Abstract
We used a baculovirus-based system to prepare structural proteins of hepatitis C virus (HCV) genotype 1a. Binding of this preparation to cultured human hepatic cells was both dose dependent and saturable. This binding was decreased by calcium depletion and was partially prevented by ligands of the asialoglycoprotein receptor (ASGP-R), thyroglobulin, asialothyroglobulin, and antibody against a peptide in the carbohydrate recognition domain of ASGP-R but not preimmune antibody. Uptake by hepatocytes was observed with both radiolabeled and dye-labeled HCV structural proteins. With hepatocytes expressing the hH1 subunit of the ASGP-R fused to green fluorescent protein, we could show by confocal microscopy that dye stain cointernalized with the fusion protein in an area surrounding the nucleus. Internalization was more efficient with a preparation containing p7 than with one that did not. The two preparations bound to transfected 3T3-L1 cells expressing either both (hH1 and hH2) subunits of the ASGP-R (3T3-22Z cells) or both hH1 and a functionally defective variant of hH2 (3T3-24X cells) but not to parental cells. Additionally, uptake of dye-labeled preparation containing p7 was observed with 3T3-22Z cells but not with 3T3-L1 or 3T3-24X cells or with the preparation lacking p7, suggesting that p7 regulates the internalization properties of HCV structural proteins. Our observations suggest that HCV structural proteins bind to and cointernalize with the ASGP-R in cultured human hepatocytes.
Hepatitis C virus (HCV) infection has become a major health problem affecting an estimated 170 million people worldwide. Persistent infection occurs in more than 70% of people infected with HCV, which may be complicated by cirrhosis and/or hepatocellular carcinoma (24, 33). Despite highly competitive and extensive research in this field, a highly effective treatment is not yet available. The mainstay of anti-HCV therapy, alpha interferon (IFN-α) or pegylated IFN-α, together with ribavirin leads, at best, to viral clearance in ca. 41 to 54% of patients infected with HCV genotype 1a (34, 38). Since mechanisms of HCV infection remain unclear, characterization of these mechanisms is now a major issue for the development of new strategies for anti-HCV treatment and prevention.
HCV is a member of the Flaviviridae family with a positive-strand RNA of ∼9.6 kb. More than six distinct genotypes exist among different HCV isolates; however, genotypes 1a and 1b are the most prevalent worldwide (8). The viral genome is translated into a single polyprotein of ∼3,000 amino acids in host cells (for a review, see reference 6). The amino-terminal part is cleaved by host cell proteases and its products, core and envelope (E1 and E2) proteins, are believed to be the major constituents of HCV particles (virions). E1 and E2 are glycosylated and are associated in two types of complexes: (i) heterodimers stabilized by noncovalent bonds, which probably represent the prebudding form of the viral envelope, and (ii) high-molecular-mass disulfide-bonded aggregates representing the misfolded protein (10, 14, 19). Both types of complexes are retained in the endoplasmic reticulum, the proposed site for HCV assembly and budding. In most virus strains, a cleavage occurs between E2 and p7, a hydrophobic domain found at the carboxy terminus of E2. In some virus strains, however, the cleavage is incomplete, resulting in the production of two E2 species, E2/p7(+) and E2/p7(−) (36). The function of p7 remains unclear, as well as the significance of its cleavage.
The precise mechanisms of early steps of HCV infection remain largely unknown. Attempts to elucidate those mechanisms have been hampered by the difficulties to obtain a sufficient amount of free virion from the plasma of infected individuals and to establish a robust in vitro system for virus propagation. Nevertheless, it is generally accepted that HCV envelope proteins (E1 and E2), as with other enveloped viruses, may play a major role in virus binding and entry into target cells. Hepatocytes represent the primary site of HCV replication in vivo, although the HCV genome has also been found in lymphoid cells, in particular B cells (70), and perhaps dendritic cells (46, 56). Infection of these latter types of cells has been implicated in extrahepatic manifestations of HCV infection such as mixed cryoglobulinemia and B-lymphocyte proliferative disorders (2, 57).
The processing of the HCV structural proteins after viral infection is poorly understood. Additionally, it is unclear whether this processing plays a major role in the infectious process and in triggering a proper immune response. Experimentally, HCV E1/E2 envelope proteins have been shown to induce a protective immune response against homologous viral challenge in the chimpanzee (9), and high titers of anti-envelope antibodies have been shown to correlate with the natural resolution of chronic hepatitis C (26, 30). These data suggest that the presentation of envelope antigens by target cells is probably a critical event that leads to recognition by the immune system and triggers the appropriate response. Nevertheless, viral antigen presentation does not lead to significant production of neutralizing anti-HCV antibodies in chronic hepatitis C infections or to a sustained, vigorous cytotoxic-T-lymphocyte (CTL) response (35, 51).
In the present study, our aim, using HCV structural proteins (HCV-SP) derived from strain H77 (1a genotype), was to identify the additional and/or distinct cellular surface protein used by HCV-SP for attachment and entry. HCV-SP were obtained by expressing the core, E1, and E2 proteins in a baculovirus-based system, as previously described for HCV 1b genotype (7). Cultured primary human hepatocytes, HepG2 cells, and Molt-4 cells were used to characterize HCV-SP-cell interactions. We then attempted to identify HCV-SP putative receptor(s) by using competitors to the binding of HCV-SP. Our data provide evidence that binding of HCV-SP to primary human hepatocytes and HepG2 cells, as well as internalization of HCV-SP, involves, at least in part, the asialoglycoprotein receptor (ASGP-R). Supporting this conclusion, cotransfection of a nonpermissive mouse fibroblast (3T3-L1) cell line with human hepatocyte ASGP-R cDNAs conferred HCV-SP binding and entry.
(This work was presented in part at the 7th International Meeting on Hepatitis C Virus and Related Viruses, Gold Coast, Australia, December 3 to 7, 2000 [abstr. A024].)
MATERIALS AND METHODS
Reagents, antibodies, virus, and cell lines.
Bovine serum albumin (BSA), GNA (lectin from Galanthus nivalis), asialo-GM1 (asialoganglioside), and asialofetuin were obtained from Sigma (St. Louis, Mo.). Coomassie Plus protein assay reagent was from Pierce (Rockford, Ill.). Enzyme-linked immunosorbent assay (ELISA) plates (Immulon-4) were obtained from Dynex Technologies (Chantilly, Va.). The protein G-Sepharose column and ELISA reader were from Bio-Rad (Hercules, Calif.). The mammalian expression vectors pcDNA3.1 and pcDNA3.1/NT-green fluorescent protein (GFP)-Topo, as well as zeocin, were from Invitrogen (Carlsbad, Calif.). Lipophilic dye (CellTracker CM-DiI, the chloromethylbenzamido derivative of DiI [1,1′-dioctadecyl-3,3,3′,3′-tetramethyl-indocarbocyanine iodine]) was from Molecular Probes (Eugene, Oreg.), and sterile glass chamber slides (Lab-Tek II) were from Nalge Nunc International (Rochester, N.Y.). Hybridoma cells expressing anti-E1, A4, and anti-core (C1) monoclonal antibodies (MAbs) (20) were gifts from H. B. Greenberg (Stanford Medical School, Stanford, Calif.), and the anti-E2 MAbs AP33 and ALP98 (47) were from A. H. Patel (Institute of Virology, Glasgow, United Kingdom), respectively. The peroxidase-labeled goat anti-mouse and anti-human immunoglobulin G (IgG), fluorescein isothiocyanate (FITC)-labeled goat anti-mouse IgG, and ABTS [2,2′azinobis(3-ethylbenzthiazolinesulfonic acid)] Microwell peroxidase substrate system were obtained from Kirkegaard & Perry Laboratories (Gaithersburg, Md.). The plasmid DNA containing the infectious HCV clone of H77 strain, p90/HCV. FL-long pU (32), was a gift from C. M. Rice (Rockefeller University) and S. M. Feinstone (Food and Drug Administration, Bethesda, Md.). Flow cytometric analysis was performed on a FACSCalibur (Becton Dickinson).
Primary cultured human hepatocytes, hepatocyte culture medium (HCM), and SingleQuots were obtained from Clonetics (BioWhittaker, Inc., Walkersville, Md.). Human hepatoma cell lines (HepG2), the human T-cell line Molt-4, and a mouse fibroblast cell line (3T3-L1) were obtained from the American Type Culture Collection (Rockville, Md.).
Recombinant baculovirus constructs.
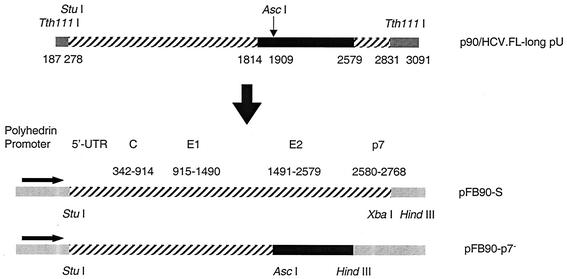
p90/HCV.FL-long pU was used as a template to generate two recombinant baculoviruses coding for the structural HCV proteins: core, E1, and either E2/p7(+) (Bac.HCV.1a-S) or E2/p7(−) (Bac.HCV.1a-p7−). Bac.HCV.1a-S has an additional 63 nucleotides (nt) of the amino-terminal part of NS2. This plasmid was digested with StuI and TthIII I, releasing a DNA fragment (nt 278 to 2831) corresponding to the core, E1, and E2/p7(+) proteins that was subcloned between the StuI-XbaI sites of a pFastBac plasmid, allowing its expression under the control of a polyhedrin promoter (pFB90-S). A second DNA fragment (nt 1814 to 2579) was generated from p90/HCV.FL-long pU; PCR was performed with Pfu DNA polymerase and the two following primers: 5′-AAG ACC TTG TGG CAT TGT GC-3′ (sense) and 5′-TCG AAA GCT TAC GCC TCC GCT TGG GAT ATG AGT-3′ (antisense). For cloning purposes, a HindIII site (underlined) was introduced into this amplimer. The 775-bp PCR product was subcloned into the SmaI site (blunt end) of pUC19 vector (pUC775). pUC775 and pFB90-S plasmids were digested with AscI and HindIII, respectively, to obtain a 671-bp DNA fragment (nt 1909 to 2579) and to remove a fragment (nt 1909 to 2831) of pFB90-S. The 671-bp fragment was then ligated with the truncated plasmid (pFB90-p7−) that encodes for the E2/p7(−) protein. Plasmids pFB90-S and pFB90-p7− were used to generate recombinant baculoviruses, Bac.HCV.1a-S and Bac.HCV.1a-p7−, respectively, by using the BAC-to-BAC Baculovirus Expression System (Gibco-BRL/Life Technologies, Gaithersburg, Md.) according to the manufacturer's protocols. The schematic diagram of the cloning procedures is shown in Fig. 1. The nucleotide sequences of the recombinant baculoviruses were verified by restriction enzyme analysis and DNA sequencing. Virus titer was determined by BacPAK Baculovirus Rapid titer kit (Clontech, Palo Alto, Calif.). Expression of core, E1, and E2 proteins of the recombinant baculoviruses in Sf9 cells (from Spodoptera frugiperda) was analyzed by indirect immunofluorescence.
FIG. 1.
Diagram of recombinant Bac-HCV.1a-S and Bac-HCV.1a-p7− constructs. Two recombinant baculoviruses encoding for the HCV-SP of genotype 1a (H77 strain): core, E1, and E2/p7 proteins (Bac-HCV.1a-S) or that of lacking the p7 protein (Bac.HCV.1a-p7−) were generated, as described in Materials and Methods. UTR, untranslated region.
Expression and purification of protein preparations obtained with Bac.HCV.1a-S.
Sf9 cells grown at 27°C in Sf900 medium (Gibco-BRL/Life Technologies) were infected with recombinant baculovirus at a multiplicity of infection (MOI) of 5 in a 500-ml Erlenmeyer flask, and cells were harvested at 3 days postinfection. All purification steps were carried out at 4°C on ice. Cells were harvested (3,000 rpm for 15 min), washed once in 10 mM Tris-HCl (pH 7.4)-150 mM NaCl-1 mM CaCl2 (TNC) buffer containing 1 mM Pefabloc SC and a cocktail of EDTA-free protease inhibitors (Roche, Indianapolis, Ind.), and finally resuspended at 107 cells/ml in TNC buffer containing 0.25% digitonin and protease inhibitors (cf. above). Cells were homogenized, placed on ice for 4 h with gentle agitation, and centrifuged at 30,000 × g for 45 min. The supernatant was collected, precipitated with 10% polyethylene glycol 8000 and 0.15 M NaCl for 2 h, and pelleted at 10,000 rpm for 30 min at 4°C. The pellet was resuspended in TNC buffer and briefly homogenized. Then, 100 to 200 μl of homogenized suspension was applied to 10.5 ml of 20 to 60% sucrose gradient and centrifuged at 156,000 × g for 16 h. Fractions (1 ml) were collected from the top of the tube and were tested for E1, E2, and core proteins by ELISA and Western blotting. Fractions containing recombinant proteins produced by Bac.HCV.1a-S [HCV-SP/p7(+)] were pooled, diluted with TNC buffer and pelleted at 100,000 × g for 3 h. Pellets containing HCV-SP were resuspended in TNC buffer and stored at −70°C. Protein concentration was determined with Coomassie Plus protein assay reagent (Pierce) with BSA as the protein standard. A similar method was used to express and purify proteins produced with Bac.HCV.1a-p7− [HCV-SP/p7(−)].
Anti-core, anti-E1, and anti-E2 antibodies.
C1, A4, AP33, and ALP 98 hybridoma cells were grown in RPMI medium supplemented with 10% fetal calf serum. To produce ascites, 2 × 106 cells in phosphate-buffered saline (PBS) were injected intraperitoneally into each of five female BALB/c mice (22). The ascitic fluid was purified through a protein G-Sepharose column, and the purified mouse anti-core, anti-E1, or anti-E2 IgGs were stored at −70°C.
E2 ELISA.
A 96-well plate was coated with 100 μl (20 μg/ml in PBS) of GNA at 37°C for 3 h. To prevent nonspecific binding, 150 μl of 4% goat serum (in 5% skim milk-PBS) was added, followed by incubation for 3 h at room temperature. Samples containing HCV-SP were diluted in 5% skim milk-PBS, added to each well, and incubated at 4°C overnight. Anti-E2 MAb (AP33; 100 μl, 6 μg/ml) was added, and the plate was incubated for 3 h at 37°C. Peroxidase-labeled goat anti-mouse IgG (at a dilution of 1/1,000) was then added, followed by incubation for 1 h at 37°C. Bound antibodies were detected by adding ABTS from the Microwell peroxidase substrate system, followed by analysis on an ELISA reader at an optical density of 405 nm. The plate was washed six times with PBS between each step and, after the addition of anti-E2 MAb, with PBS-0.05% Tween 20. All dilutions were made in PBS containing 5% skim milk.
Cell cultures.
Cryopreserved primary human hepatocytes were resuspended in HCM supplemented with HCM SingleQuots and were used on the same day after plating. HepG2 and Molt-4 cells were grown in minimal essential medium and RPMI culture media, respectively, supplemented with 10% fetal calf serum. 3T3-L1 cells were grown in Dulbecco modified Eagle medium culture medium containing 4.5 g of glucose/liter supplemented with 10% calf serum. All cell lines were grown in an incubator at 37°C with an H2O-saturated 95% air-5% CO2 atmosphere.
Binding assay.
The binding assay was performed in a U-bottom 96-well plate. All of the incubation (on a rocking platform) and centrifugation or washing steps (800 rpm, 5 min) were carried out at 4°C. All dilutions were made in ice-cold binding buffer (TNC buffer containing 1% BSA and a cocktail of EDTA-free protease inhibitors). Adherent cells were washed twice with PBS and detached with 2.5 mM EDTA (in PBS) at 37°C for 10 min prior to use. Cells were rinsed once and then resuspended in TNC buffer at 2 × 106 cells/ml, and 100 μl was added to each well. HCV-SP binding was measured by indirect labeling. Next, 0.125 to 2.5 μg of HCV-SP was incubated with cells for 2 h, and cells were washed twice to remove unbound proteins. Anti-E2 (AP33) or anti-E1 (A4) MAb was added, and the cells were incubated for 1 h, washed twice, and further incubated for 1 h with FITC-labeled goat anti-mouse IgG (4 μg/ml). Cells were washed twice and then resuspended in 150 μl of binding buffer, and bound HCV-SP was analyzed by flow cytometry. Nonspecific fluorescence was measured by adding primary and secondary antibodies in the absence of HCV-SP to cells. The mean fluorescence intensity (MFI) of bound HCV-SP was determined after subtracting the nonspecific fluorescence value.
In other experiments, cells were preincubated with various ASGP-R ligands prior to the addition of HCV-SP as described above. The 19S-thyroglobulin (Tg) fraction contains Tg dimers (apparent molecular mass of 660 kDa) that have a sedimentation coefficient of 19S as determined by ultracentrifugation. Crude Tg was extracted from the bovine thyroid gland, and 19S-Tg was purified by column chromatography, as previously described (68). Orosomucoid and 19S-Tg were incubated with agarose bead-linked neuraminidase, as recommended by the manufacturer (Sigma). After centrifugation, protein concentration of the supernatants containing asialo-orosomucoid and asialo-Tg was determined. All preincubation steps were performed for 2 h at 4°C.
GFP-hH1-transfected HepG2 cells.
GFP-ASGP-R construct was obtained by cloning the PCR amplimer coding for the ASGP-R hH1 subunit into the pcDNA3.1/NT-GFP-Topo vector. Briefly, cytoplasmic RNA extracted from HepG2 cells was subjected to reverse transcription and then to PCR with specific primers to obtain DNA fragments coding for hH1. The pcDNA3.1/NT-GFP-hH1 construct was verified by sequencing for correct sequence and alignment. A transient-transfection experiment was performed to confirm the expression of GFP-hH1 fusion protein. By laser scanning confocal microscopy (LSCM) analysis, a green fluorescent signal was detected in few cells, predominating at the levels of Golgi apparatus and plasma membrane, but was also detected in other cell structures, such as vesicles (not shown). HepG2 cells were then transfected with this plasmid construct by using Lipofectamine-Plus and, after a few days, selection antibiotic was added into the culture medium. Stable transfectants were obtained, and the most positive cells were sorted by using a Beckman-Coulter system.
Internalization assay. (i) Method with radiolabeled material.
Sf9 cells (5 × 108 cells) were infected with Bac-HCV 1a.S (MOI = 5) in Sf900 medium containing 0.5% fetal bovine serum at 27°C for 4 h. Cells were pelleted and washed once with starvation medium (Sf900 medium minus cysteine and methionine), and then cells were grown in this medium for 24 h. Next, 2 mCi of Redivue Pro-Mix 35S-labeled methionine and cysteine was added to the medium, and the cells were further incubated for 24 h. The labeling medium was discarded, and the cells were washed once and resuspended in Sf900 medium. HCV-SP were harvested at 3 days postinfection as described above. The internalization experiment was performed by incubating 100 μg of 35S-labeled HCV-SP per 2 × 108 cells/well in a six-well plate for 15, 30, or 60 min at 37°C.
(ii) Method with dye-labeled material.
HCV-SP was labeled with 4 μM CellTracker CM-DiI in TNC buffer for 1 h at 4°C in the dark. Dye-labeled HCV-SP was purified through a 30% sucrose cushion at 100,000 × g for 3 h; the pellet was resuspended in TNC buffer containing 1% BSA and protease inhibitors. HepG2 cells were seeded into sterile glass chamber slides 1 day before the assay. Cells were incubated with labeled HCV-SP in serum-free Dulbecco modified Eagle medium at 4°C for 30 min, followed by incubation at 37°C for 5, 15, or 30 min. Cells were rinsed once with ice-cold PBS and fixed with 4% paraformaldehyde in PEM buffer (80 mM PIPES-KOH, pH 6.8; 5 mM EGTA; 2 mM MgCl2) for 30 min on ice. Cells were then rinsed three times with PEM buffer, and slides were mounted with DAPI (4′,6′-diamidino-2-phenylindole)-antifade system and kept in the dark at 4°C until LSCM analysis was performed. Cells were analyzed with a laser-scanning confocal microscope (Leica; TCS SP) coupled with a DMIRBE inverted epifluorescent microscope. The wavelengths used to analyze GFP and CM-DiI staining were 499 and 553 nm for excitation and 519 and 570 nm for emission, respectively.
Stable cell line expressing recombinant human hepatic ASGP-R.
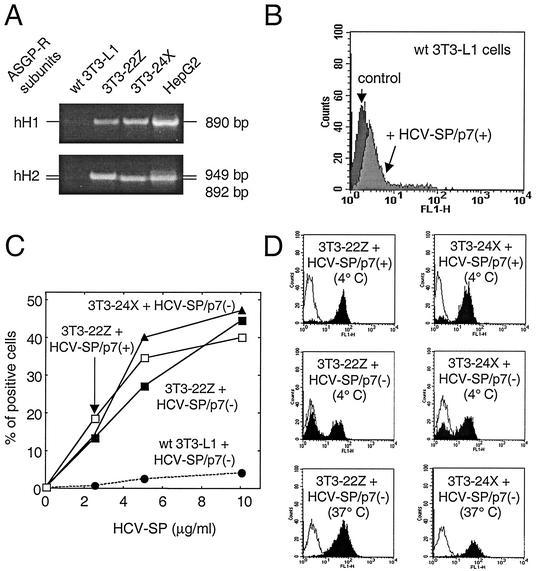
To determine whether ASGP-R can facilitate HCV-SP binding to nonpermissive cells, 3T3-L1 cells were cotransfected with plasmid constructs coding for two full-length subunits of human hepatic ASGP-R (hH1 and hH2) that have both been previously shown to be targeted to the plasma membrane in HepG2 cells (63). Briefly, cytoplasmic RNA extracted from HepG2 cells was subjected to reverse transcription and then PCR with specific primers to obtain cDNA fragments coding for hH1 and hH2. To allow simultaneous selection of stable transfected cells expressing both subunits, two mammalian expression vectors (pcDNA3.1-Zeo and pcDNA3.1-Neo) were used. Each hH1 or hH2 cDNA fragment was inserted into one distinct vector, allowing its expression under the control of a cytomegalovirus promoter. The correct sequences of both constructs were verified by sequencing. 3T3-L1 cells were then transfected with both constructs simultaneously using Lipofectamine-Plus according to a protocol provided by the manufacturer (Gibco-BRL/Life Technologies). At 3 days posttransfection, cells were passaged and grown under G418 and zeocin selection. Upon several passages, stable 3T3-L1 transfectants were obtained. Total RNA was extracted from these cells, and cDNA was synthesized by reverse transcription; PCR experiments were then performed by using the same pairs of primers as described above. One amplimer was detected for each PCR (hH1 or hH2) in these cells (3T3-22Z); agarose gel analysis showed that each amplimer had the same size as the corresponding amplimer obtained in HepG2 cells, whereas no amplimer was detected in 3T3-L1 parental cells. In addition, we obtained a variant of the full-length ASGP-R hH2 subunit lacking part of the hH2 cytoplasmic domain (nonfunctional variant) but still targeted to the plasma membrane in HepG2 cells (63). Another stable-transfected cell line coexpressing hH1 and the hH2 variant was then established (3T3-24X).
RESULTS
Characterization of HCV-SP.
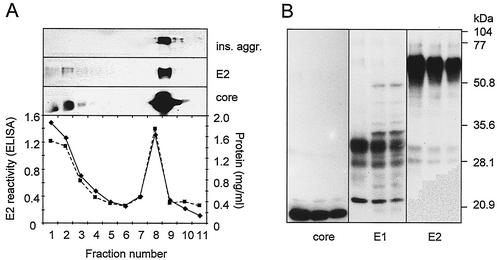
HCV-SP produced with a recombinant baculovirus-insect cell-based system were purified by sedimentation by equilibrium sucrose density centrifugation (Fig. 2A). Eleven fractions (1 ml) were collected from the top and then analyzed for the presence of E1, E2, and core proteins by both ELISA and Western blotting. ELISA results showed E2 reactivity was detected in two peaks: the lighter density (fractions 1 to 3) corresponds to a buoyant density of 1.02 to 1.10 g/ml, and the heavier density (fractions 8 to 9) corresponds to buoyant densities of 1.2 to 1.24 g/ml. Western blot analysis with anti-E2 MAbs (ALP98 or AP33) showed a group of major E2 protein bands of ∼70 kDa (Fig. 2); the core protein was detected as a band at ∼20 kDa (Fig. 2). In light fractions, two major forms of E1 (∼33 and ∼28 kDa) reflecting the different extent of N-linked glycosylation were also observed (Fig. 2B). The E2 protein of HCV-SP was recognized by conformation-sensitive anti-E2, H2, and H53 MAbs (a gift from J. Dubuisson, Institut Pasteur, Lille, France [data not shown]), suggesting that the E2 protein of HCV-SP assumes a proper conformation (66). The purification procedure was reproduced in several independent experiments.
FIG. 2.
Characterization of HCV-SP. Insect cells were infected with recombinant Bac.HCV.1a-S and were harvested at 3 days postinfection. HCV-SP were purified by equilibrium sucrose gradient centrifugation. (A) Profile of HCV-SP/p7(+) after equilibrium sucrose gradient centrifugation. In the bottom panel, 1-ml fractions were collected from the top, and the protein concentration was measured (squares, dashed line); 50 μl of each fraction was tested for E2 reactivity with AP33 MAb by ELISA (diamonds, solid line). A similar pattern was observed for HCV-SP/p7(−) (not shown). In the top panels, 50 μl of each fraction was suspended into Laemmli buffer in denaturing conditions and analyzed by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis and then blotted onto a nitrocellulose membrane. HCV-SP was tested for E2 and core reactivity by incubating the membrane with AP33 and C1 MAbs, respectively; antigen-antibody complexes were revealed by incubating the membrane with horseradish peroxidase-coupled anti-mouse antibody and then subjected to enhanced chemiluminescence and autoradiography; the reactivity against insoluble aggregates (ins. aggr.) is shown on the top panel. (B) Immunoblot analysis of light fractions of HCV-SP/p7(+). Proteins were prepared, analyzed, and blotted onto a polyvinylidene difluoride membrane, and the immunoreactivity with anti-core, anti-E1, and anti-E2 antibodies was determined as described above. The electrophoretic mobilities of the bands that displayed the highest core and E2 reactivity, respectively, were identical to those of positive controls with recombinant proteins expressed in mammalian cells (not shown).
Cell binding of HCV-SP/p7(+) and HCV-SP/p7(−).
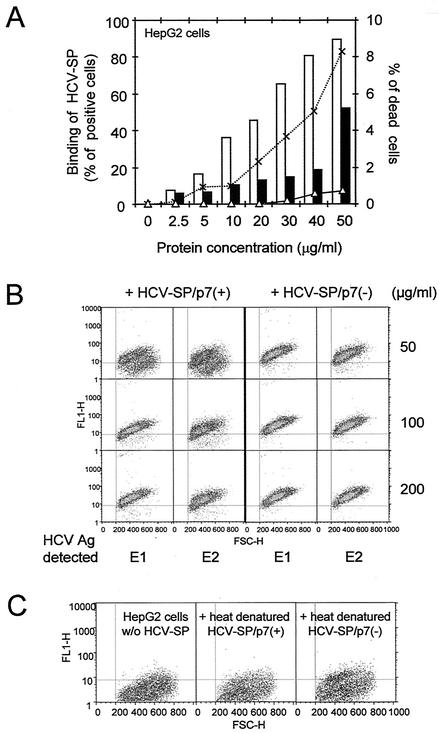
Binding of the HCV-SP preparations to HepG2 cells was performed as described in Materials and Methods. As shown in Fig. 3A, the binding of the light fraction of HCV-SP occurred in a dose-dependent manner. In contrast, very little binding was observed with the heavy fraction and only at a high concentration. In addition, a slight toxic effect on cells was observed with this latter fraction. This may be due to the presence of insoluble aggregates (Fig. 2A) that were less recognized by conformational antibodies with ELISA. It is well known that expression of E1 and E2 glycoproteins in mammalian cells also produces high-molecular-weight, disulfide-linked aggregates (10, 14, 19).
FIG. 3.
Binding of HCV-SP/p7(+) and HCV-SP/p7(−) to HepG2 cells. (A) Binding of light and heavy fractions of HCV-SP to cells. HepG2 cells were incubated with HCV-SP derived from HCV strain 1a as described in Materials and Methods; both light (open bars) and heavy (full bars) fractions were tested. Cell-bound HCV-SP was detected by incubating cells with anti-E2 MAb, followed by FITC-labeled goat anti-mouse IgG and subjecting them to fluorescence-activated cell sorting (i.e., with a FACscan). Nonspecific fluorescence was measured by adding primary and secondary antibodies in the absence of HCV-SP to cells. The cytotoxicity of both light (▵) and heavy (×) fractions was indirectly evaluated by the shift of cell size and the granularity to the bottom left corner of scattered plots. (B) Saturability of the binding of HCV-SP to HepG2 cells. The indicated amounts of proteins from the light fractions of HCV-SP/p7(+) and HCV-SP/p7(−) preparations were incubated with HepG2 cells, and binding was evaluated by using anti-E1 or anti-E2 MAbs with a FACscan as described above. The results are presented on scattered plots: cell granularity is plotted on x axis (FSC-H), whereas the fluorescence intensity is plotted on the y axis (FL1-H); the MFI is calculated for each plot, and the percentages of positive cells were determined according to the threshold values (vertical and horizontal lines) established after the control (in the absence of primary antibody). (C) Inhibition of the binding of HCV-SP to HepG2 cells by heat denaturation. Proteins from the light fractions of HCV-SP/p7(+) and HCV-SP/p7(−) preparations were heated at 90°C for 10 min in the binding buffer; 50 μg/ml was then incubated with HepG2 cells, and the extent of binding was measured as in panel B.
The binding of HCV-SP/p7(+) and HCV-SP/p7(−) preparations were compared. Binding was observed with the lighter fractions of both preparations (Fig. 3B), whereas heavy fractions of both HCV-SP/p7(+) and HCV-SP/p7(−) displayed a much lower lever of binding activity and only at a much higher concentration (≥50 μg/ml). Thereafter, we only used HCV-SP from light fractions for our further studies. In HepG2 cells, maximum binding was generally observed starting within a range of 50 to 100 μg of protein/ml for HCV-SP/p7(+) and 30 to 50 μg of protein/ml for HCV-SP/p7(−) and grossly corresponded to a similar HCV-SP immunoreactivity, as evaluated by ELISA or Western blotting (not shown).
In addition, we studied the binding of both HCV-SP preparations to HepG2 cells after protein heat denaturation at 90°C for 10 min (in the absence of both disulfide bond reducing reagent and sodium dodecyl sulfate); we subsequently observed an almost complete inhibition of HCV-SP binding to HepG2 cells (Fig. 3C). This result indicates that the level of HCV-SP nonspecific binding is probably extremely low and certainly does not account for the binding of HCV-SP that we report here with HCV-SP from lighter fractions. It further suggests that asialated termini of carbohydrate structures on HCV-SP molecules are not enough to mediate HCV-SP binding to ASGP-R.
Binding of HCV-SP to primary human hepatocytes, HepG2, and Molt-4 cells.
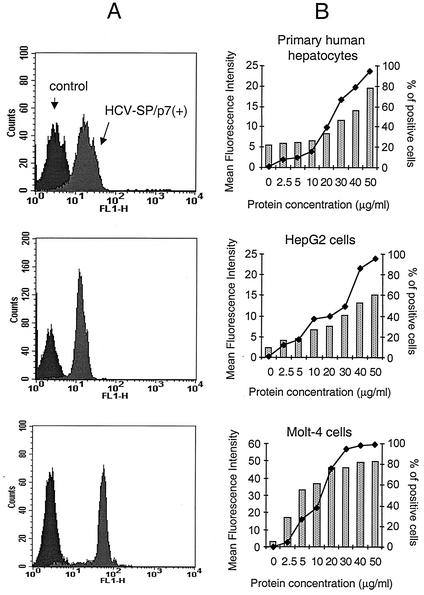
The ability of HCV-SP to bind various target cells was analyzed by flow cytometry (Fig. 4). The binding of HCV-SP to target cells occurred in a concentration-dependent manner in various cell types (Fig. 4B). Specific HCV-SP binding was found in human hepatic cells (primary human hepatocytes and HepG2 cells) and human T cells (Molt-4 cells; Fig. 4A) but not in mouse fibroblasts (3T3-L1 cells; see Fig. 9A).
FIG. 4.
Binding of HCV-SP/p7(+) to primary human hepatocytes, HepG2, and Molt-4 cells. Cells of various types were incubated with HCV-SP from the light fractions as described in Materials and Methods; cell-bound HCV-SP and nonspecific fluorescence were measured as in Fig. 2. (A) Histogram patterns of HCV-SP binding to the target cells. (B) Quantified results and percentage of positive cells (♦). Cells were considered positive when they displayed fluorescence with a value above that of the nonspecific fluorescence threshold. The MFI values were determined for each cell after subtraction of the nonspecific fluorescence value. The results shown represent the mean values obtained from three independent experiments.
FIG. 9.
Binding of HCV-SP to ASGP-R-transfected 3T3-L1 cells. (A) 3T3-L1 cells were transfected to stably coexpress two subunits of human liver ASGP-R (hH1 and hH2). Clone 3T3-22Z coexpressed both full-length hH1 and hH2, whereas clone 3T3-24X coexpressed full-length hH1 with a truncated variant of hH2 defective for internalization but functional for binding. Total RNA was extracted from parental (wt) 3T3-L1 cells, from clones 3T3-22Z and 3T3-24X, or from HepG2 cells and subjected to reverse transcription; these cDNAs were then used to amplify by PCR a DNA fragment corresponding to either ASGP-R hH1 or hH2 subunits, as indicated. (B) Mouse fibroblasts (3T3-L1 cells) were incubated with 10 μg of HCV-SP/p7(+)/ml and subjected to the same detection protocol as in Fig. 2. (C) Both 3T3-22Z (squares) and 3T3-24X (triangles) cells, as well as parental 3T3-L1 cells (circles), were challenged with various amounts (2.5 to 10 μg/ml) of either HCV-SP/p7(+) (open symbols) or HCV-SP/p7(−) (solid symbols) and incubated for 2 h at 4°C. Cell-bound HCV-SP was detected by flow cytometry as in Fig. 2. (D) Histograms of the binding of either HCV-SP/p7(+) (top panels, 4°C) or HCV-SP/p7(−) (middle, 4°C, and bottom, 37°C, panels) to 3T3-22Z (right panels) and 3T3-24X (left panels) cells are presented.
With this method of detection using antibodies, we could not compare the Kd values for HCV-SP/p7(+) or HCV-SP/p7(−) binding to the various cell types. Thus, the affinity of the primary antibody may be different for HCV-SP/p7(+) and HCV-SP/p7(−) preparations and, moreover, this method relies not only on the binding of HCV-SP to cells but also on the binding of primary and secondary antibodies to cell-bound HCV-SP. In fact, it is unclear how much these two latter steps may influence the outcome and, therefore, the calculation of the Kd. We have evaluated the Kd elsewhere by using another method of purification and labeling that skipped the steps with antibodies (67).
Effect of calcium and ASGP-R ligands on HCV-SP binding.
As the structure of the HCV virion remains elusive, it has been previously shown that recombinant HCV envelope glycoproteins are asialated when produced in mammalian cells (61). Glycosylation of neosynthesized envelope proteins is critical for proper folding and assembly of HCV-SP in the endoplasmic reticulum. We therefore sought to determine whether this asialated trait could play a role in the binding of HCV-SP to hepatic cells. The ASGP-R is a C-type (calcium-dependent) lectin that is most commonly found in the liver, although it is also expressed in other tissues (62). It has been implicated in the clearance of asialoglycoproteins, i.e., desialated or galactose-terminal glycoproteins, from the circulation by receptor-mediated endocytosis. ASGP-R was also shown to mediate entry of hepatitis A virus-IgA complexes into hepatocytes (18) and has been proposed as a potential target for gene delivery into hepatocytes, with the goal of inducing a CTL response against viral alloantigens (13). This receptor consists of a heteromultimer of two homologous subunits, hH1 and hH2 (41, 62). Each subunit is subdivided into four functional domains: the cytosolic domain, the transmembrane domain, the stalk, and the carbohydrate recognition domain (CRD). The CRD of hH1 requires three calcium ions for proper binding conformation and sugar binding (41).
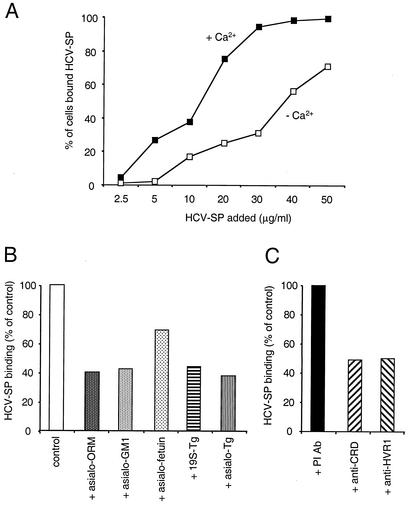
Since ASGP-R binding is calcium sensitive, we first sought to determine whether HCV-SP binding to cells occurs in a calcium-dependent manner. A positive result would only be consistent with such a possibility, whereas a negative result would argue against such a possibility not only for ASGPR but also for other calcium-dependent receptors that might be involved in HCV-SP binding. Indeed, the simultaneous removal of calcium from the binding medium, together with the addition of a 5 mM concentration of the calcium chelator EGTA, reduced HCV-SP binding to HepG2 cells (Fig. 5A). To test more directly whether the ASGP-R might mediate HCV-SP binding to hepatic cells, primary human hepatocytes and HepG2 cells were preincubated with several ASGP-R ligands. Asialo-orosomucoid, a high-affinity ligand of the ASGP-R in the liver (68), inhibited HCV-SP binding to HepG2 cells in a concentration-dependent manner (not shown). As shown in Fig. 5B, asialo-orosomucoid, as well as asialoganglioside and asialofetuin, both reported to bind ASGP-R in the liver, partially inhibited the binding of HCV-SP to HepG2 cells, the latter being, however, less effective. Tg has been previously reported to bind the ASGP-R (11, 16, 43, 48, 68). 19S-Tg and its desialated form (asialo-Tg) both inhibited HCV-SP binding to HepG2 cells. At lower concentrations, asialo-Tg (0.4 mg/ml) exhibited the same or greater inhibitory effects on HCV-SP binding as that of 19S-Tg (at 1 mg/ml). Desialated Tg is indeed known to have a higher affinity to the ASGP-R than 19S-Tg (11, 68).
FIG. 5.
HCV-SP binding to HepG2 cells is inhibited by ligands of ASGP-R. (A) HCV-SP binding to HepG2 cells is calcium dependent. Cells and HCV-SP were suspended either in binding buffer in the presence of CaCl2 (▪) or in binding buffer containing 5 mM EGTA in the absence of CaCl2 (□), and a binding assay was performed as in Fig. 2. (B) Effect of ASGP-R ligands on HCV-SP binding in HepG2 cells. Cells were preincubated in binding buffer (with CaCl2) at 4°C either with buffer alone (control), with 5 mg of asialo-orosomucoid (asialo-ORM)/ml, with 200 μg of asialoganglioside (asialo-GM1)/ml, with 2 mg of asialofetuin/ml, with 1 mg of 19S-Tg/ml, or with 0.4 mg of asialo-19S-Tg/ml (asialo-Tg); an HCV-SP binding assay was then performed. (C) Effect of antibodies on HCV-SP binding in HepG2 cells. Cells were preincubated in binding buffer at 4°C with preimmune antibody (PI Ab; 1/100), polyclonal antibody recognizing a peptide in the CRD of ASGP-R (anti-CRD; 1/100), or antibody directed against HVR1 of the E2 envelope protein (anti-HVR1; 1/100); an HCV-SP binding assay was then performed with anti-E1 antibody.
More importantly, preincubation of cells with polyclonal antibody against a peptide of the CRD of the hH1 subunit of the ASGP-R resulted in a decrease in HCV-SP binding to HepG2 cells (Fig. 5C) and primary hepatocytes (not shown); this decrease was not observed with preimmune antibody (Fig. 5C). Additionally, an antibody directed against hypervariable region 1 (HVR1) of E2 envelope protein was as effective as the former antibody in inhibiting HCV-SP binding to HepG2 cells. The partial inhibition observed with the anti-HVR1 antibody suggests that additional binding sites of HCV-SP might exist that are neither competed for by ASGP-R ligands nor sensitive to calcium, perhaps on the E1 envelope protein.
Internalization of radiolabeled HCV-SP in HepG2 cells.
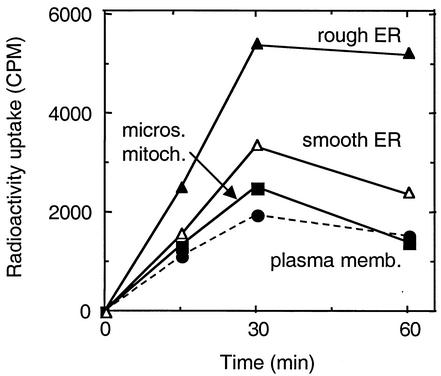
We next addressed the question of whether, after it binds to the cell surface receptor, HCV-SP could be internalized into human hepatic cells. For that, we first infected insect cells with recombinant baculovirus and then incubated them with a [35S]methionine and [35S]cysteine mix. HCV-SP was prepared and purified as described above, and radiolabeled material was incubated with HepG2 cells at 37°C. Cells were harvested, disrupted, and submitted to cell fractionation with sucrose gradient ultracentrifugation, as described previously (20), resulting in four fractions corresponding to four membrane-enriched cell compartments. Figure 6 shows that radioactivity was detected in the various compartments even after a short incubation with cells. After 15 min, the increasing amount of radioactivity was observed in all cellular compartments (plasma membrane < microsome-mitochondrion < smooth endoplasmic reticulum [SER] < rough endoplasmic reticulum [RER]), suggesting that the incorporation of labeled HCV-SP occurred in this order. After 30 min of incubation, the amount of radioactivity had reached a steady state in the RER, whereas it started to decrease in the other intracellular compartments, suggesting that the majority of radiolabeled HCV-SP has reached the rough endoplasmic reticulum-enriched compartment.
FIG. 6.
Internalization of radiolabeled HCV-SP/p7(+) in HepG2 cells. Sf9 insect cells were infected with recombinant Bac.HCV.1a-S baculovirus and then incubated with 35S-labeled methionine-cysteine mix. HCV-SP was prepared as described previously and then purified, and radiolabeled material (50 μg/ml) was incubated with HepG2 cells at 37°C for the indicated times. Cells were harvested, disrupted, and subjected to cell fractionation. Four membrane fractions were isolated, each enriched in either plasma (•), microsomial/mitochondrion (▪), smooth endoplasmic reticulum (▵), or rough endoplasmic reticulum (▴) membranes. Radioactivity uptake was quantified by liquid scintillation counting.
Internalization of dye-labeled HCV-SP in HepG2 cells and colocalization with ASGP-R GFP-hH1.
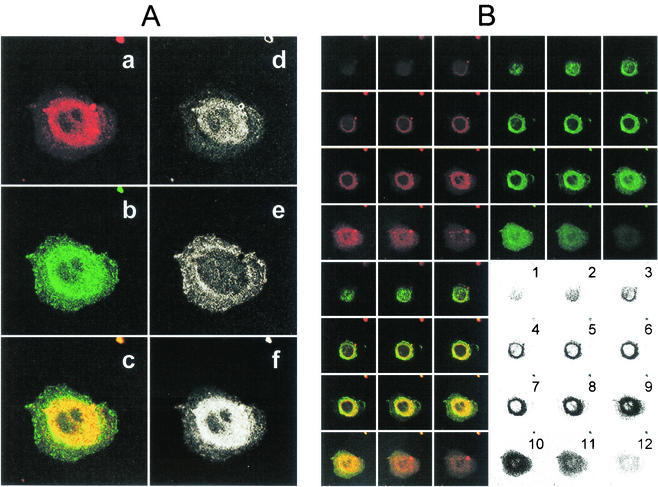
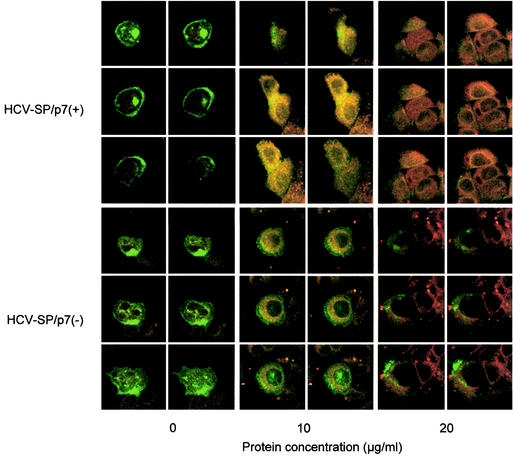
Since several experiments suggested that ASGP-R was involved in the binding of HCV-SP to HepG2 cells, we then sought to determine whether ASGP-R was involved in internalization. For this purpose, we established a clone of stable transfected HepG2 cells expressing a fusion protein between the hH1 subunit of ASGP-R and the GFP (GFP-hH1/HepG2 cells). In basal conditions, some GFP signal was visible in the endoplasmic reticulum area, but most was seen in the Golgi apparatus area, suggesting that the GFP-hH1 subunit was properly glycosylated before targeting to the plasma membrane. After incubation of cells with CM-DiI-labeled HCV-SP (red), colocalization was analyzed by LSCM. We observed that, after uptake, this material accumulated in the cell area surrounding the nucleus. Moreover, by superimposing the pictures obtained in green and red channels, we observed a clear yellow signal in the perinuclear area. Lack of strong red (d) or green (e) signals in the superimposition picture (c) suggest that most CM-DiI-labeled HCV-SP and GFP-hH1 subunits colocalized (f) in this area (Fig. 7A). This suggests that HCV-SP not only entered HepG2 cells but also that it was targeted toward an area surrounding the nucleus, simultaneously with the hH1 subunit of ASGP-R.
FIG. 7.
Colocalization of dye-labeled HCV-SP and ASGP-R GFP-hH1 in the perinuclear area. HepG2 cells expressing a fusion protein between GFP and ASGP-R hH1 subunit (GFP-hH1-HepG2 cells) were seeded into sterile glass eight-chamber slides 1 day before the assay. HCV-SP was dye (CM-DiI) labeled and purified as described in Materials and Methods. GFP-hH1-HepG2 cells were incubated with 10 μg of CM- DiI-labeled HCV-SP/ml for 60 min at 37°C; the cells were then rinsed and fixed with 4% paraformaldehyde, and the slides were mounted with DAPI-antifade system and kept in the dark at 4°C until they were analyzed by LSCM in both green (GFP) and red (CM-DiI) wavelength channels. (A) Colocalization pattern in a confocal horizontal section of a single cell. In the left column are shown the signals obtained in red (a) and green (b) wavelength channels, and superimposition of a + b signals (c). In the right column, patterns of red (d), green (e), and yellow (f) pixels can be detected in the superimposition picture (c). Most pixels in the perinuclear area are yellow (i.e., green plus red = colocalization). (B) Serial horizontal sections of the same cell (from top to bottom = numbers 1 to 12). Sections were obtained in both green and red wavelength channels (top panels) and superimposed (bottom left panel); areas displaying colocalization are shown in the bottom right panel. A threshold was applied to keep only the most significant pixels; darkness increases with the intensity of the colocalized signals.
Furthermore, as shown in Fig. 8, the incubation at 37°C of dye-labeled HCV-SP with GFP-hH1/HepG2 cells was followed by a concentration-dependent uptake of the labeled material. The intensity of HCV-SP/p7(−) uptake was less than that observed with HCV-SP/p7(+) preparation (Fig. 8). Finally, no uptake of dye-labeled HCV-SP was observed in a cell line of human thyrocytes (Aro cells; data not shown) or a cell line of mouse fibroblasts (3T3-L1 cells; see Fig. 10) that do not express ASGP-R transcripts (Fig. 9A). In addition, by using a dye-labeled control preparation obtained by expressing recombinant β-glucuronidase with a baculovirus construct (bac-GUS), no uptake was observed in HepG-2 or HuH-7 cells (67), both well known to express ASGP-R at a high level.
FIG. 8.
Internalization of HCV-SP into GFP-hH1-transfected HepG2 cells. GFP-hH1-HepG2 cells were first incubated with 10 or 20 μg of dye labeled HCV-SP/p7(+) (top panels) or HCV-SP/p7(−) (bottom panels)/ml, or without, as indicated in the figure, in serum-free medium at 4°C for 30 min. This step was followed by further incubation at 37°C for 60 min. The cells were then subjected to the same procedure as in Fig. 7 and submitted to LSCM analysis in both green (GFP) and red (CM-DiI) wavelength channels; horizontal sections (six per cell or group of cells are shown) obtained in both green and red wavelength channels were superimposed. Areas displaying colocalization appear yellow.
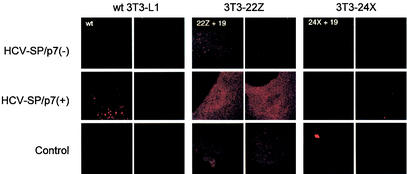
FIG. 10.
Internalization of labeled HCV-SP into ASGP-R-transfected 3T3-L1 cells. ASGPR hH1/hH2-dual-transfected 3T3-L1 cells (clone 3T3-22Z [= 22Z + 19] or clone 3T3-24X [= 24X + 19]) or wild-type 3T3-L1 cells (wt) were incubated in the presence of 10 μg of labeled HCV-SP/p7(+) or HCV-SP/p7(−)/ml for 30 min at 37°C. The cells were subjected to the same procedure as in Fig. 7 and subjected to LSCM analysis in the red (CM-DiI) wavelength channel. Sections of two distinct cells are shown for each condition.
Binding of HCV-SP to transfected 3T3-L1 cells expressing the human liver ASGP-R subunits.
3T3-L1 cells, a nonpermissive cell line of mouse fibroblasts, was chosen to express the human hepatic ASGP-R (subunits hH1 and hH2). Stable ASGP-R dually transfected cells were obtained, and two cell clones (3T3-22Z and 3T3-24X) were isolated (Fig. 9A) and tested for HCV-SP binding. Clone 3T3-22Z coexpressed both full-length hH1 and hH2, whereas clone 3T3-24X coexpressed full-length hH1 with a variant of hH2 (hH2′) that has a truncated cytoplasmic domain: the absence of this domain impairs cell trafficking of the hH2′ subunit but does not affect its binding domain or properties (63). As shown in Fig. 9B, parental cells did not bind HCV-SP; this was tested in the range of 10 to 50 μg/ml of HCV-SP for both preparations (not shown). In Fig. 9C, either type of HCV-SP preparation (added at a low concentration [2.5 to 10 μg/ml] onto 104 cells) bound to the ASGP-R-expressing cells in a dose-dependent manner (13.23 to 44.46% of positive cells). The histograms in Fig. 9D suggest that some cells did not bind HCV-SP/p7(−) at 4°C in both 3T3-22Z and 3T3-24X cell clones, at variance with HCV-SP/p7(+); this difference disappeared at 37°C. A difference of ASGP-R subunits expressed at the cell surface between 4 and 37°C in a proportion of transfected cells could affect the binding of HCV-SP/p7(−) more than that of HCV-SP/p7(+). This could be due to either a difference of glycosylation pattern of ASGP-R subunits present at the cell surface or their organization.
Internalization of HCV-SP into transfected 3T3-L1 cells expressing the human liver ASGP-R.
Parental 3T3-L1 cells and cell clones 3T3-22Z and 3T3-24X were used to study whether the expression of ASGP-R not only allowed nonpermissive cells to bind HCV-SP but also renders them permissive for HCV-SP internalization. Figure 10 shows that, as expected, parental 3T3-L1 cells (wild type) did not uptake dye-labeled HCV-SP/p7(+) or HCV-SP/p7(−). Interestingly, although 3T3-22Z cells do bind both HCV-SP/p7(+) and HCV-SP/p7(−), only dye-labeled HCV-SP/p7(+) uptake was observed (Fig. 10). This finding correlates with the lesser uptake of HCV-SP/p7(−) observed in HepG2 cells compared to HCV-SP/p7(+). Finally, 3T3-24X cells that also bind both HCV-SP/p7(+) and HCV-SP/p7(−) did not take up any of the two dye-labeled HCV-SP (Fig. 10). This was expected because 3T3-24X cells express the ASGP-R hH2′ subunit whose internalization is greatly impaired in HepG2 cells (63).
DISCUSSION
We sought to determine whether HCV structural proteins (HCV-SP) produced in insect cells could bind and enter into cultured human hepatocytes. Our results provide several lines of evidence indicating that HCV-SP binds to the ASGP-R in these cells and that ASGP-R may in part be responsible for their internalization into cultured hepatocytes.
The arguments for the involvement of the ASGP-R in HCV-SP binding are as follows. First, the binding of HCV-SP was inhibited by anti-ASGP-R CRD peptide antibody but not by preimmune antibody. The CRD is the extracellular part of the ASGP-R responsible for binding terminal nonreducing galactose residues and N-acetylgalactosamine residues of desialated N-linked tri- or tetra-antennary glycans, conferring part of its specificity to this receptor (41). The CRD of hH1 requires calcium for proper binding conformation and sugar binding (41). Consistent with that, calcium chelation by EGTA inhibited the binding of HCV-SP to cells. Second, the binding of HCV-SP to cells was partially inhibited by ASGP-R ligands such as asialo-orosomucoid (63) and Tg. Tg is the major component of colloid substance in the thyroid and is a large glycoprotein involved in thyroid hormone synthesis. Tg has been reported to bind an ASGP-R-related receptor in the thyroid (11, 16, 43, 48, 68). Finally, the transfection of a nonpermissive cell line in order to express both subunits of the human liver ASGP-R conferred HCV-SP binding to these cells (3T3-22Z cells). Altogether, these results suggest that HCV-SP may also bind to ASGP-R in hepatocytes.
Several mechanisms may be relevant to explain the binding of HCV-SP to ASGP-R. First, recombinant glycoproteins produced in insect cells generally lack sialic acid residue at the extremity of their mature carbohydrate domain (27, 39). It is thus likely that HCV-SP lack sialic acid, as already reported for recombinant HCV envelope proteins produced in mammalian cells (61). Moreover, asialo-Tg was more potent than Tg in inhibiting the binding of HCV-SP to human hepatocytes. Evidence that the penultimate galactose and N-acetylglucosamine residues of the complex asialated carbohydrate of Tg bind to ASGP-R in thyrocytes has been previously established (11, 12, 60, 68). Therefore, the simplest explanation for Tg-induced inhibition of HCV-SP binding is the presence of asialocarbohydrates on the Tg molecule. Interestingly, uptake of circulating desialated Tg by hepatocytes has previously been described (45). It is indeed well known that a small proportion of Tg is released in the bloodstream (49), albeit the role is not yet understood.
Although largely mediated by their sugar moiety, the binding of various ligands to ASGP-R occurs within a broad range of affinity and with a certain degree of specificity (62). Thus, asialo-orosomucoid and asialo-Tg have very different binding affinities in liver and thyroid (11). In this respect, modifications in either number or position of antennas in the carbohydrate moiety of asialoglycoproteins are monitored by drastic changes in their affinity of binding to ASGP-R (62). This has raised the possibility that determinants other than asialation of the carbohydrate termini are involved in binding to ASGP-R. In fact, several reports suggest that maturation of the carbohydrate structure of asialoglycoproteins expressed in insect cells differ from that observed in mammals (39). It is therefore highly probable that the carbohydrate structures of HCV-SP and asialo-Tg are different. Moreover, the idea that the carbohydrate domain is the sole determinant of binding to the ASGP-R is no longer operative (17). Thus, point mutations in the protein frame of ASGP-R ligands, even if still asialated, lead to a dramatic decrease of their binding affinity. This may perhaps be due to modifications in maturation of their carbohydrate domain or change of conformation of that domain but may also relate to the importance of other determinants on the protein core (17, 41, 62). In the thyroid, the Tg content in iodotyrosine residues plays a critical role in Tg binding (59). We therefore think that HCV-SP binding to ASGP-R may not be solely determined by its carbohydrate structure and may have some pathophysiological relevance.
Additional arguments suggest that the binding to ASGP-R is followed by other events and that the ASGP-R is involved in the internalization of HCV-SP as well. First, transfection of nonpermissive cells expressing both subunits of the human liver ASGP-R (3T3-22Z cells) conferred HCV-SP/p7(+) binding and entry into these cells. As expected, entry was not observed with a dual-transfectant cell line expressing a function defective variant of the hH2 subunit of ASGP-R (3T3-24X cells), suggesting that full ASGP-R functionality was required. The reason why 3T3-22Z cells did not uptake HCV-SP/p7(−) remains unknown. However, in a similar manner, HCV-SP/p7(−) uptake appeared not to be as effective as for HCV-SP/p7(+) in HepG2 cells. It has been hypothesized that the lack of p7 may lead to conformational changes of envelope proteins (36), as reported for closely related virus (21), that are perhaps important for cell binding and entry of the HCV virion. In fact, the discrepancy between the two types of HCV-SP preparations observed in HepG2 cells could in part be due to their differential affinity for ASGP-R and/or a conformational issue important to trigger internalization. This discrepancy could be even more obvious with 3T3-22Z cells that, in fact, express less ASGP-R than do HepG2 cells (Fig. 9B). This view would also be consistent with an involvement of noncarbohydrate determinants in the mechanism of entry of HCV-SP after binding to ASGP-R.
In GFP-hH1-transfected HepG2 cells, both HCV-SP and GFP-ASGP-R fusion protein cointernalized and then colocalized in a region surrounding the nucleus. It has been previously reported that, after internalization, asialo-orosomucoid is targeted to the lysosomial compartment in an acidification-dependent manner in HepG2 cells (63). Additionally, a recycling of ASGP-R to the cell surface after ligand delivery has been suggested (for a review, see reference 62). In contrast to these studies, we observed here that both types of HCV-SP were targeted with ASGP-R to an area surrounding the nucleus, likely within the rough endoplasmic reticulum and/or nuclear envelope compartments. Consistent with this hypothesis we detected, after its uptake by nontransfected HepG2 cells, radiolabeled HCV-SP mainly in an endoplasmic reticulum membrane-enriched compartment. It is well known that newly synthesized misfolded proteins in the endoplasmic reticulum are targeted toward the proteasome system for protein degradation. Variants of the hH2 and hH1 subunits of ASGP-R, hH2a and hH1i5, respectively, have also been suggested to undergo such a processing (29, 58). However, if this were the case for GFP-hH1 as well, this subunit would also be targeted toward the endoplasmic reticulum compartment in basal conditions, a hypothesis that, in fact, our observations do not support. We, therefore, think that targeting of the GFP-ASGP-R fusion protein toward the endoplasmic reticulum compartment triggered upon HCV-SP binding may be specific.
ASGP-R binding and pathway of entry may perhaps constitute an alternative pathway to induce an immune response against HCV. Several arguments suggest an involvement of ASGP-R in immune responses. First, ASGP-R has been previously suggested as being one potential target in chronic hepatic autoimmune disease in humans (40) and, experimentally, antibodies against ASGP-R are produced in animals challenged with woodchuck hepatitis virus (15). Second, HCV is known to favor the occurrence of autoantibodies, such as those observed in autoimmune thyroid diseases (55). The existence of a negative correlation between the occurrence of various autoantibodies and anti-ASGP-R autoantibodies has been reported in chronically HCV infected patients (25). Thus, a balance is observed between hepatic autoimmune response and other autoimmune disorders upon HCV infection. Third, recognition of complex N-linked glycans by lectins is probably important for the development of an appropriate immune response. Thus, altered protein glycosylation is involved in triggering autoimmune diseases in animal models, although the mechanism remains poorly understood as yet (37). This suggests that ASGP-R plays some role in regulating the immune system.
ASGP-R expression has also been described in dendritic cells (69). These cells play a pivotal role in the immune response against infectious pathogens (23, 44), including the induction of appropriate CTL response against dengue virus, another member of the flavivirus group (50). Several arguments suggest that dendritic cells could be also involved in the clearance of HCV infection. There is growing evidence that dendritic cells of chronically HCV infected patients present with an impaired antigen processing and presentation, leading to incomplete activation of HCV-specific T cells (4, 5, 28, 31). This phenomenon has also been linked to the observation that HCV could directly infect dendritic cells (46, 56). In addition, several type II C-type (calcium-dependent) lectins are expressed at the surface of dendritic cells, which have been implicated in antigen uptake and targeting toward proteasome or endoplasmic reticulum compartments. Antigen processing and association with molecules of the major histocompatibility complex class I occur in the endosplasmic reticulum before presentation to the immune system at the cell surface (23). It is thought that the CTL response is usually triggered via such a mechanism. Although no direct evidence is available so far, our present data are consistent with the fact that HCV-SP/GFP-hH1 complexes could undergo processing in the endoplasmic reticulum compartment in hepatocytes. It is therefore tempting to speculate that HCV-SP entry through the ASGP-R pathway may trigger the processing of HCV-SP-derived peptides through the major histocompatibility complex class I processing pathway and lead to antigen presentation to the immune system by hepatocytes or dendritic cells.
Certainly, other factors or receptors may also be involved in HCV-SP binding and entry into HepG2 cells. CD81 was recently reported to bind recombinant HCV E2 envelope protein (52). However, no expression of CD81 was detected by reverse transcription-PCR in the HepG2 cells that we used, whereas a clear expression was detected in Molt-4 cells (not shown). This, obviously, precluded CD81 from taking any part in HCV-SP binding or entry in HepG2 cells. The low-density lipoprotein receptor (LDL-R) has been implicated in the binding and entry of HCV into cells (1, 42) and also requires calcium for ligand binding (3). The HCV virion has been reported to complex with low-density lipoprotein and very-low-density lipoprotein in the serum (54, 64, 65). This binding of HCV to lipoproteins in the plasma has even been proposed as an explanation for the lack of detection of viral envelope proteins with various antibodies (53). We are currently not sure whether LDL-R is a direct or indirect target for HCV to enter into cells and whether HCV-SP directly interacts with this receptor or not. We also do not know whether, in addition to ASGP-R, another receptor is involved in HCV-SP/p7(−) internalization that would not be required in that of HCV-SP/p7(+). Nevertheless, the sum of our observations suggest that ASGP-R is involved in the binding and uptake of both HCV-SP preparations by human hepatocytes and that this ASGP-R should be considered a potential component of this process.
Acknowledgments
We thank C. M. Rice (Rockefeller University, New York, N.Y.) and Stephen M. Feinstone (Food and Drug Administration, Bethesda, Md.) for the gift of a plasmid containing an infectious clone of HCV and Harry B. Greenberg (Stanford University, Stanford, Calif.) and Arvind H. Patel (Institute of Virology, Glasgow, United Kingdom) for the gifts of antibodies and hybridomas. We thank Dinah S. Singer (Experimental Immunology Branch, National Cancer Institute, Bethesda, Md.) for continuous support, Edward Berger (LVD, NIAID, Bethesda, Md.), and T. Jake Liang and Theo Heller (Liver Diseases Section, NIDDK, Bethesda, Md.) for support and constructive comments. We are also grateful to Tatiana Karpova and Jim McNally (LRBGE, National Cancer Institute, Bethesda, Md.) for allowing us to use their facilities for confocal microscopy.
This work was supported in part by the Institut National de la Santé et de la Recherche Médicale (INSERM, Paris, France).
REFERENCES
- 1.Agnello, V., G. Abel, M. Elfahal, G. B. Knight, and Q.-X. Zhang. 1999. Hepatitis C virus and other flaviviridae enter cells via low density lipoprotein receptor. Proc. Natl. Acad. Sci. USA 96:12766-12771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agnello, V., R. T. Chung, and L. M. Kaplan. 1992. A role for hepatitis C virus infection in type II cryoglobulinemia. N. Engl. J. Med. 327:1490-1495. [DOI] [PubMed] [Google Scholar]
- 3.Atkins, A. R., I. M. Brereton, P. A. Kroon, H. T. Lee, and R. Smith. 1998. Calcium is essential for the structural integrity of the cysteine-rich, ligand-binding repeat of the low-density lipoprotein receptor. Biochemistry 37:1662-1670. [DOI] [PubMed] [Google Scholar]
- 4.Auffermann-Gretzinger, S., E. B. Keeffe, and S. Levy. 2001. Impaired dendritic cell maturation in patients with chronic, but not resolved, hepatitis C virus infection. Blood 97:3171-3176. [DOI] [PubMed] [Google Scholar]
- 5.Bain, C., A. Fatmi, F. Zoulim, J. P. Zarski, C. Trepo, and G. Inchauspe. 2001. Impaired allostimulatory function of dendritic cells in chronic hepatitis C infection. Gastroenterology 120:512-524. [DOI] [PubMed] [Google Scholar]
- 6.Bartenschlager, R., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631-1648. [DOI] [PubMed] [Google Scholar]
- 7.Baumert, T. F., S. Ito, D. Wong, and T. J. Liang. 1998. Hepatitis C structural proteins assemble into viruslike particles in insect cells. J. Virol. 72:3827-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bukh, J., R. H. Miller, and R. H. Purcell. 1995. Genetic heterogeneity of hepatitis C virus: quasispecies and genotypes. Semin. Liver Dis. 15:41-62. [DOI] [PubMed] [Google Scholar]
- 9.Choo, Q.-L., G. Kuo, R. Ralston, A. Weiner, D. Chien, G. Van Nest, J. Han, K. Berger, K. Thudium, C. Kuo, J. Kansopon, J. McFraland, A. Tabrizi, K. Ching, B. Moss, L. B. Cummins, M. Houghton, and E. Muchmore. 1994. Vaccination of chimpanzees against infection by the hepatitis C virus. Proc. Natl. Acad. Sci. USA 91:1294-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cocquerel, L., J.-C. Meunier, A. Pillez, C. Wychowski, and J. Dubuisson. 1998. A retention signal necessary and sufficient for endoplasmic reticulum localization maps to the transmembrane domain of hepatitis C virus glycoprotein E2. J. Virol. 72:2183-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Consiglio, E., G. Salvatore, J. E. Rall, and L. D. Kohn. 1979. Thyroglobulin interactions with thyroid plasma membranes: the existence of specific receptors and their potential role. J. Biol. Chem. 254:5065-5076. [PubMed] [Google Scholar]
- 12.Consiglio, E., S. Shifrin, Z. Yavin, F. S. Ambesi-Impiombato, J. E. Rall, G. Salvatore, and L. D. Kohn. 1981. Thyroglobulin interactions with thyroid membranes: relationship between receptor recognition of N-acetylglucosamine residues and the iodine content of thyroglobulin preparations. J. Biol. Chem. 256:10592-10599. [PubMed] [Google Scholar]
- 13.Cruz, P. E., P. L. Khalil, T. D. Dryden, H. C. Chiou, P. S. Fink, S. J. Berberich, and N. J. Bigley. 1999. A novel immunization method to induce cytotoxic-T-lymphocyte responses (CTL) against plasmid-encoded herpes simplex virus type-1 glycoprotein D. Vaccine 17:1091-1099. [DOI] [PubMed] [Google Scholar]
- 14.Deleersnyder, V., A. Pillez, C. Wychowski, K. Blight, J. Xu, Y. S. Hahn, C. M. Rice, and J. Dubuisson. 1997. Formation of native hepatitis C virus glycoprotein complexes. J. Virol. 71:697-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diao, J., N. D. Churchill, and T. I. Michalak. 1998. Complement-mediated cytotoxicity and inhibition of ligand binding to hepatocytes by woodchuck hepatitis virus-induced autoantibodies to asialoglycoprotein receptor. Hepatology 27:1623-1631. [DOI] [PubMed] [Google Scholar]
- 16.Di Jeso, B., S. Formisano, and E. Consiglio. 1999. Depletion of divalent cations within the secretory pathway inhibits the terminal glycosylation of complex carbohydrates of thyroglobulin. Biochimie 81:497-504. [DOI] [PubMed] [Google Scholar]
- 17.Dodd, R. B., and K. Drickamer. 2001. Lectin-like proteins in model organisms: implications for evolution of carbohydrate-binding activity. Glycobiology 11:71R-79R. [DOI] [PubMed] [Google Scholar]
- 18.Dotzauer, A., V. Gebhardt, K. Bieback, U. Göttke, A. Kracke, J. Mages, S. M. Lemon, and A. Vallbracht. 2000. Hepatitis A virus-specific immunoglobulin A mediates infection of hepatocytes with hepatitis A virus via the asialoglycoprotein receptor. J. Virol. 74:10950-10957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dubuisson, J. 2000. Folding, assembly and subcellular localization of hepatitis C virus glycoproteins. Curr. Top. Microbiol. Immunol. 242:135-148. [DOI] [PubMed] [Google Scholar]
- 20.Dubuisson, J., H. H. Hsu, R. C. Cheung, H. B. Greenberg, D. G. Russell, and C. M. Rice. 1994. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J. Virol. 68:6147-6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harada, T., N. Tautz, and H.-J. Thiel. 2000. E2-p7 region of the bovine viral diarrhea virus polyprotein: processing and functional studies. J. Virol. 74:9498-9506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual, p. 274-275. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 23.Hartgers, F. C., C. G. Figdor, and G. J. Adeama. 2000. Towards a molecular understanding of dendritic cell immunobiology. Immunol. Today 21:542-545. [DOI] [PubMed] [Google Scholar]
- 24.Hoofnagle, J. H. 1997. Hepatitis C: the clinical spectrum of disease. Hepatology 26:15S-20S. [DOI] [PubMed] [Google Scholar]
- 25.Husa, P., P. Chalupa, H. Stroblova, L. Husova, P. Slesinger, and J. Zajic. 2001. Autoantibodies to asialoglycoprotein receptor in chronic hepatitis C patients. Acta Virol. 45:7-11. [PubMed] [Google Scholar]
- 26.Ishii, K., D. Rosa, Y. Watanabe, T. Katayama, H. Harada, C. Wyatt, K. Kiyosawa, H. Aizaki, Y. Matsuura, M. Houghton, S. Abrignani, and T. Miyamura. 1998. High titers of antibodies inhibiting the binding of envelope to human cells correlate with natural resolution of chronic hepatitis C. Hepatology 28:1117-1120. [DOI] [PubMed] [Google Scholar]
- 27.Jarvis, D. L., Z. S. Kawar, and J. R. Hollister. 1998. Engineering N-glycosylation pathways in the baculovirus-insect cell system. Curr. Opin. Biotechnol. 9:528-533. [DOI] [PubMed] [Google Scholar]
- 28.Kakumu, S., S. Ito, T. Ishikawa, Y. Mita, T. Tagaya, Y. Fukuzawa, and K. Yoshioka. 2000. Decreased function of peripheral blood dendritic cells in patients with hepatocellular carcinoma with hepatitis B and C virus infection. J. Gastroenterol. Hepatol. 15:431-436. [DOI] [PubMed] [Google Scholar]
- 29.Kamhi-Nesher, S., M. Shenkman, S. Tolchinsky, S. V. Fromm, R. Ehrlich, and G. Z. Lederkremer. 2001. A novel quality control compartment derived from the endoplasmic reticulum. Mol. Biol. Cell 10:1711-1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanto, T., N. Hayashi, T. Takehara, H. Hagiwara, E. Mita, M. Naito, A. Kasahara, H. Fusamoto, and T. Kamada. 1995. Density analysis of hepatitis C particle population in the circulation of infected hosts: implications for virus neutralization or persistence. J. Hepatol. 22:440-448. [DOI] [PubMed] [Google Scholar]
- 31.Kanto, T., N. Hayashi, T. Takehara, T. Tatsumi, N. Kuzushita, A. Ito, Y. Sasaki, A. Kasahara, and M. Hori. 1999. Impaired allostimulatory capacity of peripheral blood dendritic cells recovered from hepatitis C virus-infected individuals. J. Immunol. 162:5584-5591. [PubMed] [Google Scholar]
- 32.Kolykhalov, A. A., E. V. Agapov, K. J. Blight, K. Mihalik, S. M. Feinstone, and C. M. Rice. 1997. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science 277:570-574. [DOI] [PubMed] [Google Scholar]
- 33.Lauer, G. M., and B. D. Walker. 2001. Hepatitis C virus infection. N. Engl. J. Med. 345:41-52. [DOI] [PubMed] [Google Scholar]
- 34.Layden, T. J., B. Mika, and T. E. Wiley. 2000. Hepatitis C kinetics: mathematical modeling of viral response to therapy. Semin. Liver Dis. 20:173-183. [DOI] [PubMed] [Google Scholar]
- 35.Lechner, F., J. Sullivan, H. Spiegel, D. F. Nixon, B. Ferrari, A. Davis, B. Borkowsky, H. Pollack, E. Barnes, G. Dusheiko, and P. Klenerman. 2000. Why do cytotoxic T lymphocytes fail to eliminate hepatitis C virus? Lessons from studies using major histocompatibility complex class I peptide tetramers. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355:1085-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin, C., B. D. Lindenbach, B. M. Pragai, D. W. McCourt, and C. M. Rice. 1994. Processing in the hepatitis C virus E2-NS2 region: identification of p7 and two distinct E2-specific products with different C termini. J. Virol. 68:5063-5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lowe, J. B. 2001. Glycosylation, immunity, and autoimmunity. Cell 104:809-812. [DOI] [PubMed] [Google Scholar]
- 38.Manns, M. P., J. G. McHutchison, S. C. Gordon, V. K. Rustgi, M. Shiffman, R. Reindollar, Z. D. Goodman, K. Koury, M. Ling, and J. K. Albrecht. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958-965. [DOI] [PubMed] [Google Scholar]
- 39.Marchal, I., D. L. Jarvis, R. Cacan, and A. Verbert. 2001. Glycoproteins from insect cells: sialylated or not? Biol. Chem. 382:151-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McFarlane, B. M., C. G. McSorley, D. Vergani, I. G. McFarlane, and R. Williams. 1986. Serum autoantibodies reacting with the hepatic asialoglycoprotein receptor protein (hepatic lectin) in acute and chronic liver disorders. J. Hepatol. 3:196-205. [DOI] [PubMed] [Google Scholar]
- 41.Meier, M., M. D. Bider, V. N. Malashkevich, M. Spiess, and P. Burkhard. 2000. Crystal structure of the carbohydrate recognition domain of the H1 subunit of the asialoglycoprotein receptor. J. Mol. Biol. 300:857-865. [DOI] [PubMed] [Google Scholar]
- 42.Monazahian, M., I. Böhme, S. Bonk, A. Koch, C. Scholz, S. Grethe, and R. Thomssen. 1999. Low-density lipoprotein receptor as a candidate receptor for hepatitis C virus. J. Med. Virol. 57:223-229. [DOI] [PubMed] [Google Scholar]
- 43.Montuori, N., F. Pacifico, S. Mellone, D. Liguoro, B. Di Jeso, S. Formisano, F. Gentile, and E. Consiglio. 2000. The rat asialoglycoprotein receptor binds the amino-terminal domain of thyroglobulin. Biochem. Biophys. Res. Commun. 268:42-46. [DOI] [PubMed] [Google Scholar]
- 44.Moser, M., and K. M. Murphy. 2000. Dendritic cell regulation of TH1-TH2 development. Nat. Immunol. 1:199-205. [DOI] [PubMed] [Google Scholar]
- 45.Muhle, T., K. W. Wenzel, and E. Hofmann. 1987. Binding and uptake of asialothyroglobulin by isolated rat hepatocytes. Biomed. Biochim. Acta 46:479-486. [PubMed] [Google Scholar]
- 46.Navas, M. C., A. Fuchs, E. Schvoerer, A. Bohbot, A. M. Aubertin, and F. Stoll-Keller. 2002. Dendritic cells susceptibility to hepatitis C virus genotype 1 infection. J. Med. Virol. 67:152-161. [DOI] [PubMed] [Google Scholar]
- 47.Owsianka, A., R. F. Clayton, L. D. Loomis-Price, J. A. McKeating, and A. H. Patel. 2001. Functional analysis of hepatitis C virus E2 glycoproteins and virus-like particles reveals structural dissimilarities between different forms of E2. J. Gen. Virol. 82:1877-1883. [DOI] [PubMed] [Google Scholar]
- 48.Pacifico, F., D. Liguoro, R. Acquaviva, S. Formisano, and E. Consiglio. 1999. Thyroglobulin binding and TSH regulation of the RHL-1 subunit of the asialoglycoprotein receptor in rat thyroid. Biochimie 81:493-496. [DOI] [PubMed] [Google Scholar]
- 49.Pacini, F., and A. Pinchera. 1999. Serum and tissue thyroglobulin measurement: clinical applications in thyroid disease. Biochimie 81:463-467. [DOI] [PubMed] [Google Scholar]
- 50.Palucka, A. K. 2000. Dengue virus and dendritic cells. Nat. Med. 6:748-749. [DOI] [PubMed] [Google Scholar]
- 51.Pape, G. R., T. J. Gerlach, H. M. Diepolder, N. Gruner, M. Jung, and T. Santantonio. 1999. Role of the specific T-cell response for clearance and control of hepatitis C virus. J. Viral Hepat. 6:36-40. [DOI] [PubMed] [Google Scholar]
- 52.Pileri, P., Y. Uematsu, S. Campagnoli, G. Galli, F. Falugi, R. Petracca, A. Weiner, M. Houghton, D. Rosa, G. Grandi, and S. Abrignani. 1998. Binding of hepatitis C virus to CD81. Science 282:938-941. [DOI] [PubMed] [Google Scholar]
- 53.Prince, A. M. 1994. Challenges for development of hepatitis C virus vaccines. FEMS Microbiol. Rev. 14:273-277. [DOI] [PubMed] [Google Scholar]
- 54.Prince, A. M., T. Huima-Byron, T. S. Parker, and D. M. Levine. 1996. Visualization of hepatitis C virions and putative defective interfering particles isolated from low-density lipoproteins. J. Viral Hepat. 3:11-17. [DOI] [PubMed] [Google Scholar]
- 55.Rocco, A., S. Gargano, A. Provenzano, M. Nardone, G. M. De Sanctis, N. Altavilla, L. V. Chircu, and F. Grimaldi. 2001. Incidence of autoimmune thyroiditis in interferon-alpha treated and untreated patients with chronic hepatitis C virus infection. Neuroendocrinol. Lett. 22:39-44. [PubMed] [Google Scholar]
- 56.Sarobe, P., J. J. Lasarte, N. Casares, A. Lopez-Diaz de Cerio, E. Baixeras, P. labarga, N. Garcia, F. Borras-Cuesta, and J. Prieto. 2002. Abnormal priming of CD4+ T cells by dendritic cells expressing hepatitis C virus core and E1 proteins. J. Virol. 76:5062-5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmidt, W. N., J. T. Stapleton, D. R. LaBrecque, F. A. Mitros, K. Kirkegaard, M. J. P. Phillips, and D. Brashear. 2000. Hepatitis C virus (HCV) infection and cryoglobulinemia: analysis of whole blood and plasma HCV-RNA concentrations and correlation with liver histology. Hepatology 31:737-744. [DOI] [PubMed] [Google Scholar]
- 58.Shenkman, M., M. Ehrlich, and G. Z. Lederkremer. 2000. Masking of an endoplasmic reticulum retention signal by its presence in the two subunits of the asialoglycoprotein receptor. J. Biol. Chem. 275:2845-2851. [DOI] [PubMed] [Google Scholar]
- 59.Shifrin, S., E. Consiglio, P. Laccetti, G. Salvatore, and L. D. Kohn. 1982. Bovine thyroglobulin: 27S iodoprotein interactions with thyroid membranes and formation of a 27S iodoprotein in vitro. J. Biol. Chem. 257:9539-9547. [PubMed] [Google Scholar]
- 60.Shifrin, S., E. Consiglio, and L. D. Kohn. 1983. Effect of the complex carbohydrate moiety on the structure of thyroglobulin. J. Biol. Chem. 258:3780-3786. [PubMed] [Google Scholar]
- 61.Spaete, R. R., D. Alexander, M. E. Rugroden, Q. L. Choo, K. Berger, K. Crawford, C. Kuo, S. Leng, C. Lee, R. Ralston, et al. 1992. Characterization of the hepatitis C virus E2/NS1 gene product expressed in mammalian cells. Virology 188:819-830. [DOI] [PubMed] [Google Scholar]
- 62.Stockert, R. J. 1995. The asialoglycoprotein receptor: relationships between structure, function, and expression. Physiol. Rev. 75:591-609. [DOI] [PubMed] [Google Scholar]
- 63.Stoorvogel, W., H. J. Geuze, and G. J. Strous. 1987. Sorting of endocytosed transferrin and asialoglycoprotein occurs immediately after internalization in HepG2 cells. J. Cell Biol. 104:1261-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thomssen, R., S. Bonk C. Propfe, K. H. Heermann, H. G. Köchel, and A. Uy. 1992. Association of hepatitis C virus in human sera with beta-lipoprotein. Med. Microbiol. Immunol. 181:293-300. [DOI] [PubMed] [Google Scholar]
- 65.Thomssen, R., S. Bonk, and A. Thiele. 1993. Density heterogeneities of hepatitis C virus in human sera due to the binding of beta-lipoproteins and immunoglobulins. Med. Microbiol. Immunol. 182:329-334. [DOI] [PubMed] [Google Scholar]
- 66.Triyatni, M., J. Vergalla, A. R. Davis, K. G. Hadlock, S. K. Foung, and T. J. Liang. 2002. Structural features of envelope proteins on hepatitis C virus-like particles as determined by anti-envelope monoclonal antibodies and CD81 binding. Virology 298:124-132. [DOI] [PubMed] [Google Scholar]
- 67.Triyatni, M., B. Saunier, P. Maruvada, A. R. Davis, L. Ulianich, T. Heller, A. Patel, L. D. Kohn, and T. J. Liang. 2002. Interaction of hepatitis C virus-like particles and cells: a model system for studying viral binding and entry. J. Virol. 76:9335-9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ulianich, L., K. Suzuki, A. Mori, M. Nakazato, M. Pietrarelli, P. Goldsmith, F. Pacifico, E. Consiglio, S. Formisano, and L. D. Kohn. 1999. Follicular thyroglobulin (TG) suppression of thyroid-restricted genes involved the apical membrane asialoglycoprotein receptor and TG phosphorylation. J. Biol. Chem. 274:25099-25107. [DOI] [PubMed] [Google Scholar]
- 69.Valladeau, J., V. Duvert-Frances, J. J. Pin, M. J. Kleijmer, S. Ait-Yahia, O. Ravel, et al. 2001. Immature human dendritic cells express asialoglycoprotein receptor isoforms for efficient receptor-mediated endocytosis. J. Immunol. 167:5767-5774. [DOI] [PubMed] [Google Scholar]
- 70.Zignego, A. L., M. De Carli, M. Monti, G. Careccia, G. La Villa, C. Giannini, M. M. D'Elios, G. Del Prete, and P. Gentilini. 1995. Hepatitis C virus infection of mononuclear cells from peripheral blood and liver infiltrates in chronically infected patients. J. Med. Virol. 47:58-64. [DOI] [PubMed] [Google Scholar]