Abstract
The Cre-lox system is an important tool for genetic manipulation. To promote integrative reactions, two strategies using mutant lox sites have been developed. One is the left element/right element (LE/RE)-mutant strategy and the other is the cassette exchange strategy using heterospecific lox sites such as lox511 or lox2272. We compared the recombination efficiencies using these mutant lox sites in embryonic stem (ES) cells, and found that the combination of the LE/RE mutant and lox2272 showed high recombination efficiency and stability of the recombined structure. Taking advantage of this stability, we successfully integrated the cre gene into the mutant lox sites by Cre-mediated recombination. Germ line chimeric mice were produced from the cre-integrated ES cell clones, and Cre-expressing mouse lines were established. The inserted cre gene was stably maintained through the generations. This cre knock-in system using the LE/RE-lox2272 combination should be useful for the production of various cre mice for gene targeting or gene trapping.
INTRODUCTION
The Cre-lox recombination system has proven to be the most useful tool for genetic manipulation of mammalian cells (1–3) and mice (4,5). Cre recombinase catalyzes reciprocal site-specific recombination between two loxP sites. The loxP sequence is composed of an asymmetric 8 bp spacer flanked by 13 bp inverted repeats (6), and Cre protein binds to the 13 bp repeat mediating the recombination within the 8 bp spacer (7). Cre can excise any intervening sequence flanked by loxP sites at high efficiency. The combination of the Cre-lox system and gene targeting has provided a means to achieve conditional gene knockout or activation (2,8,9).
The Cre-lox system has also been used for site-specific integration or replacement of transfected DNA into a chromosomally positioned lox site(s) (10–16). The integration reaction is inefficient with wild-type loxP sites due to the re-excision of the recombined product, and therefore mutant lox sites have been developed to increase the efficiency of Cre-mediated insertion or replacement. We previously demonstrated targeted integration using the left element/right element (LE/RE) mutant lox sites in embryonic stem (ES) cells (11). The LE mutant lox, lox71, has 5 bp mutated in the left 13 bp repeat, and the RE mutant lox, lox66, has 5 bp mutated in the right 13 bp repeat. Recombination between a chromosomally located lox71 site and a lox66 site on a targeting plasmid results in site-specific integration of the plasmid producing a LE+RE mutant lox site with mutated sequences in both the 13 bp repeats and a wild-type loxP site. Since the binding affinity of the LE+RE lox mutant site for Cre recombinase is reduced, the integrated plasmid is stably retained.
An alternative method using a different kind of mutant lox sites has also been developed and is termed recombinase mediated cassette exchange (13,15) or the double lox strategy (14). In this method, heterospecific lox sites which have mutation(s) in the 8 bp spacer region are used. The principle is that recombination does not occur between two lox sites differing in the spacer region whereas lox sites having the identical spacer region recombine efficiently (17). Several groups have used lox511, which contains a single base substitution, and demonstrated successful gene replacement in which a genomic DNA segment flanked by lox511 and loxP was replaced with another cassette flanked by lox511 and loxP located on a transfected plasmid vector (12–15). Lee and Saito (18) developed two other heterospecific mutant lox sites with two base substitutions, lox2272 and lox5171, and showed that they never recombined with the wild-type loxP site, while lox511 can recombine with loxP at low frequency using an in vitro system. Recently, successful selection marker-free replacement using lox2272 and loxP was demonstrated by Kolb (16).
The Cre-mediated site-specific targeting system has many potential uses, especially in mouse ES cells, because it allows precise and repeated knock-ins of any DNA to target lox sites introduced by gene targeting through homologous recombination. The only exception to this is the cre gene because expression of the cre gene may excise itself, and targeted integration of the cre gene into a transcriptionally active site by Cre-mediated recombination has not been reported.
In this study, we have identified the best combination of mutant lox sites to give high recombination efficiency and stability. We also demonstrate that the cassette exchange strategy using LE/RE mutant lox and heterospecific lox2272 sites is useful for knock-in of the cre gene in ES cells and the production of Cre-expressing mouse lines.
MATERIALS AND METHODS
Plasmids
The loxP, lox66, lox71, lox511 and lox2272 sequences (11,17,18) were synthesized and cloned into pBluescript (pBS)SK– (Stratagene) or pSP73 (Promega). The Flp recognition target (FRT) sequence was excised from pOG45 (Stratagene) and cloned into pBS-KS+. Plasmid p5F2 was constructed by inserting a FRT fragment and lox2272 fragment into pSP73-lox511. The pCAG71bs5F2 was constructed by introducing a fragment of CAG-lox71-bsr-pA from pCAGlox71bsr (11) into p5F2.
Replacement plasmids were derived from ploxPNZneo and plox66NZneo (11). The lox511, lox2272 or lox511-FRT-lox2272 fragment was inserted into the multicloning site of the 5′ end of MC1NeopolyA. The pCreneo was constructed by replacing the NLSlacZ gene of p66-2272 with the cre gene.
The recombination monitor plasmid, pCAG71bsr66NZ, was constructed by replacing the lox511-FRT-lox2272 sequence of pCAG71bs5F2 into the lox66-NLS-lacZ gene excised from p66-511. The plasmid was purified using a QIAquick Nucleotide Removal Kit (Qiagen) before microinjection.
The sequences of all the lox sites contained in these plasmids were confirmed by DNA sequencing. The Cre-expression vector, pCAGGS-Cre, was described previously (3).
Cell culture and electroporation
The ES cell line, TT2 (19), was grown as described (20) except for the use of G418 or Blasticidin S (BlaS)-resistant primary mouse embryo fibroblasts as feeder layers.
Establishment of the cell lines carrying target lox sites was performed (11) with 50 µg of SpeI-digested pCAG71bs5F2. For the co-electroporation experiments, a targeting plasmid and pCAGGS-Cre were used together in their circular forms. The cells (1 × 107 cells/0.8 ml in PBS) were electroporated at 200 V and 960 µF, and after 24 or 48 h they were selected with G418 at 200 µg/ml for 7 days. Colonies were stained with 5-bromo-4-chloro-3-indolyl β-D-galactopyranoside (X-gal) (21) or picked and expanded for DNA analyses.
Southern hybridization and PCR
Six micrograms of genomic DNA was digested with appropriate restriction enzymes, electrophoresed on a 0.9% agarose gel and blotted onto a nylon membrane. Hybridization was performed using a DIG DNA Labeling and Detection Kit (Roche Diagnostics GmbH). For PCR analysis, DNA (0.05–0.1 µg) was subjected to 30 cycles of amplification (each cycle consisted of 1 min at 94°C, 1.5 min at 58°C and 1.5 min at 72°C) using AmpliTaq polymerase (Perkin-Elmer). The sequences of the primers used in the experiments were as follows; AG2, 5′-CTGCTAACCATGTTCATGCC-3′; LZUS3, 5′-GCGCATCGTAACCGTGCAT-3′; Cre2, 5′-TAACCAGTGAAACAGCATTGC-3′.
Production of chimeric mice and microinjection
Chimeric mice were produced by aggregation of ES cells with eight-cell embryos of CD1 mice (Charles Liver) (19). Chimeric male mice were mated with C57BL/6J females (Nippon Clea) to obtain F1 heterozygotes.
For microinjection of pCAG71bsr66NZ, superovulated BDF1 (Charles Liver) females were mated with F1 heterozygous males. Fertilized eggs were collected and pronuclear injection of circular pCAG71bsr66NZ plasmid (5 ng/µl) was performed according to the standard procedure (22). The injected eggs were cultured for 48 or 72 h and stained with X-gal (23).
Statistical analyses
The recombination efficiencies were evaluated by non-repeated measures ANOVA. Where a significant difference (P <0.05) was identified, the differences were analyzed further with SNK tests for making multiple comparisons.
RESULTS
Experimental design for comparison of recombination efficiency
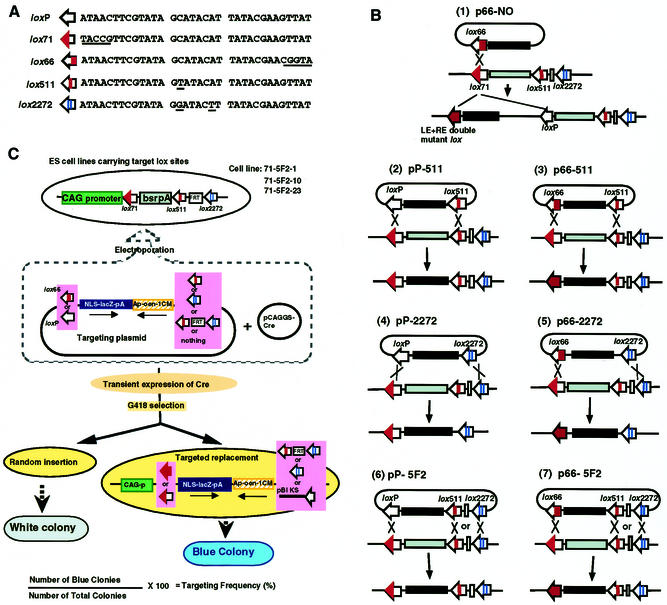
The lox sequences used in this experiment are shown in Figure 1A. Seven targeting plasmids carrying different combinations of the mutant lox sites were constructed (Fig. 1B) and the recombination efficiencies were compared. As a control, we used p66-NO, which contains only one lox66 site [Fig. 1B, (1)], to obtain the efficiency of targeted integration through single recombination between lox66 and lox71. The other plasmids contained heterospecific lox sites, and targeted cassette exchange, in which a DNA segment flanked by heterospecific lox sites on genomic DNA is replaced by another DNA segment flanked by heterospecific lox sites on the targeting plasmid, takes place through double reciprocal recombination [Fig. 1B, (2)–(7)].
Figure 1.
Experimental strategy for comparison of recombination efficiency. (A) Nucleotide sequences of lox sites. Striped boxes representing each lox sequence are indicated after the name. The mutated nucleotides are underlined. (B) Targeting plasmids used and the predicted recombination reactions. The target structure on genomic DNA is shown in the middle, and the resulting recombinant structures are shown at the bottom. (C) Three ES cell lines carrying a single copy of the CAG-lox71-bsr-pA-lox511-FRT-lox2272 fragment were established. The cell lines were co-electroporated with the Cre-expression vector and the targeting plasmids carrying the promoter-less NLSlacZ gene, and then selected with G418. Since the targeting plasmids contain a complete neo gene cassette (MC1neopA), random integrants can also appear. In targeted recombination, colonies are positively stained with X-gal, because the NLSlacZ gene is joined to the CAG promoter, whereas colonies with random integration are not stained because there is no promoter for the NLSlacZ gene. The percentage of blue colonies represents the frequency of targeted recombination.
The experimental design outlined in Figure 1C was used to assess the recombination efficiencies. As a chromosomal target, we constructed pCAG71bs5F2 that contained a cassette of CAG-lox71-bsr-pA-lox511-FRT-lox2272. The CAG promoter is quite a strong promoter based on the chicken β-actin promoter (24), and the bsr gene is a selection marker gene against BlaS. Three ES cell lines, 71-5F2-1, 71-5F2-10 and 71-5F2-23, carrying a single copy of the cassette were established through BlaS selection. A targeting plasmid together with a Cre-expression vector, pCAGGS-Cre, was introduced into the three ES cell lines by co-electroporation. The targeting plasmids contained the promoter-less lacZ gene fused with the nuclear localizing signal (NLSlacZ) and the complete neo gene with the MC1 promoter and poly(A) signal. In this system, both random and targeted recombinants became G418 resistant, but only the targeted colonies were stained blue with X-gal since the NLSLacZ gene was inserted downstream of the CAG promoter through targeted recombination. The percentage of blue colonies represented the frequency of targeted recombination. By using three independently established cell lines, we examined the recombination efficiencies at three chromosomal positions.
Targeted recombination frequency
The ES cell lines were electroporated with 20 µg of each targeting plasmid and pCAGGS-Cre. After G418 selection for 1 week, the colonies were stained and the percentage of blue colonies was scored. The total numbers of colonies ranged from 100 to 300. Each electroporation was repeated at least three times on independent days.
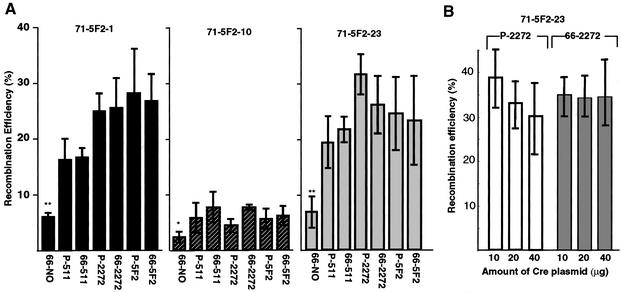
The results are shown in Figure 2A. The targeted recombination efficiencies of the control p66-NO were the lowest, ranging from 3 to 8%. The efficiencies of cassette exchange recombination with all the other plasmids carrying heterospecific lox sites showed 2.5–4 times higher efficiencies than p66-NO. However, the combination of lox66/71 and heterospecific lox sites did not confer any promoting effect on the recombination efficiency using loxP and heterospecific lox sites under these conditions.
Figure 2.
Targeting recombination efficiencies. (A) Effects of mutant lox sites. Twenty micrograms of each replacement plasmid and the Cre-expressing vector were co-electroporated into the 71-5F2-1 line (left), 71-5F2-10 line (middle) or 71-5F2-23 line (right). After 24 h the cells were selected with G418 for 7 days. Colonies were stained with X-gal, and the percentage of positive colonies was scored as the recombination efficiency. The means of at least three independent electroporations are shown with the standard deviation and evaluation by ANOVA. The 71-5F2-1 and 71-5F2-23 lines showed significant differences at P <0.01, and 71-5F2-10 line at P <0.05. The differences among individual means were analyzed further by the SNK test. *, P <0.05 with respect to 66-511, 66-2272 and 66-5F2; **, P <0.01 with respect to all other plasmids. (B) Effect of amount of Cre-expression plasmid. The 71-5F2-23 line was co-electroporated with 20 µg of the replacement plasmid (pP-2272 or p66-2272) and 10, 20 or 40 µg of the Cre-expressing vector. After 48 h the cells were selected with G418 for 7 days. The set of six electroporations was performed at the same time and repeated four times on different days.
To compare the efficiency between lox511 and lox2272, relative efficiencies to that of p66-NO were calculated. As shown in Table 1, the lox2272-plasmids (pP-2272 and p66-2272) gave approximately 1.5 times higher efficiency than the lox511 plasmids (pP-511 and p66-511) in the two cell lines in which the recombination efficiencies were relatively high.
Table 1. Relative recombination efficiency of lox511 plasmids and lox2272 plasmids.
| Integration with lox66/71 | Cassette exchange with heterospecific lox | ||
|---|---|---|---|
| Cell line | p66-NO | lox511 (pP-511, p66–511) | lox2272 (pP-2272, p66–2272) |
| 5F2-1** | 1.0 ± 0.13 (n = 3) | 2.7 ± 0.45a (n = 6) | 4.2 ± 0.70a (n = 7) |
| 5F2-10* | 1.0 ± 0.44 (n = 3) | 2.9 ± 1.14b (n = 6) | 2.6 ± 0.86b (n = 6) |
| 5F2-23** | 1.0 ± 0.42 (n = 3) | 3.0 ± 0.51a (n = 6) | 4.2 ± 0.73a (n = 8) |
**, P <0.01; *, P <0.05 (ANOVA).
aP <0.01 among the three efficiencies (SNK).
bP <0.05 versus p66-NO (SNK).
In the LE/RE mutant system, we observed instability of the recombination products when an increased amount of the Cre-expression vector was used (11). In order to compare the stability of the integration product between pP-2272 and p66-2272, we analyzed recombination frequencies by increasing the amount of Cre-expression plasmid. As shown in Figure 2B, there was a tendency for stable integration when the p66-2272 plasmid was used, even at a high dose of the Cre-expression plasmid. However, there was no statistically significant difference.
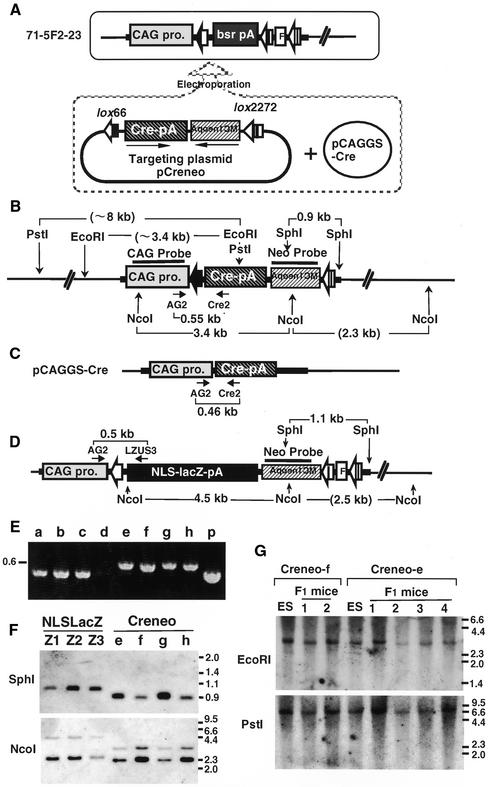
We examined the integration pattern of the cassette of NLSLacZ and neo genes in three blue clones (Z1, Z2 and Z3 in Fig. 3F) obtained by using the pP-5F2 and 71-5F2-23 lines. As shown in Figure 3D, the 5′ junction of the recombination can be amplified using the primer pairs AG2 and LZUS3, and the all blue clones gave a band of the expected size (data not shown). The 3′ junction of the recombination can be detected by Southern blotting with neo probe. The three blue clones exhibited the predicted 1.1 kb fragments with SphI digestion (Fig. 3D and F). The absence of the bsr gene and the vector sequence of the replacement plasmids was also confirmed by PCR (data not shown). These results demonstrate that the clones stained with X-gal were precisely targeted recombinants.
Figure 3.
(Following page) Targeted replacement of the bsr gene with the cre gene through Cre-mediated recombination. (A) Schematic diagram of the replacement plasmid, pCreneo. The 71-5F2-23 line was co-electroporated with 20 µg of the Cre-expression vector and the targeting vector with lox66-cre-pA-MC1neopA-lox2272, and then selected with G418. The small horizontal arrows indicate the gene directions. (B) The expected DNA structure for site-specific replacement of the cre gene. Restriction enzyme sites are indicated by vertical arrows and the name of the enzymes, and the sizes of the expected bands are shown. The sizes in parentheses are the observed specific band sizes for the 71-5F2-23 line. (C) The structure of the Cre-expression vector, pCAGGS-Cre. The small horizontal arrows indicate the positions and directions of the PCR primers used in (E). (D) The DNA structure for site-specific replacement of the NLSLacZ gene. The size in parentheses is the observed specific band size. (E) PCR analysis. a–h, Creneo clones; p, pCAGGS-Cre used as a positive control. (F) Southern blot analysis of SphI- or NcoI-digested genomic DNAs with a Neo probe. Z1–Z3, pP5F2 blue clones; e–h, targeted Creneo ES clones. (G) Southern blot analysis of Creneo F1 mice. Genomic DNAs were digested with EcoRI or PstI and hybridized with a CAG probe. Size markers are shown on the right.
Targeted integration of the cre gene by Cre recombinase
The results in Figure 2B encouraged us to insert the cre gene using the LE/RE-2272 double lox system. 71-5F2-23 cells were electroporated with the Cre-expression vector and cre gene targeting plasmid (pCreneo) which carries lox66-cre gene-pA-MC1neopA-lox2272 (Fig. 3A). Of 129 G418r colonies, 36 were randomly picked and analyzed for targeting replacement (Fig. 3B). PCR analysis identified seven positive clones, but three clones (Fig. 3E, a–c) showed a shorter band than the other four (e–h). Sequence analyses of these bands revealed that the shorter band was derived from randomly integrated Cre-expression plasmid (Fig. 3C). Such random integration of a Cre-expression plasmid was also reported by Soukharev et al. (14). In order to confirm the 3′ junction, the Creneo clones (e–h) and the NLSLacZ clones (Z1–Z3) obtained using pP-5F2 and the same parental cell line 71-5F2-23 were analyzed by Southern blotting with the neo probe. The expected structures of targeted replacement and band sizes are shown in Figure 3B and D. The four Creneo clones and the three NLSLacZ clones exhibited the predicted 0.9 kb and 1.1 kb fragments, respectively, with SphI digestion. Since both the Creneo and NLSLacZ cassettes were inserted into the same chromosomal position, the junction fragments in the Creneo clones should be 0.2 kb shorter than those of the NLSLacZ clones, corresponding to the size of the FRT-lox511 length (Fig. 3D). In agreement with this hypothesis, the junction fragments obtained by NcoI digestion were 2.3 and 2.5 kb in the Creneo and NLSLacZ clones, respectively. These results confirmed that exact replacement of the bsr gene by the Creneo cassette occurred in clones e–h.
Establishment of Cre mice using the replaced clones
Eleven chimeric mice were successfully obtained from two (e and f) of the four clones. All were germ line chimeras even after two electroporations. In order to prove the stability of the inserted cre gene during germ line transmission, we examined the genomic DNA of the F1 mice by Southern blotting with a CAG probe, a probe outside of the replaced region (Fig. 3B). As shown in Figure 3G, the band patterns of the Creneo F1 mice were identical to the original ES clones, demonstrating the stability of the recombinant product.
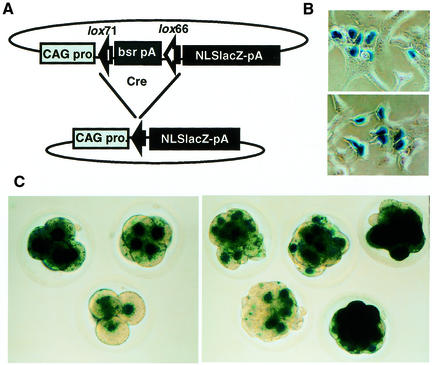
It is possible that the integration is stable due to inactivation of the cre gene through methylation during the establishment of the F1 mice. To rule out this possibility, the activity of the inserted cre gene was examined using a monitor plasmid, which contains a cassette of CAG-lox71-bsr-pA-lox66-NLSLacZ-pA (Fig. 4A). If the cre gene is expressed in ES cells or fertilized eggs obtained from Creneo mice, recombination should occur in the monitor plasmid leading to deletion of the bsr gene and expression of NLSLacZ.
Figure 4.
Recombination activity in Creneo ES cells and pre-implantation embryos from Creneo mice. (A) Schematic diagram of the recombination monitor plasmid, pCAG71bsr66NZ. Before recombination, the bsr gene is expressed but the NLSLacZ gene is not. Cre-mediated recombination excises the bsr gene, resulting in the expression of the NLSLacZ gene. (B) Cre activity in Creneo ES clones. Creneo-e (upper) and Creneo-f (lower) ES clones (1 × 107 cells/ 0.8 ml in PBS) were electroporated with 20 µg of pCAG71bsr66NZ at 200 V and 960 µF and plated onto gelatin-coated dishes. After 48 h the cells were stained with X-gal. (C) Cre activity in pre-implantation embryos from Creneo-e mice. Fertilized eggs from superovulated females mated with F1 from superovulated females mated with F1 Creneo-e males were microinjected with circular pCAG71bsr66NZ plasmid, cultured for 48 (left) or 72 h (right), and stained with X-gal. Since the β-gal protein was fused with the NLS, the nuclei of the cells were strongly stained.
We introduced the monitor plasmid in its circular form into the Creneo-e and -f ES clones and TT2 ES cells as a control, and stained the cells with X-gal 48 h after the electroporation. Creneo clones (Fig. 4B) but not TT2 cells were stained with X-gal (data not shown). Fertilized Creneo eggs microinjected with the monitor plasmid were cultured and examined for NLSLacZ expression. As shown in Figure 4C and Table 2, ∼40% of the Creneo embryos were stained, whereas normal embryos were never stained. This is consistent with the hypothesis that half the embryos should carry the integrated cre gene. Thus, the cre gene integration is stable even if the cre gene is expressed under a strong promoter.
Table 2. Cre-mediated β-galactosidase activation by microinjection of the monitor plasmid.
| No. of eggs injected | No. of eggs stained | |
|---|---|---|
| Creneo eggs stained after 48 h | 36 | 13 (36%) |
| Creneo eggs stained after 72 h | 38 | 17 (45%) |
| Normal eggs stained after 72 h | 77 | 0 (0%) |
DISCUSSION
We have shown here that the combination of the LE/RE mutant lox and lox2272 showed high recombination efficiency and the resulting recombined structure was stable. Using this combination, the cre gene itself can be inserted successfully and transmitted stably through the germ line. The targeting frequency of the Creneo cassette was lower than that of the NLSLacZ cassette (11.1 versus 26%). At present, it is not clear whether the difference is statistically significant, because only 36 clones were examined in the experiment of targeted integration of the Creneo cassette. As far as we know, targeted integration of the cre gene using the Cre-lox system has not been reported. This should be quite useful for production of various Cre mice.
We found that the targeting frequencies with lox2272 are higher than those with lox511. Kolb (16) also performed targeted gene replacement using lox511-loxP or lox2272-loxP. Although the recombination frequencies in the two combinations were not exactly evaluated, better results were obtained with lox2272 than with lox511. Kolb found that lox511 was recombined with loxP at a low frequency, and proposed that this might be the reason for the lower efficiency in the lox511-loxP combination. However, in our result, pP-5F2 showed better efficiency than pP-511 in spite of the identical recombination product with the two plasmids (see Fig. 1B), indicating that the excisive reaction between lox511 and loxP is not the main cause for the lower efficiency with pP-511. Lee and Saito (18) mentioned that lox2272 possibly has higher efficiency of recombination than lox511. Our result in which the plasmids pP-5F2 and p2272-5F2 showed comparable efficiencies to those of the lox2272-plasmids (see Fig. 2A) indicates that the observed difference between lox511 and lox2272 may be due to the higher recombination efficiency of lox2272 rather than lox511.
The targeting frequencies in the combination with heterospecific lox sites are 2.5–4 times higher than the frequencies of targeted integration with the use of p66-NO, single recombination between lox66 and lox71. This suggests that re-excision may occur between the LE+RE mutant lox and wild-type loxP on the recombination product with p66-NO even at a low efficiency, but rarely occurs between heterospecific lox sites in cassette exchange recombination. In addition, the two exchanged molecules, genomic DNA and plasmid DNA, may be rapidly separated, and therefore re-exchange between these molecules rarely occurs even when the loxP site is used.
The results in Figure 2B indicate that the amount of 10 µg of the Cre-expression plasmid is enough for efficient recombination between heterospecific lox sites. In our preliminary experiments, the use of 5 µg of the Cre-expression plasmid still showed high recombination efficiency (30–40%) with the plasmid p66-2272 and 5F2-23 line (data not shown). Since the optimal amount of the plasmids in the single recombination between lox66 and lox71 was 20 µg of each (11), the kinetics of the recombination in the cassette exchange recombination with heterospesific lox sites seems to be different from that in the single recombination with the LE/RE mutant lox sites. Bethke and Sauer (12) also reported a similar observation, the lower requirement for Cre recombinase in the double lox replacement system with lox511 than the single-crossover integration system using wild-type loxP. Although the reason is not clear, the stability of the recombination product may be related to the optimum conditions.
Targeted integration of the cre gene using the Cre-lox system has not previously been reported. This system is useful for the production of various Cre mice. Gene targeting or gene trapping with the neo or β-geo cassette flanked by lox71 and lox2272 can be performed as a first step, and then the cassette can be replaced by the cre gene flanked by lox66 and lox2272 through Cre-mediated recombination. Since the inserted cre gene is driven by the endogenous promoter, the expression of the cre gene would be exactly controlled in a spatiotemporal manner. The combination of the LE/RE mutant and lox2272 will be a powerful tool in genetic manipulation in ES cells including chromosomal rearrangement or integration of yeast or bacterial artificial chromosomes.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank Ms Y. Kiyonaga, H. Niizato, Y. Soejima, R. Minato and Y. Tsuruta for technical assistance. This work was supported by grants from the Ministry of Education, Science, Culture and Sports of Japan, by a grant from the Osaka Foundation for Promotion of Clinical Immunology, and a grant from the Science and Technology Agency.
REFERENCES
- 1.Sauer B. and Henderson,N. (1988) Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc. Natl Acad. Sci. USA, 85, 5166–5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gu H., Zou,Y.R. and Rajewsky,K. (1993) Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell, 73, 1155–1164. [DOI] [PubMed] [Google Scholar]
- 3.Araki K., Imaizumi,T., Okuyama,K., Oike,Y. and Yamamura,K. (1997) Targeted integration of DNA using mutant lox sites in embryonic stem cells. J. Biochem. (Tokyo), 122, 977–982.9443813 [Google Scholar]
- 4.Lakso M., Sauer,B., Mosinger,B.,Jr, Lee,E.J., Manning,R.W., Yu,S.H., Mulder,K.L. and Westphal,H. (1992) Targeted oncogene activation by site-specific recombination in transgenic mice. Proc. Natl Acad. Sci. USA, 89, 6232–6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rossant J. and Nagy,A. (1995) Genome engineering: the new mouse genetics. Nature Med., 1, 592–594. [DOI] [PubMed] [Google Scholar]
- 6.Hoess R.H., Ziese,M. and Sternberg,N. (1982) P1 site-specific recombination: nucleotide sequence of the recombining sites. Proc. Natl Acad. Sci. USA, 79, 3398–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee L. and Sadowski,P.D. (2001) Directional resolution of synthetic Holliday structures by the Cre recombinase. J. Biol. Chem., 276, 31092–31098. [DOI] [PubMed] [Google Scholar]
- 8.Dragatsis I. and Zeitlin,S. (2001) A method for the generation of conditional gene repair mutations in mice. Nucleic Acids Res., 29, e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sauer B. (1998) Inducible gene targeting in mice using the Cre/lox system. Methods, 14, 381–392. [DOI] [PubMed] [Google Scholar]
- 10.Fukushige S. and Sauer,B. (1992) Genomic targeting with a positive-selection lox integration vector allows highly reproducible gene expression in mammalian cells. Proc. Natl Acad. Sci. USA, 89, 7905–7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Araki K., Araki,M. and Yamamura,K. (1997) Targeted integration of DNA using mutant lox sites in embryonic stem cells. Nucleic Acids Res., 25, 868–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bethke B. and Sauer,B. (1997) Segmental genomic replacement by Cre-mediated recombination: genotoxic stress activation of the p53 promoter in single-copy transformants. Nucleic Acids Res., 25, 2828–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bouhassira E.E., Westerman,K. and Leboulch,P. (1997) Transcriptional behavior of LCR enhancer elements integrated at the same chromosomal locus by recombinase-mediated cassette exchange. Blood, 90, 3332–3344. [PubMed] [Google Scholar]
- 14.Soukharev S., Miller,J.L. and Sauer,B. (1999) Segmental genomic replacement in embryonic stem cells by double lox targeting. Nucleic Acids Res., 27, e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng Y.Q., Seibler,J., Alami,R., Eisen,A., Westerman,K.A., Leboulch,P., Fiering,S. and Bouhassira,E.E. (1999) Site-specific chromosomal integration in mammalian cells: highly efficient CRE recombinase-mediated cassette exchange. J. Mol. Biol., 292, 779–785. [DOI] [PubMed] [Google Scholar]
- 16.Kolb A.F. (2001) Selection-marker-free modification of the murine β-casein gene using a lox2272 [correction of lox2722] site. Anal. Biochem., 290, 260–271. [DOI] [PubMed] [Google Scholar]
- 17.Hoess R.H., Wierzbicki,A. and Abremski,K. (1986) The role of the loxP spacer region in P1 site-specific recombination. Nucleic Acids Res., 14, 2287–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee G. and Saito,I. (1998) Role of nucleotide sequences of loxP spacer region in Cre-mediated recombination. Gene, 216, 55–65. [DOI] [PubMed] [Google Scholar]
- 19.Yagi T., Tokunaga,T., Furuta,Y., Nada,S., Yoshida,M., Tsukada,T., Saga,Y., Takeda,N., Ikawa,Y. and Aizawa,S. (1993) A novel ES cell line, TT2, with high germline-differentiating potency. Anal. Biochem., 214, 70–76. [DOI] [PubMed] [Google Scholar]
- 20.Niwa H., Araki,K., Kimura,S., Taniguchi,S., Wakasugi,S. and Yamamura,K. (1993) An efficient gene-trap method using poly A trap vectors and characterization of gene-trap events. J. Biochem. (Tokyo), 113, 343–349. [DOI] [PubMed] [Google Scholar]
- 21.Gossler A. and Zachgo,J. (1993) Gene and enhancer trap screens in ES cell chimeras. In Joyner,A.L. (ed.), Gene Targeting–A Practical Approach. IRL Press, Oxford, UK, pp. 181–214.
- 22.Hogan B., Beddington,R., Costantini,F. and Lacy,E. (1994) Manipulating The Mouse Embryo. A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 23.Araki K., Araki,M., Miyazaki,J. and Vassalli,P. (1995) Site-specific recombination of a transgene in fertilized eggs by transient expression of Cre recombinase. Proc. Natl Acad. Sci. USA, 92, 160–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niwa H., Yamamura,K. and Miyazaki,J. (1991) Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene, 108, 193–199. [DOI] [PubMed] [Google Scholar]