Abstract
By different approaches, we characterized the birnavirus blotched snakehead virus (BSNV). The sequence of genomic segment A revealed the presence of two open reading frames (ORFs): a large ORF with a 3,207-bp-long nucleotide sequence and a 417-nucleotide-long small ORF located within the N-terminal half of the large ORF, but in a different reading frame. The large ORF was found to encode a polyprotein cotranslationally processed by the viral protease VP4 to generate pVP2 (the VP2 precursor), a 71-amino-acid-long peptide ([X]), VP4, and VP3. The two cleavage sites at the [X]-VP4 and VP4-VP3 junctions were identified by N-terminal sequencing. We showed that the processing of pVP2 generated VP2 and several small peptides (amino acids [aa] 418 to 460, 461 to 467, 468 to 474, and 475 to 486). Two of these peptides (aa 418 to 460 and 475 to 486) were positively identified in the viral particles with 10 additional peptides derived from further processing of the peptide aa 418 to 460. The results suggest that VP4 cleaves multiple Pro-X-Ala↓Ala motifs, with the notable exception of the VP4-VP3 junction. Replacement of the members of the predicted VP4 catalytic dyad (Ser-692 and Lys-729) confirmed their indispensability in the polyprotein processing. The genomic segment B sequence revealed a single large ORF encoding a putative polymerase, VP1. Our results demonstrate that BSNV should be considered a new aquatic birnavirus species, slightly more related to IBDV than to IPNV.
The family Birnaviridae includes three genera: Aquabirnavirus with the infectious pancreatic necrosis virus (IPNV) species, Avibirnavirus with the infectious bursal disease virus (IBDV) species, and Entomobirnavirus with the Drosophila X virus (DXV) species. Birnavirus particles are single-shelled unenveloped viruses with T=13 icosahedral capsids and are about 60 nm in diameter. VP2 and VP3 form the outer and inner layers, respectively, of the virions, which contain several VP1 molecules and the bisegmented double-stranded RNA genome (5, 13, 14, 27). Both genomic segments A and B of DXV and of a large number of IPNV and IBDV strains have been cloned and sequenced (1, 3, 4, 6, 7, 9-11, 15-17, 19, 21, 22, 24, 28-31, 34-36). Segment B codes for a putative RNA-dependent RNA polymerase, VP1. Segment A contains two overlapping reading frames (ORFs). In IPNV and IBDV, the smallest ORF is 5′ proximal and encodes VP5, a nonstructural polypeptide, whereas for DXV, the small ORF resides in the 3′-half of the segment. The large ORF encodes a 110-kDa polyprotein (NH2-pVP2-VP4-VP3-COOH). The polyprotein is cotranslationally processed through the proteolytic activity of VP4 to generate pVP2, VP4, and VP3. Cleavage sites at pVP2-VP4 and VP4-VP3 junctions have been identified for IPNV, IBDV, and DXV (11, 26, 32, 33). For IBDV, the processing of pVP2 (amino acids [aa] 1 to 512) generates VP2 and four small peptides (aa 442 to 487 [M. Skinner, personal communication], 488 to 494, 495 to 501, and 502 to 512) (12). At least three of these peptides (aa 442 to 487, 488 to 494, and 502 to 512) are associated with the viral particles (12). This maturation cleavage process of pVP2 requires assembly of viral capsids (8). The IBDV and IPNV VP4 proteases have been shown to use a serine-lysine catalytic dyad to control the processing of the polyprotein (2, 26, 32).
The blotched snakehead virus (BSNV) was isolated from a cell line derived from the blotched snakehead fish (Channa lucius). Although BSNV has been proposed to belong to the family Birnaviridae on the basis of biochemical characteristics, cross-neutralization assays have established the serological distinctness of BSNV from IPNV (20). In this study, we further characterized BSNV.
Molecular cloning and sequence of the BSNV genomic segments A and B.
The BSNV used in this study (kindly provided by W. Starkey, Stirling University, Stirling, United Kingdom) was propagated onto a cell line derived from Ophicephalus striatus by W. Wattanavijarn (Veterinary Faculty, Chulalongkorn University, Bangkok, Thailand) and provided by P. de Kinkelin (Institut National de la Recherche Agronomique, Jouy-en-Josas, France). Once the cytopathic effect was complete, virus was purified by CsCl gradient centrifugation as previously described (12). The viral RNA was extracted following sodium dodecyl sulfate (SDS)-proteinase K treatment and recovered following phenol-chloroform extraction and ethanol precipitation. For the first-strand cDNA synthesis, the RNA was denatured with hydroxymethyl mercury and incubated with the primers 5′-ACACTACCAGCAGGTCTCTATGCACTGAACGGA and 5′-GTTCTTGAGGAGCTCTGGGTTTGGGATCAGCTC, the sequences of which were derived from a consensus between the VP2 nucleotide sequences of IPNV, IBDV, and DXV. The viral RNA was reverse transcribed, and DNA amplification was carried out with rtTh (Perkin-Elmer) by standard techniques and procedures. A 680-bp PCR product was cloned and sequenced. Comparison to known sequences in data banks was carried out with BLAST programs. Significant homology was detected at the amino acid level between the encoded protein and the VP2 of birnaviruses sequenced so far (data not shown). By using specific primers, the complete coding region of segment A was then recovered by reverse transcription-PCR (RT-PCR) as eight overlapping cDNA clones. Additionally, the 5′ and 3′ ends of the segment were recovered by 5′ rapid amplification of cDNA ends (RACE) with specific primers. All of these clones were sequenced to ascertain that no artifactual sequences were added during the amplification process. Compilation of the sequencing data (EMBL database accession no. AJ459382) allowed the determination of the sequence of the BSNV genomic segment A containing two overlapping ORFs on the same strand. The large ORF of 1,069 codons is preceded by a 165-nucleotide 5′-untranslated region and is capable of encoding a long polypeptide and thus was designated as the polyprotein ORF. The small ORF of 139 codons started at nucleotide 452.
Several clones specific to segment B were also isolated. The cloning of the complete ORF was then carried out with specific primers. A compilation of the sequence data obtained from seven independent clones allowed the identification of a unique ORF (EMBL database accession no. AJ459383). This ORF encodes an 867-aa-long protein that shares homologies with other birnavirus VP1 polymerases.
N-terminus determination of two processed products of the polyprotein, VP4 and VP3.
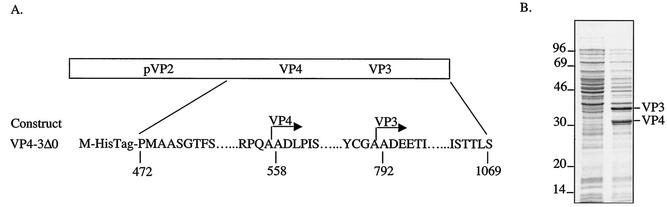
Since we previously observed that the IBDV and IPNV VP4 proteases are active in Escherichia coli (26, 32), we chose this expression system to determine the N termini of VP4 and VP3, which are derived from BSNV polyprotein processing. For this purpose, we engineered into the pET-28b vector (Novagen) a VP4-3Δ0 gene construct encoding a 5′-truncated form of the polyprotein, overlapping all its putative primary cleavage sites (Fig. 1A). The VP4-3Δ0 polypeptide was expressed in E. coli by a procedure previously described (26), and bacterial extracts were submitted to SDS-polyacrylamide gel electrophoresis (PAGE) analysis according to the method of Laemmli (25) (Fig. 1B). Two proteins were specifically induced, suggesting that cleavages occurred efficiently on the VP4-3Δ0 polypeptide. Direct sequence analyses of their first 7 aa were carried out as described previously (32), and N termini were determined as Ala-Asp-Leu-Pro-Ile-Ser-Leu and Ala-Asp-Glu-Glu-Thr-Ile-Glu. The resulting amino acid sequences were identical to the polyprotein sequence starting at Ala-558 (N terminus of VP4) and Ala-792 (N terminus of VP3), respectively, showing that cleavages occurred between the two alanine dipeptides located at positions 557 to 558 and 792 to 793. On the purified virus, we found that the N termini of VP3 and VP2 were blocked (data not shown).
FIG. 1.
Identification of the N termini of the VP3 and VP4 proteins. (A) Schematic representation of the VP4-3Δ0 construct expressed in E. coli. The numbers indicate the amino acid position in the polyprotein, and the horizontal arrows indicate the N termini of VP3 and VP4. (B) SDS-PAGE analysis of the bacterial extracts following (+) or not following (−) induction with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). The two induced bands were excised and submitted to seven Edman degradation cycles.
The pVP2 processing generates peptides present on the virus particles.
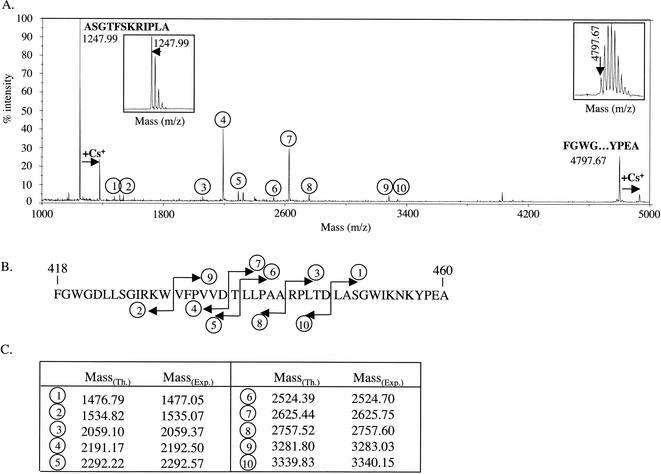
We previously showed that the processing of the pVP2 of IBDV generated VP2 and four small peptides associated with the viral particles and derived from the C-terminal domain of pVP2 (12). The processing of the BSNV pVP2 precursor was analyzed by carrying out an N-terminal sequencing of the purified virus. Two sequences were identified: NH2-FGWGDLLSGIRKWVFPVVDTLLPAA and NH2-AXXTFXKRIPLA. The first peptide started at Phe-418 of the polyprotein, and the second started at Ala-475. To further demonstrate the presence of these peptides on the virions, mass spectrometry analysis of the viral particles was carried out as previously described (12). A peptide with an [M+H]+ mass of 4,797.67 Da was indeed detected (Fig. 2A). No other peptide with the same range of mass was detected. This [M+H]+ mass fitted well the theoretical mass of a peptide extending from Phe-418 to Ala-460 (4,797.59 Da). A second peptide with an [M+H]+ mass of 1,247.99 Da was also detected. This [M+H]+ mass fitted well the theoretical mass of the peptide extending from Ala-475 to Ala-486 (1,247.71 Da) which was identified by N-terminal sequencing. Ten additional peptides were identified by mass spectrometry in the mass range between 1,500 and 3,400 Da (Fig. 2A). All of them appeared to be the cleaved products of the peptide (aa 418 to 460), since we identified, among the 10 peptides, five peptide couples for which the sum of the masses corresponded to the mass of the peptide (aa 418 to 460) minus the mass of a water molecule (Fig. 2B and C). Analysis of these five cleavage sites suggests that the specificity of the endopeptidase involved in this process is very broad and that a cellular protease instead of VP4 might be associated with these cleavage events. On the viral particles, we did not detect any products possibly derived from the 14-aa domain located between the peptides of aa 418 to 460 and 475 to 486 by N-terminal sequencing nor by mass spectrometry. However, we believe that the presence of two peptides (each 7 aa long) derived from this domain on virus particles cannot be ruled out. From these results, we assumed that pVP2 and VP2 extend from aa 1 to 486 and 1 to 417, respectively. Thus, a 71-aa-long domain, which we named [X], was defined between the C terminus of pVP2 and the N terminus of VP4 located on Ala-558.
FIG. 2.
Characterization of peptides present in BSNV particles. (A) Mass spectrometry analysis of BSNV particles. Two main signals were identified at mass/charge (m/z) ratios of 1,247.99 and 4,797.67. Magnified signals showing the isotopic pattern are inserted. Two Cs+ adducts were identified for these peptides. Ten additional signals (circles 1 to 10) were identified on the mass/charge window ranging from 1,000 to 5,000. (B) Proposed assignment for the 10 peaks as cleaved products of the peptide (aa 418 to 460). (C) Comparison of the experimental (Exp.) and predicted (theoretical [Th.]) masses of the 10 peptides.
Primary processing of the BSNV polyprotein.
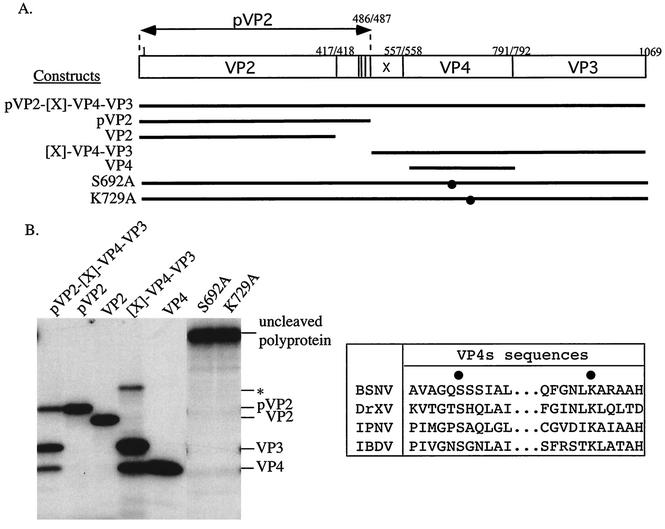
To analyze the primary processing of the BSNV polyprotein, pVP2-[X]-VP4-VP3, we engineered a complete cDNA clone encoding the complete polypeptide and constructs driving the expression of pVP2 (aa 1 to 486), VP2 (aa 1 to 417), VP4 (aa 558 to 792), and [X]-VP4-VP3 (aa 487 to 1069) (Fig. 3A). The complete polyprotein ORF was amplified by RT-PCR and cloned into the BamHI site of pSP73 (Promega) under the control of the T7 promoter to generate pSP73-BSNA. To obtain the reading frames of pVP2, VP2, [X]-VP4-VP3, and VP4 under the control of the T7 promoter, oligonucleotides were designed to introduce when necessary an initiation codon and a stop codon upstream and downstream of the reading frame, respectively. The corresponding RT-PCR products were cloned into pcDNA3 (Invitrogen) or pET-28b+. The in vitro processing of the different engineered constructs was then analyzed with the TNT coupled transcription/translation system (Promega) (Fig. 3B). Processing of the full-length BSNV polyprotein yielded three main bands, which were found to comigrate with pVP2, VP4, and VP3. Thus, the [X] domain was not evidenced as a C- or N-terminal extension of pVP2 or VP4, respectively. (The X peptide could not be visualized because it lacks internal methionine.) We conclude that [X] was cleaved on both of its sides during the primary polyprotein processing. As shown previously with IPNV and IBDV (23, 26, 32), BSNV pVP2 was not further processed to VP2 when the polyprotein was expressed in vitro. A faint band of about 65 kDa was present when [X]-VP4-VP3 was expressed. To determine if this band corresponded to the uncleaved form, [X]-VP4-VP3, we engineered a mutation (S692A) that inactivated the VP4 catalytic site (defined below) in the construct driving its expression. The electrophoretic mobility of the 65-kDa band was compared with those obtained with the catalytically inactive construct. The 65-kDa band had a higher mobility than the mutated construct (data not shown), suggesting that this band corresponded to an internal initiation inside the VP4 reading frame.
FIG. 3.
Assignment of the BSNV polyprotein processing products and of the VP4 protease active site. (A) Schematic representation of the BSNA polyprotein and the derived constructs produced by expression of plasmids. The position of the mutated VP4 catalytic residues is indicated (•). (B) (Left panel) In vitro expression of the wild type (pVP2-[X]-VP4-VP3) and of the derived constructs was carried out with a rabbit reticulocyte expression system. Expression products were analyzed by SDS-PAGE (10% polyacrylamide), and gels were processed for autoradiography. The locations of the viral polypeptides are indicated on the right. The asterisk indicates a 65-kDa band. (Right panel) Comparison including the amino sequences of VP4 of IPNV, IBDV, and DXV (shown as DrXV). The active site residues (Ser-692 and Lys-729) of the BSNV VP4 are indicated (•).
The catalytic site of the BSNV VP4 protease.
An alignment of the domains surrounding the VP4 catalytic residues of IPNV, IBDV (2, 26, 32), DXV, and BSNV shows that members of the VP4 catalytic dyad are conserved in the BSNV VP4 sequence (Fig. 3B). To confirm their critical importance, the identified residues, serine 692 and lysine 729, were substituted for with alanine by site-directed mutagenesis with Pfu DNA polymerase by using the QuickChange site-directed mutagenesis kit (Stratagene). The full-length segment A polyproteins carrying the mutations were expressed with an in vitro T7-driven expression system, and their processing was analyzed by PAGE (Fig. 3B). Processing of the wild-type protein yielded the expected cleavage products, with pVP2, VP3, and VP4 and without an uncleaved precursor. The replacement of serine 692 and lysine 729 completely inactivated the polyprotein processing. We thus further confirmed the existence of the Ser-Lys catalytic dyad of the birnavirus VP4 proteases.
Cleavage sites.
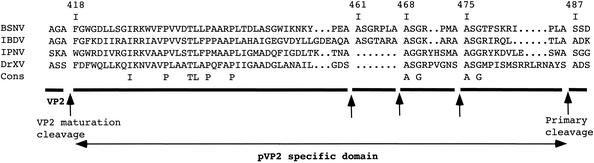
All cleavage sites could be defined by the Pro-X-Ala↓(Ala/Ser) motif with two exceptions: the VP4-VP3 junction is defined by the sequence Cys-Gly-Ala↓Ala, and the cleavage site between aa 417 and 418 is defined by the sequence Ala-Gly-Ala↓Phe. The BSNV cleavage motif thus defined was different from the cleavage sites of IPNV, IBDV, and DXV defined by the (Ser/Thr)-X-Ala↓(Ser/Ala)-Gly, (Thr/Ala)-X-Ala↓Ala, and (Ala/Gly)-X-Ser↓Ala motifs, respectively (11, 26, 32, 33). Concerning the most proximal cleavage site, which generates the mature BSNV VP2 (P1-P′1 position, 417-418) (Fig. 4), we noted that the P′1 homologs in each birnavirus are aromatic (and hydrophobic) residues that are never present in other cleavage sites. This observation raises the question of whether the VP4 protease is also involved in this final maturation cleavage.
FIG. 4.
The pVP2-specific domain of different birnavirus polyproteins. A sequence alignment with the IBDV, IPNV, and DXV pVP2-specific domains is shown. The alignment is anchored to the multiple cleavage sites identified on BSNV and IBDV or proposed for IPNV and DXV (shown as DrXV) (12). Residues conserved in the four sequences are indicated at the consensus sequence line (Cons).
BSNV genetic organization.
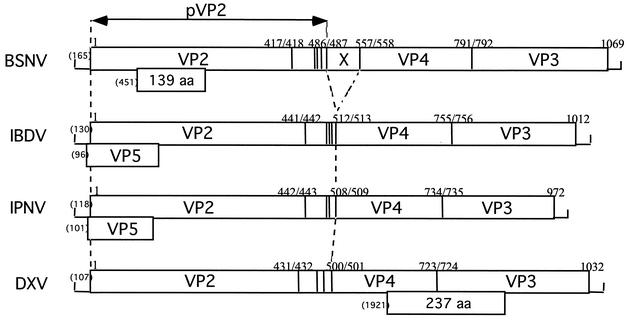
The genetic organization of BSNV genome segments A and B is similar to those of other birnaviruses (IPNV, IBDV, and DXV) (Fig. 5). However, we identified two notable exceptions. Whereas a small ORF overlaps the 5′ end of the polyprotein ORF, as in the cases of IPNV and IBDV or the VP4-VP3 junction in DXV, a BSNV small ORF is located inside the VP2-encoding sequence. In addition, the putative product of this ORF would have no (or weak) sequence similarities with those of the other three birnaviruses. In addition, the N-terminal amino acid sequence determination of VP4 and the identification of the structural peptides derived from the C terminus of pVP2 allowed the identification of a 71-aa-long peptide that we named [X], located between the pVP2 and VP4 coding regions. This peptide does not have any counterpart in other birnaviruses.
FIG. 5.
Schematic representation of the gene arrangement of genome segment A of BSNV and comparison with its IBDV, IPNV, and DXV homologs. The polyprotein BSNV ORF encompasses most of the genome segment and contains the 5′-pVP2-[X]-VP4-VP3 sequence. The cleavage sites of the protease are indicated by vertical lines, and their locations are given by the P1/P′1 amino acid number. The locations and sizes of the small ORFs are indicated below the polyprotein ORFs. Numbers in parentheses at the 5′ ends indicate the first nucleotide involved in the initiation codon.
Protein sequence comparison between BSNV and other birnaviruses.
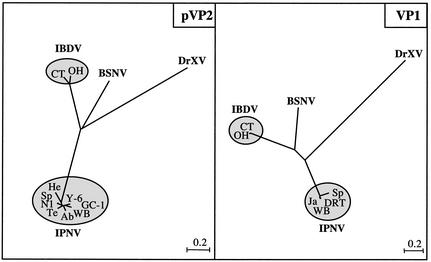
Pairwise alignment of the polymerase and the polyprotein sequence with those of other birnaviruses revealed only 18 to 49% identity, depending on the protein analyzed. As previously mentioned, between IPNV, IBDV, and DXV (11, 13), there is still very little sequence identity between the VP4 protease sequences. Even the residues surrounding its catalytic site are not conserved, with the exception of the GxS motif, a signature of serine hydrolases. We believe that the VP4 primary sequence drift reflects different strategies of birnaviruses to regulate their protease activity. Between the two capsid proteins VP2 and VP3, the VP2 sequence is the most conserved, whereas this protein is external and submitted to immune response pressure. However, the central region of VP2 between residues 185 and 340 is highly divergent, a domain that has been proposed to represent the group- and type-specific antigenic sites (1, 17, 28). The BSNV pVP2-specific domain (from residues 418 to 486) seems to be more related to IBDV than to other birnaviruses, since, like IBDV, four peptides were delineated as processed products of this domain (Fig. 4). The VP1 polymerase sequence was compared with its birnavirus homologs. Even if several polymerase motifs were conserved in the three sequences, the GDD motif was absent in the BSNV VP1 sequence, as previously noted with its IPNV and DXV homologs (10, 16, 34). Phylogenetic trees were constructed with the polypeptides derived from the pVP2 and the polymerase reading frames of DXV, two strains of IBDV (belonging to the serotypes 1 and 2), and strains representative of the different genogroups of aquatic birnaviruses (3) (Fig. 6). The branching structure of the two phylogenetic trees in conjunction with the differences in the genetic organization and protease specificity strongly suggests that BSNV is a new birnavirus species, slightly more related to IBDV than to IPNV and DXV. We propose that it could be considered as representative of a new genus of the family Birnaviridae rather than belonging to the genus defined by the IPNV species.
FIG. 6.
Phylogenetic analysis of the main birnavirus proteins pVP2 and VP1. The analysis was carried out with the Clustal W program (18) and drawn with the njplot software. The nucleotide sequence accession numbers are as follows: IPNV strains, West Buxton (WB), AF342727 and AF078669; Sp, AF342728 and P22174; He, AF342730; N1, P22495; Te, AF342731; Ab, AF342729; GC-1, AY064396; Y-6, AB006783; Ja, M58756; DRT, D26527; and IBDV strains, CT (serotype 1), U56907 and M58757; OH (serotype 2), U30819 and P27276; and DrXV, U60650 and AF196645. Estimation of branch lengths was based on uncorrected amino acid substitutions and on distances corrected for multiple substitutions (PAM distances): 1 U =100 × PAM.
Acknowledgments
We thank W. Starkey (Stirling University), who provided the BSNV virus; P. de Kinkelin (INRA, Jouy-en-Josas, France) for the cell line derived from Ophicephalus striatus; and Jean Lepault and Michel Bremont for critical reading of the manuscript.
Footnotes
This work is dedicated to the memory of Ove Noren.
REFERENCES
- 1.Bayliss, C. D., U. Spies, K. Shaw, R. W. Peters, A. Papageorgiou, H. Muller, and M. E. Boursnell. 1990. A comparison of the sequences of segment A of four infectious bursal disease virus strains and identification of a variable region in VP2. J. Gen. Virol. 71:1303-1312. [DOI] [PubMed] [Google Scholar]
- 2.Birghan, C., E. Mundt, and A. E. Gorbalenya. 2000. A non-canonical lon proteinase lacking the ATPase domain employs the Ser-Lys catalytic dyad to exercise broad control over the life cycle of a double-stranded RNA virus. EMBO J. 19:114-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blake, S., J.-Y. Ma, D. A. Caporale, S. Jairath, and B. L. Nicholson. 2001. Phylogenetic relationships of aquatic birnaviruses based on deduced amino acid sequences of genome segment A cDNA. Dis. Aquat. Org. 45:89-102. [DOI] [PubMed] [Google Scholar]
- 4.Boot, H. J., A. A. H. M. ter Huurne, A. J. W. Hoekman, B. P. H. Peeters, and A. L. J. Gielkens. 2000. Rescue of very virulent and mosaic infectious bursal disease virus from cloned cDNA: VP2 is not the sole determinant of the very virulent phenotype. J. Virol. 74:6701-6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Böttcher, B., N. A. Kiselev, V. Y. Stel'Mashchuk, N. A. Perevozchikova, A. V. Borisov, and R. A. Crowther. 1997. Three-dimensional structure of infectious bursal disease virus determined by electron cryomicroscopy. J. Virol. 71:325-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandt, M., K. Yao, M. Liu, R. A. Heckert, and V. N. Vakharia. 2001. Molecular determinants of virulence, cell tropism, and pathogenic phenotype of infectious bursal disease virus. J. Virol. 75:11974-11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, M. D., and M. A. Skinner. 1996. Coding sequences of both genome segments of a European "very virulent' infectious bursal disease virus. Virus Res. 40:1-15. [DOI] [PubMed] [Google Scholar]
- 8.Chevalier, C., J. Lepault, I. Erk, B. Da Costa, and B. Delmas. 2002. The maturation process of pVP2 requires assembly of infectious bursal disease virus capsids. J. Virol. 76:2384-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung, H. K., S. H. Lee, H. H. Lee, Y. C. Ha, D. S. Lee, and Y. S. Kim. 1994. Nucleotide sequence analysis of the VP2-NS-VP3 genes of infectious pancreatic necrosis virus DRT strain. Mol. Cells 4:349-354. [Google Scholar]
- 10.Chung, H. K., S. H. Lee, S. Y. Kim, and H. H. Lee. 1994. Nucleotide sequence analysis of the RNA-dependent RNA polymerase gene of infectious pancreatic necrosis virus DRT strain. J. Microbiol. Biotechnol. 4:264-269. [Google Scholar]
- 11.Chung, H. K., S. Kordyban, L. Cameron, and P. Dobos. 1996. Sequence analysis of the bicistronic Drosophila X virus genome segment A and its encoded polypeptides. Virology 225:359-368. [DOI] [PubMed] [Google Scholar]
- 12.Da Costa, B., C. Chevalier, C. Henry, J.-C. Huet, S. Petit, J. Lepault, H. Boot, and B. Delmas. 2002. The capsid of infectious bursal disease virus contains several small peptides arising from the maturation process of pVP2. J. Virol. 76:2393-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dobos, P. 1995. The molecular biology of infectious pancreatic necrosis virus (IPNV). Annu. Rev. Fish Dis. 5:25-54. [Google Scholar]
- 14.Dobos, P., B. J. Hill, R. Hallett, D. T. C. Kells, H. Becht, and D. Teninges. 1979. Biophysical and biochemical characterization of five animal viruses with bisegmented double-stranded RNA genomes. J. Virol. 32:593-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duncan, R., and P. Dobos. 1986. The nucleotide sequence of infectious pancreatic necrosis virus (IPNV) dsRNA segment A reveals one large ORF encoding a precursor polyprotein. Nucleic Acids Res. 14:5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duncan, R., C. L. Mason, E. Nagy, J. A. Leong, and P. Dobos. 1991. Sequence analysis of infectious pancreatic necrosis virus genome segment B and its encoded VP1 protein: a putative RNA-dependent RNA polymerase lacking the Gly-Asp-Asp motif. Virology 181:541-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Havarstein, L. S., K. H. Kalland, K. E. Christie, and C. Endresen. 1990. Sequence of the large double-stranded RNA segment of the N1 strain of infectious pancreatic necrosis virus: a comparison with other Birnaviridae. J. Gen. Virol. 71:299-308. [DOI] [PubMed] [Google Scholar]
- 18.Higgins, D., J. Thompson, T. Gibson, J. D. Thompson, D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hudson, P. J., N. M. McKern, B. E. Power, and A. A. Azad. 1986. Genomic structure of the large RNA segment of infectious bursal disease virus. Nucleic Acids Res. 14:5001-5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.John, K. R., and R. H. Richards. 1999. Characteristics of a new birnavirus associated with a warm-water fish cell line. J. Gen. Virol. 80:2061-2065. [DOI] [PubMed] [Google Scholar]
- 21.Kibenge, F. S., D. J. Jackwood, and C. C. Mercado. 1990. Nucleotide sequence analysis of genome segment A of infectious bursal disease virus. J. Gen. Virol. 71:569-577. [DOI] [PubMed] [Google Scholar]
- 22.Kibenge, F. S., P. K. McKenna, and J. K. Dybing. 1991. Genome cloning and analysis of the large RNA segment (segment A) of a naturally avirulent serotype 2 infectious bursal disease virus. Virology 184:437-440. [DOI] [PubMed] [Google Scholar]
- 23.Kibenge, F. S., B. Qian, E. Nagy, J. R. Cleghorn, and D. Wadowska. 1999. Formation of virus-like particles when the polyprotein gene (segment A) of infectious bursal disease virus is expressed in insect cells. Can. J. Vet. Res. 63:49-55. [PMC free article] [PubMed] [Google Scholar]
- 24.Kordyban, S., G. Magyar, H. K. Chung, and P. Dobos. 1997. Incomplete dsRNA genomes in purified infectious pancreatic necrosis virus. Virology 239:62-70. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 26.Lejal, N., B. Da Costa, J.-C. Huet, and B. Delmas. 2000. Role of Ser-652 and Lys-692 in the protease activity of infectious bursal disease virus VP4 and identification of its substrate cleavage sites. J. Gen. Virol. 81:983-992. [DOI] [PubMed] [Google Scholar]
- 27.Leong, J. C., D. Brown, P. Dobos, F. S. B. Kibenge, J. E. Ludert, H. Müller, E. Mundt, and B. Nicholson. 2000. Family Birnaviridae, p. 481-490. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, D. J. McGeoch, J. Maniloff, M. A. Mayo, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Seventh Report of International Committee on the Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 28.Liao, L., and P. Dobos. 1995. Mapping of a serotype specific epitope of the major capsid protein VP2 of infectious pancreatic necrosis virus. Virology 209:684-687. [DOI] [PubMed] [Google Scholar]
- 29.Lim, B.-L., Y. Cao, T. Yu, and C.-W. Mo. 1999. Adaptation of very virulent infectious bursal disease virus to chicken embryonic fibroblasts by site-directed mutagenesis of residues 279 and 284 of viral coat protein VP2. J. Virol. 73:2854-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morgan, M. M., I. G. Macreadie, V. R. Harley, P. J. Hudson, and A. A. Azad. 1988. Sequence of the small double-stranded RNA genomic segment of infectious bursal disease virus and its deduced 90-kDa product. Virology 163:240-242. [DOI] [PubMed] [Google Scholar]
- 31.Mundt, E., and H. Muller. 1995. Complete nucleotide sequences of 5′- and 3′-noncoding regions of both genome segments of different strains of infectious bursal disease virus. Virology 209:10-18. [DOI] [PubMed] [Google Scholar]
- 32.Petit, S., N. Lejal, J.-C. Huet, and B. Delmas. 2000. Active residues and viral substrate cleavage sites of the protease of the birnavirus infectious pancreatic necrosis virus. J. Virol. 74:2057-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanchez, A. B., and J. F. Rodriguez. 1999. Proteolytic processing in infectious bursal disease virus: identification of the polyprotein cleavage sites by site-directed mutagenesis. Virology 262:190-199. [DOI] [PubMed] [Google Scholar]
- 34.Shwed, P. S., P. Dobos, L. A. Cameron, V. N. Vakharia, and R. Duncan. 2002. Birnavirus VP1 proteins form a distinct subgroup of RNA-dependent RNA polymerases lacking a GDD motif. Virology 296:241-250. [DOI] [PubMed] [Google Scholar]
- 35.Spies, U., H. Muller, and H. Becht. 1989. Nucleotide sequence of infectious bursal disease virus genome segment A delineates two major open reading frames. Nucleic Acids Res. 17:7982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yao, K., and V. N. Vakharia. 1998. Generation of infectious pancreatic necrosis virus from cloned cDNA. J. Virol. 72:8913-8920. [DOI] [PMC free article] [PubMed] [Google Scholar]