Abstract
Venezuelan equine encephalitis virus (VEEV) is an important human and equine pathogen in the Americas, with widespread reoccurring epidemics extending from South America to the southern United States. Most troubling, VEEV has been made into a weapon by several countries and is currently restricted by the Centers for Disease Control and Prevention as a potential biological warfare and terrorism agent. To facilitate the development of antiviral compounds, the structure of the nucleocapsid isolated from VEEV has been determined by electron cryomicroscopy and image reconstruction and represents the first three-dimensional structure of a nucleocapsid isolated from a single-stranded enveloped RNA virus. The isolated VEEV nucleocapsid undergoes significant reorganization relative to its structure within VEEV. However, the isolated nucleocapsid clearly exhibits T=4 icosahedral symmetry, and its characteristic nucleocapsid hexons and pentons are preserved. The diameter of the isolated nucleocapsid is ∼11.5% larger than that of the nucleocapsid within VEEV, with radial expansion being greatest near the hexons. Significantly, this is the first direct structural evidence showing that a simple enveloped virus undergoes large conformational changes during maturation, suggesting that the lipid bilayer and the transmembrane proteins of simple enveloped viruses provide the energy necessary to reorganize the nucleocapsid during maturation.
Venezuelan equine encephalitis virus (VEEV; family Togaviridae, genus Alphavirus) is a single-stranded enveloped RNA virus (10, 18, 31) that is transmitted between vertebrate hosts by infected mosquitoes (25). VEEV (and related western and eastern equine encephalitis viruses) is one of the leading causes of viral encephalitis in humans and horses in the Americas. Human infection is highly debilitating, and neurological disease develops in approximately 10% of human infections; the overall mortality rate is ∼0.5%, typically due to fatal encephalitis in children. VEEV was under active biological warfare and terrorist development in several countries and is currently restricted by the Centers for Disease Control and Prevention as a “select agent” with potential biological warfare and terrorist concerns. No antiviral drugs or widely available human vaccine exists to combat this pathogen. Thus, an understanding of the structural changes that accompany VEEV maturation may help in the development of novel antiviral agents that interfere with the assembly and disassembly of VEEV and other pathogenic enveloped viruses.
In addition to being a significant health threat, VEEV and related alphaviruses are model systems for studying enveloped virus structure, replication, and pathogenesis (4, 10, 15, 18, 19, 25, 31). Alphaviruses are simple T=4 icosahedral enveloped viruses (4, 15, 18, 19) whose genome encodes four nonstructural and three main structural proteins (25). In the mature virus, the dominant surface features are trimeric spikes consisting of the envelope proteins and arranged on a T=4 lattice (4, 8, 14, 18, 19). The capsid protein is arranged into hexons and pentons on a T=4 lattice that complements the organization of the envelope glycoproteins (4, 15, 18, 19). Before forming the mature virus, the capsid protein interacts with viral RNA to self-assemble nascent nucleocapsids in the cytosol of the cell (6, 23), while the envelope proteins are targeted to the plasma membrane. Specific interactions at the plasma membrane between the nucleocapsid and the E2 envelope protein (11, 27, 36) have been proposed to drive the budding of mature virus from an infected cell (26). The mature virus can then infect a host cell, after which fusion of the viral and cell membranes uncoats the virus and releases the nucleocapsid into the cytosol. Although the released nucleocapsid may exist as an intact structure in the cytosol, it must allow for efficient disassembly of its structure and the introduction of the viral genome into the cytosol. The molecular details of this cycle of assembly, budding, infection, uncoating, and genome release remain largely unknown. However, it is becoming clear that many viruses undergo significant conformational rearrangements during their life cycle, and direct structural evidence of capsid reorganization during maturation has been obtained for several nonenveloped icosahedral viruses (3, 12, 20, 24), enveloped nonicosahedral retroviruses (30), bacteriophages (2), and herpesvirus (16, 29).
We report here the first determination of the structure of the isolated nucleocapsid of a single-stranded enveloped RNA virus, VEEV, by electron cryomicroscopy (cryo-EM) and three-dimensional image reconstruction. Significantly, the isolated nucleocapsid of VEEV exhibits T=4 icosahedral symmetry similar to the nucleocapsid symmetry observed in the mature virus. However, the hexons and pentons are significantly reorganized in the isolated nucleocapsid structure relative to the nucleocapsid structure in the mature virus.
Purification of VEEV and nucleocapsids.
VEEV is an ideal model system for structural studies of enveloped RNA virus assembly and maturation. High concentrations (∼1012 to 1013 PFU ml−1) of VEEV were easily and reproducibly obtained as described previously (18). In order to perform the described experiments under biosafety level 2 conditions, attenuated vaccine strain TC-83 (13), derived from the Trinidad donkey strain of VEEV, was used throughout this work. Strain TC-83 and its parent differ by single amino acid changes in the nonstructural P4 and E1 proteins and five amino acid changes in the E2 protein (13); thus, the capsid and nucleocapsid structures are likely identical in these strains.
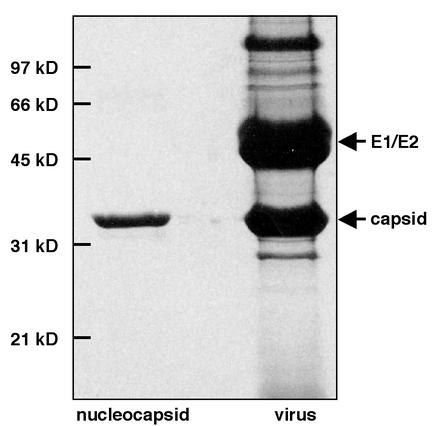
Homogeneous populations of isolated nucleocapsids were generated from purified virus particles as follows. Virus was suspended in 20 mM triethanolamine-100 mM NaCl and then incubated with Triton X-100 (2% [vol/vol]). Suspensions were applied to a 30 to 10% sucrose gradient in TEN buffer (10 mM Tris [pH 8.1], 100 mM NaCl, 1 mM EDTA) and centrifuged at 100,000 × g. Fractions containing purified nucleocapsids were pooled, concentrated on a 30% sucrose cushion in TEN buffer, and centrifuged at 110,000 × g. Purified nucleocapsids were suspended in TEN buffer for at least 24 h at 4°C. The only protein detected in preparations of purified isolated nucleocapsids was the VEEV capsid protein, as assessed by Coomassie blue staining (Fig. 1) and Western blotting (data not shown) of nucleocapsids separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis. The major bands observed after separation of the protein components of VEEV by SDS-polyacrylamide gel electrophoresis corresponded to the structural proteins of the virus (Fig. 1).
FIG. 1.
Protein compositions of purified isolated nucleocapsids and VEEV. Purified particles were denatured in SDS sample buffer, separated on a 12% polyacrylamide- SDS gel under reducing conditions, and visualized by Coomassie blue staining. The only band observed in the nucleocapsid sample corresponded to the capsid protein, while the major bands in the VEEV sample corresponded to the E1, E2, and capsid structural proteins. Protein identities were confirmed by Western blot analysis (data not shown). The minor band migrating above 97 kDa corresponded to the E2 protein, as confirmed by N-terminal sequencing, Western blot analysis, and mass spectroscopy (data not shown).
Image reconstruction of VEEV and isolated nucleocapsids.
Cryo-EM and three-dimensional image reconstruction of VEEV were done as described previously (18). Cryo-EM of both whole-virus and purified isolated nucleocapsids produced images suitable for three-dimensional computer-aided reconstructions (Fig. 2). Tobacco mosaic virus (TMV) was used as a magnification standard to accurately determine the sizes of the VEEV and isolated nucleocapsid structures. The J1 layer line of the Fourier transform of TMV images, located at 22.9 Å, was used to determine the exact magnification of the electron micrographs of the VEEV and isolated nucleocapsids.
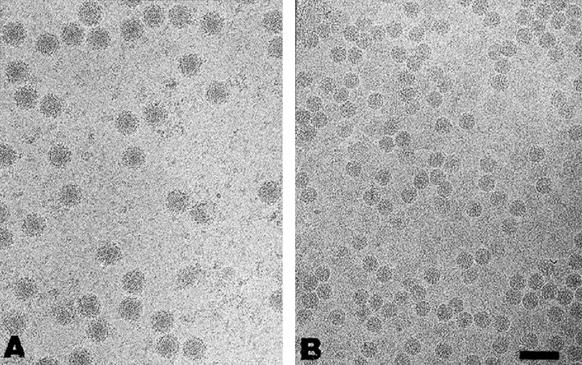
FIG. 2.
Representative micrographs from cryo-EM of purified particles suitable for three-dimensional image reconstruction. (A) Electron cryomicrograph of purified VEEV. The particles were predominantly spherical and measured ∼680 Å in diameter. A small number of virus particles (<1%) showed nonstandard conformations, in that they swelled larger than normal size (∼800 Å) or seemed to disintegrate spontaneously, releasing their nucleocapsids. These were not used in subsequent reconstructions and are typical of the purification process (17). (B) Electron cryomicrograph of purified isolated nucleocapsids from VEEV. The isolated nucleocapsid fields contained intact nucleocapsids in high concentrations and of uniform sizes and were devoid of contaminating whole virus. Images of the nucleocapsids demonstrate that the core of the virus is an approximately spherical dense body without clearly discernible features. Both VEEV and isolated nucleocapsids were embedded in vitreous ice and transferred to the cold stage of a JEOL 1200 electron cryomicroscope maintained at −164°C. Images were recorded at 100 keV. Scale bar, 100 nm.
Reconstruction of isolated nucleocapsids was performed by using hierarchical wavelet transformation and projection matching to determine particle orientations (21, 22). To prevent convergence to a local minimum, a 0.5° angular grid was used to generate the projections for matching. An assessment quality factor from the projection matching was used to retain only the best orientations for further refinement by the cross-common line method (19). Structural comparisons of whole-virus and isolated nucleocapsids were performed on reconstructions calculated at equivalent resolutions. Maps were contoured on the assumption of an average protein density of 1.325 g/cm3.
Structural plasticity of the nucleocapsid.
The isolated nucleocapsid measured ∼420 Å in diameter, ∼10% larger than the ∼384-Å diameter of the nucleocapsid determined from the three-dimensional reconstruction of infectious whole VEEV (18). These measurements are consistent with average particle diameters subsequently calculated from the reconstructions of the viral and isolated nucleocapsids. The structure of the isolated nucleocapsid clearly shows T=4 icosahedral symmetry (Fig. 3), in agreement with an earlier suggestion (7). However, the mass densities in the capsomeres of the isolated nucleocapsid are significantly rearranged relative to those in the capsomeres within the whole-virus nucleocapsid (Fig. 3). In the isolated nucleocapsid, capsid proteins are arranged into pentons and quasihexons, and this organization is a hallmark of nucleocapsid structure within mature alphaviruses (4, 15, 18, 19). The radial expansion observed in the isolated nucleocapsid occurs nonisotropically, with the hexons being pushed outward and the pentons being essentially unchanged relative to their radial positions in the whole-virus nucleocapsid (Fig. 3). This radial expansion of the hexons may allow for more rotational and lateral movements of the component capsid proteins within the isolated nucleocapsid.
FIG. 3.
Structure of VEEV nucleocapsids determined from image reconstructions of electron cryomicrographs. (A) Isosurface representation of an isolated nucleocapsid viewed along a threefold symmetry axis. (B) Isosurface representation of a mature-virus nucleocapsid viewed along a threefold symmetry axis. Images are color coded by radial depth, as shown in the linear scale bar. Isolated and mature-virus nucleocapsids were reconstructed to resolutions of 28 and 25 Å, respectively, as determined by the Fourier shell correlation coefficient, where a value of 0.5 was used to assign the resolution limit. Reconstructions were contoured on the assumption of an average protein density of 1.325 g/cm3.
In the isolated nucleocapsid, removal of the envelope proteins and lipid bilayer releases the nucleocapsid from these molecular constraints, thereby allowing it to freely expand. This expansion likely results in increased intermolecular distances between the capsid proteins that form the pentons and/or hexons (i.e., an expansion of the pentons and hexons) and/or increased distances between the pentons and the hexons that form the nucleocapsid structure. Increasing the distance between the capsid proteins likely destabilizes the isolated nucleocapsid, thus making its icosahedral symmetry less defined and its structure more plastic than those of the mature-virus nucleocapsid. However, the T=4 icosahedral symmetry observed in the isolated nucleocapsid implies that the capsid protein and viral RNA are sufficient to self-assemble into an icosahedral particle. Indeed, single-stranded RNA and purified capsid protein can self-assemble into nucleocapsid-like particles in vitro (28, 32, 34), although the structures of these particles are unknown.
The pentons retain their distinct shape in the isolated nucleocapsid, with their vertices clearly defined. Each vertex is postulated to be formed from the capsid C-terminal domain (CCD) (4). The pentons are oriented similarly in both isolated and mature-virus nucleocapsids, with their vertices directed toward the icosahedral threefold axis. The similar penton structures likely reflect the orientational constraints present in the nucleocapsid. In contrast, the hexons (which lie on a true twofold axis and adopt a quasisixfold structure) are significantly modified in the isolated nucleocapsid relative to the mature-virus nucleocapsid. Although six distinct projections (each corresponding to a CCD protein) are present in the isolated nucleocapsid, these projections are skewed from the hexon positions observed in the mature-virus nucleocapsid. In the isolated nucleocapsid, the hexon CCDs form three closely spaced projections, which are arranged on either side of a plane perpendicular to the twofold axis. This hexon skewing is similar to the “shear” dislocation observed during the maturation of herpesvirus (16) and many nonenveloped viruses, including bacteriophages and poliovirus (1, 5); in all cases, the shear occurs along an axis oblique to the icosahedral twofold axis. In addition, the density connecting the projections across this plane is narrowed relative to the density connecting the projections within this plane (Fig. 3). The CCD projections in the isolated nucleocapsid are displaced outward and above the density connecting the projections, resulting in the outer surface of the hexons adopting a trough or V-like structure. In contrast, the outer surface of the hexons in the mature-virus nucleocapsid appear relatively flat. The CCD projection that is directed toward the threefold axis in the mature-virus nucleocapsid appears to have shifted toward the pseudothreefold axis in the isolated nucleocapsid, leaving an apparent clearing around the threefold axis (Fig. 3). The hexon reorganization observed in the isolated nucleocapsid likely reflects the greater spatial freedom of the hexon capsid proteins in this nucleocapsid.
Conformational rearrangements between isolated and mature-virus nucleocapsids extend from the outer nucleocapsid surface to the interior of the nucleocapsid (Fig. 3 and 4). The base of the nucleocapsid below the CCD projections (green in Fig. 3; radial depth, ∼175 Å) is heavily fissured in the isolated nucleocapsid and relatively smooth in the mature-virus nucleocapsid, suggesting that the N-terminal region of the capsid protein undergoes numerous rearrangements upon removal of the envelope proteins and lipid bilayer. Of note are the 32-Å-diameter holes that appear at the base of the hexon centers, which are large enough to allow the passage of small proteins into the interior of the isolated nucleocapsid. Since isolated nucleocapsids are RNase sensitive, these holes may be portals through which the viral RNA genome is accessed (6; M. Lorinczi and S. J. Watowich, personal communication).
FIG. 4.
Structure of the inner surface of VEEV nucleocapsids determined from image reconstructions of electron cryomicrographs. (A) Isosurface representation of an isolated nucleocapsid viewed from the interior of the nucleocapsid along a threefold symmetry axis. (B) Isosurface representation of a mature-virus nucleocapsid viewed from the interior of the nucleocapsid along a threefold symmetry axis. Panels A and B are rotated 180° about a central horizontal axis from the views shown in Fig. 3A and B, respectively. (C) Cross-section of an isolated nucleocapsid. (D) Cross-section of a mature-virus nucleocapsid. Cross-sectional slices are ∼55 Å thick, cut perpendicular to the threefold axis, and in plane with a vertical fivefold axis. In all panels, the reconstructed electron density maps had the central disordered RNA region removed at a radial distance of 130 Å, thus exposing the inner surface of the nucleocapsid. Images are color coded by radial depth, as shown in the linear scale bar.
The inner surface of the isolated nucleocapsid (radial depth, ∼130 Å) is more knurled than the corresponding inner surface of the mature-virus nucleocapsid (Fig. 4A and B). Protuberances in the isolated nucleocapsid are more prominent, particularly in the region around the icosahedral fivefold axis, where five counterclockwise pinwheel ridges emanate. In contrast, the corresponding region in the mature-virus nucleocapsid displays a small star-like indentation and a smoother, uniform density at the inner surface. In addition, the isolated nucleocapsid has significantly less density around its inner surface threefold axis than does the mature-virus nucleocapsid. It appears that this region in the isolated nucleocapsid expands to expose holes which transverse the nucleocapsid thickness. Thus, the difference between isolated and mature-virus nucleocapsids can be viewed as a redistribution of density from the region around the icosahedral threefold axis to the region around the fivefold axis and suggests that the capsid protein and viral genome undergo large concerted rearrangements when the nucleocapsid is released from constraints imposed by the lipid bilayer.
Cross-sections through the mature-virus nucleocapsid (Fig. 4D) show a distinct angular hexagonal outline (characteristic of an icosahedral polyhedron) that delineates the inner surface of the nucleocapsid. In contrast, cross-sections through the isolated nucleocapsid (Fig. 4C) show its inner surface to have a more rounded shape. This distortion results from the nonisotropic expansion of the isolated nucleocapsid relative to the mature-virus nucleocapsid and the redistribution of density that occurs throughout the inner surface of the nucleocapsid. Interestingly, similar transitions from rounded to more angular particles have been observed during the maturation of nonenveloped bacteriophage P22 (35) and Nudaurelia capensis ω virus (3). However, for bacteriophage P22, this transition is associated with particle expansion, not compression. In addition, there is an obvious thickening of the capsid layer of the isolated nucleocapsid relative to the mature-virus nucleocapsid (Fig. 4C and D). The nucleocapsid shell, calculated from radial density plots, extends radially from 133 to 192 Å for the mature-virus nucleocapsid and 115 to 210 Å for the isolated nucleocapsid. The thickening and corresponding ∼64% increase in the shell volume of the isolated nucleocapsid relative to the mature-virus nucleocapsid likely result from a decrease in the packing density of the nucleocapsid lattice or the association of additional ordered viral RNA. Varying the contour level slightly above and below 1.325 g/cm3 did not significantly alter the observed structural differences between isolated and mature-virus nucleocapids.
In summary, we have provided the first structural insights into the disassembly mechanism of simple enveloped viruses. Removal of the envelope proteins and lipid bilayer from the enveloped virus likely mimics a metastable post-viral nucleocapsid state that exists following fusion between viral and cell endosomal membranes. Although significant rearrangements of the nucleocapsid occur following envelope protein and lipid bilayer removal, the isolated nucleocapsid clearly demonstrates that the capsid protein and viral RNA are sufficient for T=4 icosahedral symmetry. Relative to the mature-virus nucleocapsid, the isolated nucleocapsid has a less ordered, more rounded, and more expanded structure, skewed hexons, and density redistributed from the threefold to the fivefold axis. Intriguingly, these changes demonstrate that the lipid bilayer and possibly the envelope proteins provide the necessary energy to constrain and stabilize the nucleocapsid in a compressed state. Thus, removal of the lipid bilayer from the virus after its entry into a host cell releases an expanding metastable nucleocapsid into the cytosol of the cell, perhaps facilitating nucleocapsid disassembly and genome release by additional cellular factors (9, 33). In addition, similar nucleocapsid rearrangements likely occur during the maturation of newly assembled nucleocapsids and their subsequent budding as mature viruses. The rearrangements observed for VEEV are analogous to the conformational changes observed during nonenveloped virus maturation and disassembly, suggesting that diverse viruses have evolved similar strategies to regulate their maturation and thus survive the changing environments that attend their replication cycle.
Acknowledgments
We thank V. Popov, M. Kunkel, and M. Lorinczi for technical assistance; A. McGough for TMV samples; and R. Shope, R. E. Johnston, D. Brown, B. V. V. Prasad, S. Ludtke, and J. Brink for helpful discussions.
The work was supported by grants from the John Sealy Memorial Endowment Fund for Biomedical Research (to S.J.W.), the Defense Advanced Research Projects Agency and the Centers for Disease Control and Prevention (to R. Shope, S.J.W., and S.C.W.), the National Center for Macromolecular Imaging and the Robert Welch Foundation (to W.C.), the NIH (to A.P.), and the Sealy Center for Structural Biology.
REFERENCES
- 1.Belnap, D. M., D. J. Filman, B. L. Trus, N. Cheng, F. P. Booy, J. F. Conway, S. Curry, C. N. Hiremath, S. K. Tsang, A. C. Steven, and J. M. Hogle. 2000. Molecular tectonic model of virus structural transitions: the putative cell entry states of poliovirus. J. Virol. 74:1342-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butcher, S. J., T. Dokland, P. M. Ojala, D. H. Bamford, and S. D. Fuller. 1997. Intermediates in the assembly pathway of the double-stranded RNA virus φ6. EMBO J. 16:4477-4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Canady, M. A., M. Tihova, T. N. Hanzlik, J. E. Johnson, and M. Yeager. 2000. Large conformational changes in the maturation of a simple RNA virus, Nudaurelia capensis ω virus (NωV). J. Mol. Biol. 299:573-584. [DOI] [PubMed] [Google Scholar]
- 4.Cheng, R. H., R. J. Kuhn, N. H. Olson, M. G. Rossmann, H.-K. Choi, T. J. Smith, and T. S. Baker. 1995. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell 80:621-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conway, J. F., W. R. Wikoff, N. Cheng, R. L. Duda, R. W. Hendrix, J. E. Johnson, and A. C. Steven. 2001. Virus maturation involving large subunit rotations and local refolding. Science 292:744-748. [DOI] [PubMed] [Google Scholar]
- 6.Coombs, K., B. Brown, and D. T. Brown. 1984. Evidence for a change in capsid morphology during Sindbis virus envelopment. Virus Res. 1:297-302. [DOI] [PubMed] [Google Scholar]
- 7.Coombs, K., and D. Brown. 1987. Topological organization of Sindbis virus capsid protein in isolated nucleocapsids. Virus Res. 7:131-149. [DOI] [PubMed] [Google Scholar]
- 8.Fuller, S. D. 1987. The T=4 envelope of Sindbis virus is organized by interactions with a complementary T=3 capsid. Cell 48:923-934. [DOI] [PubMed] [Google Scholar]
- 9.Gaspar, L. P., A. F. Terezan, A. S. Pinheiro, D. Foguel, M. A. Rebello, and J. L. Silva. 2001. The metastable state of nucleocapsids of enveloped viruses as probed by high hydrostatic pressure. J. Biol. Chem. 276:7415-7421. [DOI] [PubMed] [Google Scholar]
- 10.Johnston, R. E., and C. J. Peters. 1996. Alphaviruses, p. 843-898. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven, New York, N.Y.
- 11.Kail, M., M. Hollingshead, W. Ansorge, R. Pepperkok, R. Frank, G. Griffiths, and D. Vaux. 1991. The cytoplasmic domain of alphavirus E2 glycoprotein contains a short linear recognition signal required for viral budding. EMBO J. 10:2343-2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.King, J., and W. Chiu. 1997. The procapsid-to-capsid transition in double-stranded DNA bacteriophages, p. 288-311. In W. Chiu, R. M. Burnett, and R. Garcia (ed.), Structural biology of viruses. Oxford University Press, New York, N.Y.
- 13.Kinney, R. M., B. J. B. Johnson, J. B. Welch, K. R. Tsuchiya, and D. W. Trent. 1989. The full-length nucleotide sequences of the virulent Trinidad donkey strain of Venezuelan equine encephalitis virus and its attenuated vaccine derivative, TC-83. Virology 170:19-30. [DOI] [PubMed] [Google Scholar]
- 14.Lescar, J., A. Roussel, M. W. Wien, J. Navaza, S. D. Fuller, G. Wengler, G. Wengler, and F. A. Rey. 2001. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105:137-148. [DOI] [PubMed] [Google Scholar]
- 15.Mancini, E. J., M. Clarke, B. E. Gowen, T. Rutten, and S. D. Fuller. 2000. Cryo-electron microscopy reveals the functional organization of an enveloped virus, Semliki Forest virus. Mol. Cell 5:255-266. [DOI] [PubMed] [Google Scholar]
- 16.Newcomb, W. W., F. L. Homa, D. R. Thomsen, F. P. Booy, B. L. Trus, A. C. Steven, J. V. Spencer, and J. C. Brown. 1996. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. J. Mol. Biol. 263:432-446. [DOI] [PubMed] [Google Scholar]
- 17.Parades, A. 1993. Ph.D.thesis. University of Texas, Austin.
- 18.Paredes, A., K. Alwell-Warda, S. C. Weaver, W. Chiu, and S. J. Watowich. 2001. Venezuelan equine encephalomyelitis virus structure and its divergence from Old World alphaviruses. J. Virol. 75:9532-9537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paredes, A. M., D. T. Brown, R. Rothnagal, W. Chiu, R. Schoepp, R. E. Johnston, and V. Prasad. 1993. Three-dimensional structure of a membrane-containing virus. Proc. Natl. Acad. Sci. USA 90:9095-9099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prasad, B. V., P. E. Prevelige, E. Marietta, R. O. Chen, D. Thomas, J. King, and W. Chiu. 1993. Three-dimensional transformation of capsids associated with genome packaging in a bacterial virus. J. Mol. Biol. 231:65-74. [DOI] [PubMed] [Google Scholar]
- 21.Saad, A., S. J. Ludtke, J. Jakana, F. J. Rixon, H. Tsuruta, and W. Chiu. 2001. Fourier amplitude decay of electron cryomicroscopic images of single particles and effects on structure determination. J. Struct. Biol. 133:32-42. [DOI] [PubMed] [Google Scholar]
- 22.Saad, A., Z. H. Zhou, J. Jakana, W. Chiu, and F. J. Rixon. 1999. Roles of triplex and scaffolding proteins in herpes simplex virus type 1 capsid formation suggested by structures of recombinant particles. J. Virol. 6821-6830. [DOI] [PMC free article] [PubMed]
- 23.Schlesinger, S., and M. J. Schlesinger. 1996. Togaviridae: the viruses and their replication., p. 825-841. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven, New York, N.Y.
- 24.Simon, L. D. 1972. Infection of Escherichia coli by T2 and T4 bacteriophages as seen in the electron microscope: T4 head morphogenesis. Proc. Natl. Acad. Sci. USA 69:907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strauss, J. H., E. G. Strauss, and R. J. Kuhn. 1995. Budding of alphaviruses. Trends Microbiol. 9:346-350. [DOI] [PubMed] [Google Scholar]
- 27.Suomalainen, M., P. Liljestrom, and H. Garoff. 1992. Spike protein-nucleocapsid interactions drive the budding of alphaviruses. J. Virol. 66:4737-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tellinghuisen, T. L., A. E. Hamburger, B. R. Fisher, R. Ostendorp, and R. J. Kuhn. 1999. In vitro assembly of alphavirus cores using nucleocapsid protein expressed in Escherichia coli. J. Virol. 73:5309-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trus, B. L., F. P. Booy, W. W. Newcomb, J. C. Brown, F. L. Homa, D. R. Thomsen, and A. C. Steven. 1996. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J. Mol. Biol. 263:447-462. [DOI] [PubMed] [Google Scholar]
- 30.Turner, B. G., and M. F. Summers. 1999. Structural biology of HIV. J. Mol. Biol. 285:1-32. [DOI] [PubMed] [Google Scholar]
- 31.Weaver, S. C., L. Dalgarno, T. K. Frey, H. V. Huang, R. M. Kinney, C. M. Rice, J. T. Roehrig, R. E. Shope, and E. G. Strauss. 2000. Family Togaviridae, p. 879-889. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: classification and nomenclature of viruses. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 32.Wengler, G., U. Boege, G. Wengler, H. Bischoff, and K. Wahn. 1982. The core protein of the alphavirus Sindbis virus assembles into core-like nucleoproteins with the viral genome RNA and with other single-stranded nucleic acids in vitro. Virology 113:401-410. [DOI] [PubMed] [Google Scholar]
- 33.Wengler, G., C. Gros, and G. Wengler. 1996. Analyses of the role of structural changes in the regulation of uncoating and assembly of alphavirus cores. Virology 222:123-132. [DOI] [PubMed] [Google Scholar]
- 34.Wengler, G., G. Wengler, U. Boege, and K. Wahn. 1984. Establishment and analysis of a system which allows assembly and disassembly of alphavirus core-like particles under physiological conditions in vitro. Virology 132:401-412. [DOI] [PubMed] [Google Scholar]
- 35.Zhang, Z., B. Greene, P. A. Thuman-Commike, J. Jakana, P. E. Prevelige, J. King, and W. Chiu. 2000. Visualization of the maturation transition in bacteriophage P22 by electron cryomicroscopy. J. Mol. Biol. 297:615-626. [DOI] [PubMed] [Google Scholar]
- 36.Zhao, H., B. Lindqvist, H. Garoff, C.-H. von Bonsdorff, and P. Liljestrom. 1994. A tyrosine-based motif in the cytoplasmic domain of the alphavirus envelope protein is essential for budding. EMBO J. 13:4204-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]