a sequence alignment between human Insig-1 and Insig-2. Most of the differences between the two proteins are found at the NH2 terminus where Insig-1 has an 83-aa extension. To determine whether this NH2-terminal extension is required for rapid degradation of Insig-1, mutant CHO cells deficient in Scap (SRD-13A cells) were transfected with cDNAs encoding full-length or NH2-terminally truncated Insig-1 tagged at the COOH terminus with six tandem repeats of a Myc epitope (pTK-Insig1-Myc). The amount of transfected cDNA was adjusted so that each version of Insig-1 protein was expressed at a similar level as judged by immunoblotting. Cells were treated with cycloheximide to inhibit protein synthesis and harvested for immunoblot analysis with anti-Myc to measure the rate of degradation of transfected Insig-1. As shown in Fig. 2A, full-length Insig-1 declined by >50% after 40 min and completely disappeared after 80 min of treatment with cycloheximide (lanes 2–4). Mutant Insig-1 with partial [Insig-1(38–277)] or complete deletion [Insig-1(84–277)] of the NH2-terminal extension declined as rapidly as wild-type Insig-1 (lanes 5–10). In contrast, the amount of Insig-2 did not drop during the course of cycloheximide treatment (lanes 11–13). Previous pulse–chase studies demonstrated that the time course of Insig-1 disappearance after cycloheximide treatment reflects its degradation rate (11). Thus, these data demonstrate that the NH2-terminal extension of Insig-1 is not required for rapid degradation.
Fig. 1.
Sequence alignment between Insig-1 and Insig-2. Identical residues are shaded. S149 in Insig-1 and S106 and E214 in Insig-2 are highlighted in yellow. The ubiquitination sites identified in Insig-1 (K156 and K158) are indicated by asterisks. Underlined are amino acids 131–207 of Insig-1 and the corresponding sequence of Insig-2.
Fig. 2.
Stability of mutant Insig-1 and chimeric proteins between Insig-1 and Insig-2. The chimera constructs between Insig-1 and Insig-2 are illustrated, with blue and red bars representing amino acid sequences derived from Insig-1 and Insig-2, respectively. SRD-13A cells were set up at 3.2 × 105 per 60-mm dish on day 0. On day 2, cells in each dish were transfected with 0.5 μg of wild-type pTK-Insig1-Myc; 0.5 μg of pTK-Insig1-Myc (38–277); 2 μg of pTK-Insig1-Myc (84–277); 1.0 μg of chimera A; 0.5 μg of chimeras B, C, and D; and 0.5 μg of pTK-Insig2-Myc, as indicated. Total plasmid concentration was adjusted to 2 μg per dish by using empty vector pcDNA3.1. On day 3, cells were treated with 50 μM cycloheximide (CHX) for the indicated time. Cells were then harvested, and cell lysate was subjected to SDS/PAGE and immunoblot analysis with anti-Myc IgG-9E10. Filters were exposed for 1 min.
To determine whether Insig-1 can be stabilized by the NH2 terminus of Insig-2, we made a cDNA encoding a fusion protein in which the NH2-terminal extension of Insig-1 was replaced by that of Insig-2 (chimera A). As shown in Fig. 2B, the fusion protein was degraded as quickly as wild-type Insig-1. Thus, the regions that display the largest differences in amino acid sequence between the two proteins, i.e., the NH2 termini, do not determine their differential stabilities.
We next made several additional chimeric proteins between Insig-1 and Insig-2. Replacing the last 69 amino acid residues in Insig-1 with the corresponding residues in Insig-2 (chimera B) did not affect the degradation of Insig-1 (Fig. 2C, lanes 4 and 5). The first positive result came when a stretch of residues in the middle of Insig-1 (amino acids 131–207) was replaced by the corresponding residues in Insig-2. These chimeric proteins (chimeras C and D) were stable (Fig. 2C, lanes 6–9).
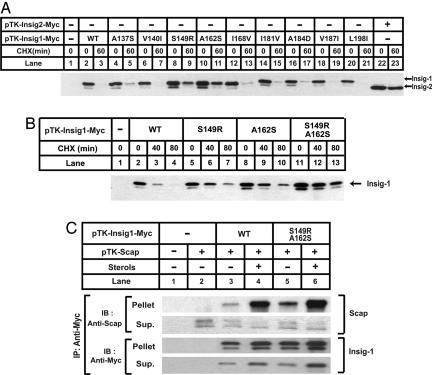
The underlined sequence in Fig. 1 shows amino acids 131–207 of Insig-1 and the corresponding sequence of Insig-2. In this region, nine amino acid residues differ between these two proteins. To determine which of these residues is responsible for the stabilization of Insig-1, we mutated these residues in Insig-1 individually to the corresponding ones in Insig-2. Among those mutations, only S149R and A162S partially stabilized Insig-1 (Fig. 3A, lanes 8–11). Insig-1 containing both S149R and A162S mutations was further stabilized. Its half life was >80 min (Fig. 3B, lanes 11–13). None of the other single mutations had any effect on Insig-1 degradation (Fig. 3A).
Fig. 3.
Insig-1 is stabilized by mutations at S149 and A162. (A and B) SRD-13A cells were set up at 3.2 × 105 per 60-mm dish on day 0. On day 2, cells in each dish were transfected with 0.5 μg of indicated plasmids. Total plasmid concentration was adjusted to 2 μg per dish by using empty vector pcDNA3.1. On day 3, cells were treated with 50 μM cycloheximide (CHX) for the indicated time. Cells were then harvested, and cell lysate was subjected to SDS/PAGE and immunoblot analysis with anti-Myc IgG-9E10. Filters were exposed for 1 min. (C) SRD-13A cells were set up at 3.2 × 105 per 60-mm dish on day 0. On day 2, cells in each dish were transfected with 1.5 μg of pTK-Scap, and 0.5 μg of wild-type or mutant pTK-Insig1-Myc, as indicated. On day 3, cells were switched to medium B containing 1% hydroxypropyl-β-cyclodextrin. After incubation for 1 h at 37°C, cells were washed twice with PBS, switched to medium B in the absence or presence of sterols (10 μg/ml cholesterol plus 1 μg/ml 25-hydroxycholesterol), and incubated for an additional 5 h at 37°C. Cells were then harvested, and cell lysate was immunoprecipitated (IP) with anti-Myc IgG-9E10 to precipitate transfected Insigs. The supernatant (Sup.) and pellet fractions of the immunoprecipitation derived from 0.1 and 0.5 dish of cells, respectively, were subjected to SDS/PAGE and immunoblot analysis with polyclonal anti-Myc and anti-Scap (R139). Filters were exposed for 1 min.
In the experiment shown in Fig. 3C, a plasmid encoding Scap was transfected into SRD-13A cells together with plasmids encoding wild-type Insig-1 or the S149R/A162S double mutant. After incubation in medium in the absence or presence of sterols, cells were harvested, and the transfected Insig-1 protein was immunoprecipitated with anti-Myc antibody. The amount of Scap coprecipitated with wild-type Insig-1 was increased when cells were incubated with sterols (lanes 3 and 4). Similar results were observed with the S149R/A162S mutant (lanes 5 and 6). These data indicate that the mutations that delay the degradation of Insig-1 do not affect its sterol-dependent association with Scap.
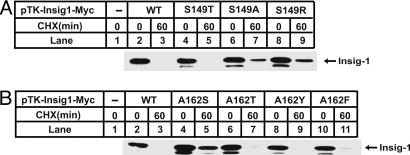
The stabilization of Insig-1 caused by S149R/A162S mutations may result from the loss of Ser-149 or Ala-162 from Insig-1, or from the gain of arginine or serine at these two positions. To differentiate between these two possibilities, we mutated S149 and A162 to other amino acids. Whereas the conservative S149T mutation still allowed Insig-1 to be rapidly degraded, the S149A mutation stabilized Insig-1 to the same extent as did the S149R mutation (Fig. 4A). In contrast, the stabilization of Insig-1 by the A162S mutation was not reproduced even by the similar A162T mutation (Fig. 4B). In fact, when we mutated A162 to all other 19 aa, only the A162S and A162G mutants were stabilized (data not shown). These data indicate that stabilization of Insig-1 is attributable to the loss of serine at residue 149 and the gain of serine at residue 162.
Fig. 4.
Stability of mutant Insig-1 containing mutations at S149 and A162. SRD-13A cells were set up, transfected with indicated plasmids, harvested, and analyzed in exactly the same way as that described in the legend for Fig. 3B.
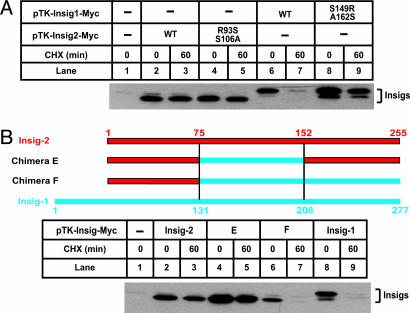
Next we attempted to identify the residues of Insig-2 that are responsible for its stability. First we mutated R93 and S106 of Insig-2 to serine and alanine, respectively. These two positions correspond to S149 and A162 in Insig-1. Surprisingly, this double mutant was as stable as wild-type Insig-2 (Fig. 5A, lanes 4 and 5), indicating that other regions of Insig-2 may be responsible for its stabilization. To identify such a region, we again made chimeric proteins between Insig-1 and Insig-2. Insig-2 remained stable when a stretch of residues in the middle of Insig-2 (amino acids 75–151) was replaced by the corresponding residues in Insig-1 (chimera E) (Fig. 5B, lanes 4 and 5). However, additional replacement of the COOH terminus of Insig-2 (amino acids 152–225) by the corresponding residues in Insig-1 (chimera F) allowed the fusion protein to be rapidly degraded (Fig. 5B, lanes 6 and 7). This finding indicates that the COOH terminus of Insig-2 contains a motif that confers stability.
Fig. 5.
Stability of mutant Insig-2 and chimeric proteins between Insig-1 and Insig-2. The chimera constructs between Insig-1 and Insig-2 are illustrated, with blue and red bars representing amino acid sequences derived from Insig-1 and Insig-2, respectively. SRD-13A cells were set up at 3.2 × 105 per 60-mm dish on day 0. On day 2, cells in each dish were transfected with 0.3 μg of wild-type or mutant pTK-Insig2-Myc, 0.5 μg of wild-type or mutant pTK-Insig1-Myc, and 0.5 μg of chimera E and F, as indicated. Total plasmid concentration was adjusted to 2 μg per dish by using empty vector pcDNA3.1. On day 3, cells were treated with 50 μM cycloheximide (CHX) for the indicated time. Cells were then harvested, and cell lysate was subjected to SDS/PAGE and immunoblot analysis with anti-Myc antibody. Filters were exposed for 1 min.
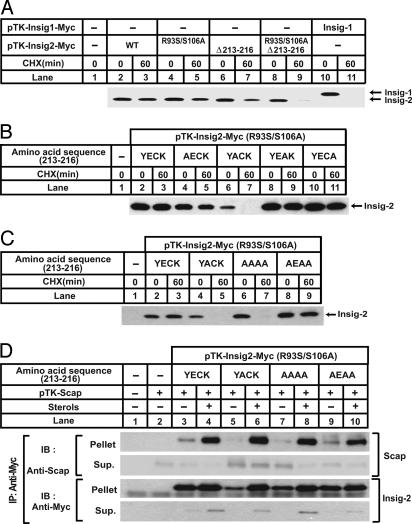
The COOH terminus of Insig-2 contains a tetrapeptide insertion (YECK) beginning at residue 213 that is absent in Insig-1 (Fig. 1). To determine whether this tetrapeptide mediates stabilization of Insig-2, we made mutants that had the sequence deleted. As shown in Fig. 6A, deletion of the tetrapeptide alone partially increased the degradation rate of Insig-2 (lanes 6 and 7). The triple mutant of Insig-2 containing both the deletion and the R93S/S106A substitutions was rapidly degraded, disappearing within 60 min after the addition of cycloheximide, just like Insig-1 (lanes 8–11).
Fig. 6.
E214 is required for the stability of Insig-2. (A–C) SRD-13A cells were set up at 3.2 × 105 per 60-mm dish on day 0. On day 2, cells in each dish were transfected with 1 μg of mutant pTK-Insig2-Myc in which the tetrapeptide sequence was changed from YECK to YACK or AAAA, or 0.5 μg of other plasmids, as indicated. Total plasmid concentration was adjusted to 2 μg per dish by using empty vector pcDNA3.1. On day 3, cells were treated with 50 μM cycloheximide for the indicated time. Cells were then harvested, and cell lysate was subjected to SDS/PAGE and immunoblot with anti-Myc IgG-9E10. Filters were exposed for 1 min. (D) SRD-13A cells were set up at 3.2 × 105 per 60-mm dish on day 0. On day 2, cells in each dish were transfected with 1.5 μg of pTK-Scap, 1.0 μg of mutant pTK-Insig2-Myc in which the tetrapeptide sequence was changed from YECK to YACK or AAAA, and 0.5 μg of other mutant pTK-Insig-2-Myc, as indicated. Total plasmid concentration was adjusted to 2.5 μg per dish by using empty vector pcDNA3.1. On day 3, cells were switched to medium B containing 1% hydroxypropyl-β-cyclodextrin. After incubation for 1 h at 37°C, cells were washed twice with PBS, switched to medium B in the absence or presence of sterols (10 μg/ml cholesterol plus 1 μg/ml 25-hydroxycholesterol), and incubated for an additional 5 h at 37°C. Cells were then harvested, and cell lysate was immunoprecipitated (IP) with anti-Myc IgG-9E10 to precipitate transfected Insigs. The supernatant (Sup.) and pellet fractions of the immunoprecipitation derived from 0.1 and 0.5 dish of cells, respectively, were subjected to SDS/PAGE and immunoblot analysis with polyclonal anti-Myc and anti-Scap (R139). Filters were exposed for 1 min.
To determine which amino acid residue within the tetrapeptide is required for the stabilization of Insig-2, we carried out an alanine scan mutagenesis of the tetrapeptide sequence. To observe a complete effect on protein degradation, the mutations were made on the background of the R93S/S106A substitution. As shown in Fig. 6B, mutant Insig-2 was rapidly degraded only when E214, the second amino acid residue in the tetrapeptide, was mutated to alanine (lanes 6 and 7). Alanine substitutions for the other three amino acids in the tetrapeptide had no effect on Insig-2 degradation (lanes 4, 5, and 8–11). The expression of the mutant that carries the YACK sequence was a little less than the other versions of Insig-2 even though cDNA encoding this mutant was transfected at a higher amount, probably due to the rapid degradation rate of this mutant. We also made a mutant in which all four residues of the tetrapeptide were mutated to alanines (AAAA). As expected, this Insig-2 was rapidly degraded (Fig. 6C, lanes 6 and 7). Remarkably, the introduction of glutamate at the second position in the AAAA sequence (AAAA to AEAA) was sufficient to stabilize Insig-2 (Fig. 6C, lanes 8 and 9). Although specific amino acids are not required at the other three positions, the stabilizing effect of E214 was lost when these three residues were deleted (data not shown), indicating that the glutamate must be in the correct position. Mutants of Insig-2 containing R93S/S106A plus various mutations in the tetrapeptide sequence bound to Scap in a sterol-dependent manner (Fig. 6D), indicating that the mutants are properly folded. Thus, the rapid degradation observed in mutant Insig-2 is unlikely due to misfolding. Similar to Insig-1 (11), the R93S/S106A/E214A mutant of Insig-2 was stabilized by sterols when SCAP was cotransfected (Fig. 6D, lanes 5–8, Insig-2 immunoblots).
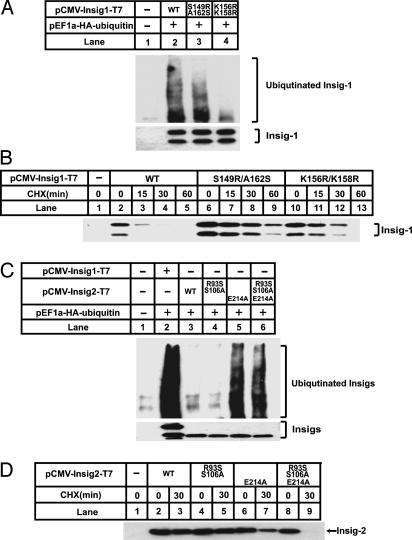
Proteasomal degradation of Insig-1 requires prior ubiquitination (11). We thus examined the ubiquitination of the mutant Insig proteins. As shown in Fig. 7A, SRD-13A cells were transfected with plasmids encoding hemagglutinin (HA)-tagged ubiquitin and wild-type or mutant versions of T7-tagged Insig-1. After treatment for 30 min with MG-132, a proteasome inhibitor that allows accumulation of ubiquitinated proteins, cells were harvested. The transfected Insig-1 was immunoprecipitated from cell lysates with anti-T7 agarose beads, and the immunoprecipitates were subjected to SDS/PAGE and immunoblot analysis with an antibody to the HA tag on ubiquitin. In control cells not expressing T7-tagged Insig-1, no ubiquitin was seen in the anti-T7 immunoprecipitates (Fig. 7A, lane 1). In cells expressing wild-type Insig-1, the Insig-1 immunoprecipitates showed the typical smear seen with polyubiquitinated protein when blotted with the antibody against the HA tag on ubiquitin (lane 2). When the previously described ubiquitination sites were mutated (K156R/K158R) (11), the amount of ubiquitinated Insig-1 was significantly reduced (lane 4). Surprisingly, the S149R/A162S mutant of Insig-1 was ubiquitinated to the same extent as wild-type Insig-1 (lane 3), even though this protein was more stable than the wild-type Insig-1 (Fig. 7B). Fig. 7A Bottom shows that equal amounts of wild-type and mutant Insig-1 were precipitated. We did not observe a higher-molecular-weight smear in this autoradiogram, which was immunoblotted with an antibody against the T7 tag on Insig-1. This is the usual finding in studies of protein ubiquitination in intact cells, and it is believed to reflect the strong activity of de-ubiquitinating enzymes (12, 13). The amount of ubiquitinated Insig-1 is sufficient for visualization when blotted for ubiquitin but not when blotted for Insig-1. Considered together, these results suggest that S149R/A162S mutation permits ubiquitination but retards the degradation of ubiquitinated Insig-1.
Fig. 7.
Ubiquitination of wild-type and mutant Insig-1 and Insig-2. SRD-13A cells were set up at 3.2 × 105 per 60-mm dish on day 0. (A) On day 2, cells in each dish were transfected with 0.2 μg of pEF1a-HA-ubiqutin, and 0.2 μg of wild-type or mutant pCMV-Insig1-T7, as indicated. Total plasmid concentration was adjusted to 2.0 μg per dish by using empty vector pcDNA3.1. On day 3, cells were treated with 10 μM MG-132 for 30 min. Cells were harvested, and cell lysate was immunoprecipitated (IP) with monoclonal anti-T7 IgG-coupled agarose beads to precipitate transfected Insigs. The pellet fractions of the immunoprecipitation were subjected to SDS/PAGE and immunoblot analysis with anti-T7 and anti-HA antibody. Filters were exposed for 30 sec. (B) On day 2, the cells in each dish were transfected with 0.2 μg of wild-type or mutant pCMV-Insig1-T7, as indicated. Total plasmid concentration was adjusted to 2.0 μg per dish by using empty vector pcDNA3.1. On day 3, cells were treated with 50 μM cycloheximide (CHX) for the indicated time. Cells were then harvested, and cell lysate was subjected to SDS/PAGE and immunoblot analysis with anti-T7 antibody. Filters were exposed for 30 sec. (C) On day 2, cells in each dish were transfected with 0.2 μg of pEF1a-HA-ubiqutin, 0.3 μg of wild-type pCMV-Insig-2-T7, 1.0 μg of mutant pCMV-Insig-2-T7 containing E214A mutation, and 0.3 μg of other mutant pCMV-Insig-2-T7, as indicated. Total plasmid concentration was adjusted to 3.0 μg per dish by using empty vector pcDNA3.1. Cells were then treated, harvested, and analyzed as described for A. (D) On day 2, cells in each dish were transfected with the same amount of wild-type or mutant pCMV-Insig2-T7 as shown for C. Total plasmid concentration was adjusted to 3.0 μg per dish by using empty vector pcDNA3.1. On day 3, cells were treated with 50 μM cycloheximide (CHX) for the indicated time. Cells were then harvested, and cell lysate was subjected to SDS/PAGE and immunoblot analysis with anti-T7 antibody. Filters were exposed for 30 sec.
Similar experiments were performed to analyze the ubiquitination and degradation of wild-type and mutant versions of Insig-2 (Fig. 7C). Wild-type Insig-2 was not ubiquitinated (Fig. 7C, lane 3), nor was it rapidly degraded (Fig. 7D, lanes 2 and 3). The R93S/S106A double mutant was also not ubiquitinated (Fig. 7C, lane 4) or rapidly degraded (Fig. 7D, lanes 4 and 5). In striking contrast, the E214A mutant was ubiquitinated (Fig. 7C, lane 5), but its degradation was only slightly increased (Fig. 7D, lanes 6 and 7). Addition of the R93S/S106A substitutions to the E214A mutation permitted the combination of ubiquitination and rapid degradation (Fig. 7 C, lane 6, and D, lanes 8 and 9). In experiments not shown, we demonstrated that ubiquitination of the R93S/S106A/E214A triple mutant of Insig-2 required the lysines at positions 100 and 102, which corresponds to K156 and K158 of Insig-1 (Fig. 1).
Discussion
Regulated degradation of Insig-1 is central to the process by which animal cells maintain cholesterol homeostasis. Rapid degradation of Insig-1 in sterol-depleted cells helps to free the Scap/SREBP complex so that the SREBP can be processed and enter the cell nucleus to activate genes involved in cholesterol synthesis and uptake (11). Upon entering the nucleus, the SREBPs also stimulate the transcription of the Insig-1 gene, thereby restoring Insig-1 protein levels. When cholesterol has been repleted, the sterol triggers the binding of Scap to the newly synthesized Insig. This binding stabilizes Insig and allows it to hold the Scap/SREBP complex in the ER, ready to respond quickly when cholesterol is subsequently depleted. We have called this process “convergent feedback inhibition” because blocking of SREBP processing requires convergence of two types of SREBP products: (i) the enzymes and receptors that replenish cholesterol and (ii) Insig-1. The second Insig protein, Insig-2, is not a target for SREBP activation. Transfected Insig-2 driven by weak herpes simplex virus thymidine kinase promoter is also not rapidly degraded even in the absence of SCAP (Fig. 2A). The slow degradation rate of transfected Insig-2 is unlikely due to artifacts of epitope tags placed at the COOH terminus of Insig-2, because both myc and T7 epitope-tagged Insig-2 had similar turnover rates (Figs. 2A and 7D). Unfortunately, we were unable to verify the observation with endogenous Insig-2, because we currently do not have an antibody sensitive enough to detect the endogenous Insig-2.
In this study, we show that the rapid degradation of Insig-1 depends on the residues at two positions, amino acids 149 and 162. A serine residue at 149 confers instability, and a serine residue at position 162 confers stability. Thus, Insig-1 protein becomes stable when serine 149 is changed to arginine or alanine, and the alanine at 162 is changed to serine. The current data are consistent with the notion that the serine residues do not control ubiquitination per se, but rather they are necessary in order for the ubiquitinated Insig-1 to be degraded by the proteasome. This follows from the observation that the S149R/A162S mutant appears to be ubiquitinated as efficiently as wild-type Insig-1 (Fig. 7A) but is not rapidly degraded (Fig. 7B). In the linear sequence, residues 149 and 162 surround the two lysines that are sites of ubiquitination (positions 156 and 158) (11). A previous study indicated that the proteasome-mediated degradation of a ubiquitinated protein is greatly accelerated if an unstructured region is present near the ubiquitination sites (14). Thus, serines at position 149 and 162 may affect the local folding of Insig-1 near the ubiquitination sites. A serine at position 149 would permit unfolding, whereas a serine at position 162 might oppose such unfolding. It is tempting to speculate that phosphorylation of these two serines may be involved in the process. However, in multiple experiments using 32P-labeling followed by immunoprecipitation, we have not been able to show that Insig proteins are phosphorylated. Moreover, the finding that the effect of serine 162 could be reproduced with glycine at this position argues against the involvement of phosphorylation.
The significance of the residues at 149 and 162 in Insig-1 was confirmed when we inserted the destabilizing residues at the corresponding positions of Insig-2. However, to destabilize Insig-2, we also had to remove the glutamic acid at position 214. This glutamic acid appeared to act as an inhibitor of ubiquitination. The effect of E214 is specific for Insig-2, because replacement of the COOH terminus of Insig-1 by the corresponding residues in Insig-2, which includes the tetrapeptide YECK, failed to stabilize Insig-1 (Fig. 2C, chimera B). It appears that the NH2-terminal 131 aa of Insig-1 rendered the protein resistant to the effect of E214, because substitution of these amino acids with corresponding residues in Insig-2 allowed the stabilization of the chimeric protein (Fig. 5B, chimera E).
When E214 was switched to alanine, Insig-2 was ubiquitinated (Fig. 7C). However, it was not rapidly degraded unless the destabilizing serines were placed in the proper position by the R93S/S106A mutations (Fig. 7D). Importantly, the R93S/S106A mutations had no effect on the degradation of Insig-2 containing E214, which prevents ubiquitination (Fig. 7 C and D). These data again suggest that the two serines function at a postubiquitination step.
If the S149R/A162S mutant were resistant to proteasomal degradation at a postubiquitination step, one might expect to observe the accumulation of the ubiquitinated Insig-1 mutant even in the absence of proteasome inhibitor MG132. However, when cells were harvested in the absence of MG132, we observed only trace amounts of ubiquitinated mutant or wild-type Insig-1 (data not shown). The failure to accumulate the ubiquitinated mutant may be explained by the de-ubiquitinating activity associated with the proteasome. It was shown previously that proteasome-associated de-ubiquitinating enzymes remove the ubiquitin moiety from ubiquitinated proteins that are not substrates for proteasome-mediated rapid degradation (14–16). According to this scenario, the S149R/A162S mutant of Insig-1 might be resistant to the proteolytic activity of the proteasome but still sensitive to its de-ubiquitinating activity. By possibly inhibiting the de-ubiquitinating activity of the proteasome, MG132 might cause the build-up of both ubiquitinated wild-type and mutant Insig-1.
Materials and Methods
Materials.
MG-132 and Nonidet P-40 alternative were obtained from Calbiochem, Fos-Choline 13 and CHAPSO detergent were obtained from Anatrace, hydroxypropyl-β-cyclodextrin was obtained from Cyclodextrin Technologies, polyclonal anti-Myc and anti-T7 IgG were obtained from Bethyl Laboratories, cycloheximide and monoclonal anti-HA IgG were obtained from Sigma, horseradish peroxidase-conjugated donkey anti-mouse and anti-rabbit IgGs (affinity-purified) were obtained from Jackson ImmunoResearch, and hybridoma cells producing IgG-9E10, a mouse monoclonal antibody against Myc tag, were obtained from American Type Culture Collection. IgG-R139, a rabbit polyclonal antibody against hamster Scap, is described in ref. 17. Lipoprotein-deficient serum (d > 1.215 g/ml) was prepared from newborn calf serum by ultracentrifugation (18). Solutions of sodium mevalonate (19), sodium compactin (19), and sodium oleate were prepared as described (19, 20).
Plasmid Constructs.
The following plasmids have been described: pEF1a-HA-ubiquitin (provided by Zhijian Chen, University of Texas Southwestern Medical Center) encodes amino acids 1–76 of human ubiquitin preceded by an epitope tag derived from the influenza HA protein (YPYDVPDY) under the control of the EF1a promoter; pTK-Scap, encoding wild-type hamster Scap under control of the herpes simplex virus thymidine kinase promoter (21); pCMV-Insig1-T7 and pCMV-Insig2-T7, encoding full-length versions of human Insig-1 and human Insig-2, respectively, followed by three tandem copies of a T7 epitope tag under control of the CMV enhancer/promoter (6); and pTK-Insig1-Myc, encoding full-length human Insig-1, followed by six tandem copies of a c-Myc epitope tag driven by herpes simplex virus thymidine kinase promoter (11). pTK-Insig2-Myc encodes full-length human Insig-2, followed by six tandem copies of a c-Myc epitope tag. It is driven by Herpes simplex virus TK promoter. This plasmid was generated by the same method used to make pTK-Insig1-Myc (11). Chimeras between human Insig-1 and human Insig-2 were generated by PCR-based approaches using pTK-Insig1-Myc and pTK-Insig2-Myc as templates. Mutant versions of pTK-Insig1-Myc and pTK-Insig2-Myc were generated with the QuikChange II XL site-directed mutagenesis kit (Stratagene). The coding regions of all plasmids were sequenced before use.
Cell Culture.
SRD-13A cells (mutant CHO cells deficient in Scap) were maintained in monolayer culture at 37°C in 8–9% CO2. Stock cultures of SRD-13A cells were maintained in medium A (1:1 mixture of Ham’s F-12 medium and DMEM containing 100 units/ml penicillin and 100 μg/ml streptomycin sulfate) supplemented with 5% (vol/vol) FCS, 5 μg/ml cholesterol, 1 mM sodium mevalonate, and 20 μM sodium oleate.
Transient Transfection of Cells and Immunoblot Analysis.
SRD-13A cells were transiently transfected with FuGENE 6 reagent (Roche Applied Science) according to the manufacturer’s protocol. After transfection, cells were incubated in either medium A supplemented with 5% (vol/vol) FCS (in most experiments) or medium B (medium A supplemented with 5% newborn calf lipoprotein-deficient serum, 50 μM sodium compactin, and 50 μM sodium mevalonate) as specified in figure legends. Duplicate dishes of cells were pooled, harvested, and lysed in 0.1 ml of buffer A (25 mM Tris·HCl, pH 7.2/0.15 M NaCl/1% Nonidet P-40/10 μg/ml leupeptin/5 μg/ml pepstatin A/10 μg/ml aprotinin/25 μg/ml N-acetyl-leucinal-leucinal-norleucinal). After centrifugation at 20,000 × g for 10 min at 4°C, the supernatant from the spin was subjected to SDS/PAGE and immunoblot analysis as described in ref. 17. Antibodies used in the current studies were IgG-R139 (5 μg/ml), IgG-9E10 (5 μg/ml), a polyclonal anti-Myc (1 μg/ml), a polyclonal anti-T7 IgG (0.2 μg/ml), a monoclonal anti-HA IgG (1:1,000 dilution), and horseradish peroxidase-conjugated donkey anti-mouse and anti-rabbit IgGs (0.2 μg/ml). Bound antibodies were visualized by chemiluminescence using the SuperSignal substrate system (Pierce) according to the manufacturer’s instructions. Filters were exposed to Kodak X-Omat Blue XB-1 films at room temperature for the indicated time.
Immunoprecipitation.
The pooled cell pellets from duplicate 60-mm dishes of cells were lysed and immunoprecipitated as described in ref. 17.
Ubiquitination of Insig-1 and Insig-2.
The ubiquitination of Insig proteins were analyzed exactly as described in ref. 11.
Acknowledgments
We thank Michael S. Brown and Joseph L. Goldstein for constant support and helpful comments, Russell A. DeBose-Boyd and George N. DeMartino for critical review of the manuscript, Lorena Avila for excellent technical assistance, and Lisa Beatty and Angela Carroll for assistance in tissue culture. This work was supported by National Institutes of Health Grant HL-20948, American Heart Association Grant 0630029N, and the Perot Family Foundation.
Abbreviations
- ER
endoplasmic reticulum
- HA
hemagglutinin.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Brown M. S., Goldstein J. L. Proc. Natl. Acad. Sci. USA. 1999;96:11041–11048. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horton J. D., Shah N. A., Warrington J. A., Anderson N. N., Park S. W., Brown M. S., Goldstein J. L. Proc. Natl. Acad. Sci. USA. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang T., Espenshade P. J., Wright M. E., Yabe D., Gong Y., Aebersold R., Goldstein J. L., Brown M. S. Cell. 2002;110:489–500. doi: 10.1016/s0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
- 4.Yabe D., Brown M. S., Goldstein J. L. Proc. Natl. Acad. Sci. USA. 2002;99:12753–12758. doi: 10.1073/pnas.162488899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldstein J. L., Brown M. S. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 6.Song B. L., Sever N., Bose-Boyd R. A. Mol. Cell. 2005;19:829–840. doi: 10.1016/j.molcel.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 7.Feramisco J. D., Goldstein J. L., Brown M. S. J. Biol. Chem. 2004;279:8487–8496. doi: 10.1074/jbc.M312623200. [DOI] [PubMed] [Google Scholar]
- 8.Sever N., Yang T., Brown M. S., Goldstein J. L., DeBose-Boyd R. A. Mol. Cell. 2003;11:25–33. doi: 10.1016/s1097-2765(02)00822-5. [DOI] [PubMed] [Google Scholar]
- 9.Sever N., Lee P. C. W., Song B. L., Rawson R. B., Bose-Boyd R. A. J. Biol. Chem. 2004;279:43136–43147. doi: 10.1074/jbc.M406406200. [DOI] [PubMed] [Google Scholar]
- 10.Lee J. N., Ye J. J. Biol. Chem. 2004;279:45257–45265. doi: 10.1074/jbc.M408235200. [DOI] [PubMed] [Google Scholar]
- 11.Gong Y., Lee J. N., Lee P. C. W., Goldstein J. L., Brown M. S., Ye J. Cell Metab. 2006;3:15–24. doi: 10.1016/j.cmet.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 12.Mimnaugh E. G., Bonvini P., Neckers L. Electrophoresis. 1998;20:418–428. doi: 10.1002/(SICI)1522-2683(19990201)20:2<418::AID-ELPS418>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 13.Sever N., Song B. L., Yabe D., Goldstein J. L., Brown M. S., Bose-Boyd R. A. J. Biol. Chem. 2003;278:52479–52490. doi: 10.1074/jbc.M310053200. [DOI] [PubMed] [Google Scholar]
- 14.Prakash S., Tian L., Ratliff K. S., Lehotzky R. E., Matouschek A. Nat. Struct. Mol. Biol. 2004;11:830–837. doi: 10.1038/nsmb814. [DOI] [PubMed] [Google Scholar]
- 15.Lam Y. A., Xu W., DeMartino G. N., Cohen R. E. Nature. 1997;385:737–740. doi: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- 16.Kenniston J. A., Sauer R. T. Nat. Struct. Mol. Biol. 2004;11:800–802. doi: 10.1038/nsmb0904-800. [DOI] [PubMed] [Google Scholar]
- 17.Sakai J., Nohturfft A., Cheng D., Ho Y. K., Brown M. S., Goldstein J. L. J. Biol. Chem. 1997;272:20213–20221. doi: 10.1074/jbc.272.32.20213. [DOI] [PubMed] [Google Scholar]
- 18.Goldstein J. L., Basu S. K., Brown M. S. Methods Enzymol. 1983;98:241–260. doi: 10.1016/0076-6879(83)98152-1. [DOI] [PubMed] [Google Scholar]
- 19.Kita T., Brown M. S., Goldstein J. L. J. Clin. Invest. 1980;66:1094–1100. doi: 10.1172/JCI109938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hannah V. C., Ou J., Luong A., Goldstein J. L., Brown M. S. J. Biol. Chem. 2001;276:4365–4372. doi: 10.1074/jbc.M007273200. [DOI] [PubMed] [Google Scholar]
- 21.Nohturfft A., Brown M. S., Goldstein J. L. Proc. Natl. Acad. Sci. USA. 1998;95:12848–12853. doi: 10.1073/pnas.95.22.12848. [DOI] [PMC free article] [PubMed] [Google Scholar]