Abstract
Hepatitis C virus (HCV) is a positive-strand RNA virus that encodes a helicase required for viral replication. Although HCV does not replicate through a DNA intermediate, HCV helicase unwinds both RNA and DNA duplexes. An X-ray crystal structure of the HCV helicase complexed with (dU)8 has been solved, and the substrate-amino acids interactions within the catalytic pocket were shown. Among these, residues W501 and V432 were reported to have base stacking interactions and to be important for the unwinding function of HCV helicase. It has been hypothesized that specific interactions between the enzyme and substrate in the catalytic pocket are responsible for the substrate specificity phenotype. We therefore mutagenized W501 and V432 to investigate their role in substrate specificity in HCV helicase. Replacement of W501, but not V432, with nonaromatic residues resulted in complete loss of RNA unwinding activity, whereas DNA unwinding activity was largely unaffected. The loss of unwinding activity was fully restored in the W501F mutant, indicating that the aromatic ring is crucial for RNA helicase function. Analysis of ATPase and nucleic acid binding activities in the W501 mutant enzymes revealed that these activities are not directly responsible for the substrate specificity phenotype. Molecular modeling of the enzyme-substrate interaction at W501 revealed a putative π-facial hydrogen bond between the 2′-OH of ribose and the aromatic tryptophan ring. This evidence correlates with biochemical results suggesting that the π-facial bond may play an important role in the RNA unwinding activity of the HCV NS3 protein.
Hepatitis C virus (HCV) is one of the major etiological agents of chronic hepatitis worldwide. Although acute hepatitis is rarely detected at the early stage of infection, approximately 80% of HCV-infected individuals develop chronic infection that can lead to liver disease, including liver cirrhosis and hepatocellular carcinoma (1). An estimated 170 million people are HCV carriers and are at risk of liver disease (42). In the United States alone, HCV is responsible for 10,000 deaths per year, and this rate is expected to increase without effective intervention (2). Currently, alpha interferon and ribavirin are the only approved HCV therapies, but only ∼40% of cases show a sustained response (5, 21).
HCV belongs to its own genus in the family Flaviviridae, which also includes the Flavivirus and Pestivirus genera of positive-strand RNA viruses (28). Six genotypes have thus far been assigned in the Hepacivirus genus, and they include more than 100 different subtypes (31). HCV contains approximately a 9.6-kb genome and encodes a long open reading frame flanked by 5′ and 3′ untranslated regions (UTRs). The 5′ UTR is 341 nucleotides long, is highly conserved (9), and contains an internal ribosome entry site for polyprotein translation (39). The 3′ UTR consists of three regions: a type-specific variable region, a poly(U) tract of variable length, and a 98-nucleotide 3′ X-tail (14, 38). On infection, the HCV genome is translated into a single polyprotein of 3010 amino acids that is processed into structural (capsid, E1, E2, and P7) and nonstructural (NS) (NS2, NS3, NS4a, NS4b, NS5a, and NS5b) proteins (27). NS3 is a bifunctional enzyme with a serine protease localized to the N-terminal 181 residues of the protein and a helicase located in the C-terminal 456 amino acids (7, 8). This enzyme is considered to be an important component of the replication complex, which includes other viral nonstructural proteins and host factor(s).
Helicases are ubiquitous enzymes that catalyze the unwinding of double-stranded DNA (dsDNA) and dsRNA during various biological processes such as replication, translation, repair, and recombination (20). A large number of helicases have been identified and divided into superfamilies based on their amino acid sequences and conserved motifs (6). Helicases are also classified as DNA or RNA helicases according to their ability to unwind dsDNA, dsRNA, or mixed duplex substrates. The HCV NS3 helicase unwinds both dsRNA and dsDNA in the 3′-to-5′ direction; thus, it is both an RNA helicase and a DNA helicase. The bovine viral diarrhea virus (BVDV) NS3 helicase, the closest relative to HCV NS3 helicase based on amino acid sequence homology, is an RNA helicase, unwinding the RNA duplex only. Generally, RNA helicases cannot be distinguished from DNA helicases simply on the basis of their amino acid sequence because the two helicases share conserved amino acid sequence motifs and have similar three-dimensional structures. As an example, the HCV helicase which unwinds both RNA and DNA duplexes is similar to bacterial DNA helicases such as PcrA and Rep (15). It is possible that the structural difference between RNA and DNA helicases could be small and confined to the substrate binding site within the catalytic pocket.
Analysis of the X-ray crystal structure of an HCV NS3 helicase revealed that the enzyme is triangular, having three nearly equal-sized domains (13, 43). Domain 1 is known as the NTPase domain and contains Walker A (264AXXXXGKS211) and B (290DEXH293) motifs. Domain 2 contains a conserved R-rich motif (460QRRGRTGR467) and was previously known as the RNA binding domain. Domain 3 has a predominantly alpha-helical structure and is referred to as the helical domain. Korolev et al. compared the structure of HCV helicase to that of the better-defined Escherichia coli Rep enzyme and found overall structural similarities despite little sequence homology between the two enzymes (15). They pointed out that the interdomain cleft in HCV helicase is similar to the one in Rep helicase; therefore, it might be important for substrate binding and catalysis. In particular, their study predicted that W501 of HCV helicase in domain 3 would be a capping residue within the catalytic pocket that is essential for substrate binding, translocation, and unwinding of the enzyme. Recently, Kim et al. determined the structure of HCV helicase complexed with a single-stranded (dU)8 oligonucleotide (13) and confirmed the prediction of Korolev et al. that the substrate binds in a groove that separates domain 3 from domains 1 and 2. In their structure, most of the interaction between the enzyme and the bound substrate involves hydrogen bonds with the phosphate backbone but not the bases of the substrate. The most significant enzyme-base interaction involves two hydrophobic stacking interactions between W501 and a base near the 3′ end of the bound substrate and between V432 and a base near the 5′ end of the substrate. Based on these findings, Kim et al. proposed a monomeric “ratchet model” for the mechanism of the HCV helicase function. Thus, both V432 and W501 define the border of the substrate binding cavity and are important for the binding, base interruption, and migration of the helicase in the 3′-to-5′ direction.
Based on these findings, we reasoned that W501 and V432 could also play an important role in determining the substrate specificity of HCV NS3 helicase. To investigate this possibility, we prepared mutant enzymes of these residues and compared RNA unwinding activity with DNA unwinding activity by using specific substrates containing HCV sequence. The results of our biochemical study demonstrate that presence of W501 residue within the catalytic site is critical to the RNA helicase function but not to the DNA helicase function of HCV NS3 protein. Further analysis of the enzyme-substrate interaction by molecular modeling suggests that the aromatic ring of W501 might form a π-facial hydrogen bond with the 2′-OH of the ribose within the RNA substrate. In this report we discuss the significance of our findings with respect to the mechanism of the unwinding function and the substrate specificity of the HCV NS3 helicase.
MATERIALS AND METHODS
Site-directed mutagenesis of NS3 helicase.
The mutant NS3 helicases were constructed from the expression vector pETbNS3H, which contains the C-terminal NS3 protein from amino acids 167 to 632 (12). Site-directed mutagenesis was carried out using a Transformer mutagenesis kit (Clontech) as specified by the manufacturer, and substituted sequences were confirmed by automatic sequencing (Applied Biosystem 373 DNA sequencer).
Expression and purification of NS3 helicase.
Wild-type and mutant NS3 helicases were expressed in E. coli BL21(DE3) and purified as described by Kim et al. (12), except for the following modifications. Briefly, E. coli cells were grown to an optical density 600 nm of 0.8 in 500 ml of Luria-Bertani broth plus 25 mg of ampicillin at 37°C and then induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 to 5 h at 25°C. All procedures for the protein purification were performed at 4°C. E. coli cells were harvested by centrifugation at 6,000 × g for 10 min. The harvested cells were resuspended with 30 ml of buffer H (20 mM Tris-HCl [pH 7.9], 0.5 M NaCl, 10% glycerol) containing 10 mM imidazole and then disrupted by sonication. The soluble fraction was obtained by centrifugation at 27,000 × g for 60 min.
The NS3 helicase was then purified from the soluble fraction by nickel affinity chromatography followed by DEAE ion-exchange chromatography. The soluble fraction was first absorbed to 0.5 ml of Ni2+-charged His-bind resin for 4 h. The resin was packed in a column and washed with 10 bed volumes of buffer H, 20 bed volumes of buffer H containing 15 mM imidazole, and finally 6 bed volumes of buffer H containing 25 mM imidazole. NS3 helicase was eluted with buffer H containing 100 mM imidazole. The eluted NS3 helicase solution was desalted with buffer P (20 mM Tris-HCl [pH 7.9], 10% glycerol) on a PD-10 column (Pharmacia). The desalted solution was further purified by high-performance liquid chromatography using a Protein-Pak 5PW (Waters; 7.5 by 75 mm) anion-exchange column. The column was eluted with a linear gradient of NaCl from 0 to 1 M in buffer P at a flow rate of 0.5 ml per min. The purity of NS3 helicase was ascertained by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with a gel stained with Coomassie brilliant blue R. The purified proteins were stored at −20°C in buffer S (20 mM Tris-Cl [pH 7.9], 0.5 M NaCl, 40% glycerol). The final protein concentration was determined by the Bradford protein assay (Bio-Rad) using bovine serum albumin (BSA) as a standard.
Preparation of helicase substrates.
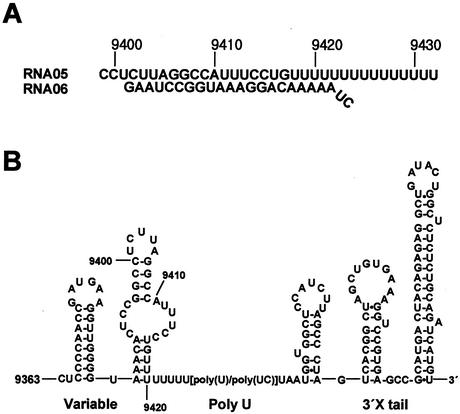
The substrates of the partially double-stranded helicase assay were obtained by annealing RNA or DNA synthetic oligonucleotides whose sequences were derived from the 3′-untranslated region of the HCV genome (see Fig. 2) (14, 38). To form a double-stranded substrate, the release strand (06) was labeled with [γ-32P]ATP by using T4 polynucleotide kinase and then both the template strand (05) and the release strand were mixed in equimolar concentrations (0.5 μM) in 20 mM Tris buffer (pH 8.0) containing 1 mM EDTA and 0.1 M NaCl. The mixture was heated at 90°C for 5 min and then cooled to 30°C for 60 min in a thermocycler. The annealed duplex was confirmed by gel analysis.
FIG. 2.
Partially double-stranded nucleic acid substrates used in the enzyme assays. (A) An RNA substrate. The template strand (05) and the release strand (06) were chemically synthesized in both ssRNA and ssDNA and annealed to form a dsRNA or a dsDNA substrate. The 5′ end of the release strand was first labeled with [γ-32P]ATP before annealing. The numbers above the substrate represent the position of the nucleotide in the HCV genome. (B) The nucleotide sequence and the secondary structure of the 3′ end of HCV genome. The portion of the sequence that was used as the substrate is numbered.
Helicase assay.
The helicase assay was performed as previously described (12) with modifications. Briefly, the assay was performed with 20 μl of reaction buffer containing 50 mM MOPS-KOH (pH 6.5), 1 mM ATP, 1 mM MgCl2, 2 mM dithiothreitol (DTT), 2 μg of BSA, 2 to 5 U of RNasin, 0.1 pmol of32P-labeled substrate, and the indicated concentrations of NS3 helicase. After preincubation for 10 min at 25°C, the unwinding reaction was carried out at 37°C for 20 min. The reaction was started by adding NS3 helicase and stopped by adding 5 ml of termination buffer (0.1 M Tris-HCl [pH 7.5], 20 mM EDTA, 0.5% SDS, 0.1% NP-40, 0.1% bromphenol blue, 0.1% xylene cyanol). The reaction product was separated on a 15% native polyacrylamide gel immediately following termination and then quantified using a BAS 2000 phosphorimager (Fuji).
ATPase activity assay.
ATPase activity was determined by monitoring the hydrolysis of [γ-32P]ATP to ADP by thin-layer chromatography as described previously (12) with the following modification. The reaction mixture (10 μl) contained 25 mM morpholinepropanesulfonic acid (MOPS)-NaOH (pH 7.0), 2 μg of BSA, 0.1 M DTT, 0.1 M ATP, 1 μCi of [γ-32P]ATP, and 1 μg of poly(U) or the test oligonuclotides (ssRNA or ssDNA) at 10 μM. The reaction mixture was preincubated at 25°C for at least 10 min. The reaction was started by adding 200 nM [in the absence of oligonucleotides or poly(U)] or 20 nM [in the presence of oligonucleotides or poly(U)] NS3 helicase and stopped with 2 μl of 0.1 M EDTA (pH 8.0) after incubation for 30 min at 37°C. The reaction product was spotted on a polyethyleneimine-cellulose plate, separated in 0.375 M potassium phosphate (pH 3.5), and quantified using a BAS 2000 phosphorimager.
Binding assay.
The gel retardation reaction was examined in a 20-μl reaction mixture containing 0.1 pmol of 32P-labeled ssRNA or ssDNA as a substrate, 50 mM MOPS-KOH (pH 6.5), 2 mM DTT, 2 μg of BSA, and 2 to 5 U of RNasin. After incubation for 30 min at 37°C, the reaction mixture was electrophoresed on a 6% native polyacrylamide gel containing 0.25× Tris-borate-EDTA. The oligonucleotide-NS3 helicase complex was quantified using a BAS 2000 phosphorimager.
Computer-aided three-dimensional analysis of the enzyme-substrate complex.
The structure of the HCV NS3 RNA helicase domain complexed with the ssDNA oligonucleotide (PDB entry 1aiv [13]) provided the initial coordinates for the molecular modeling studies. Mutations and missing residues or atoms in the protein or nucleic acid were generated and locally optimized if needed (constraining all other atoms) using SYBYL 2000 (Tripos). Water molecules and counterions were removed, and covalently modified cysteine residues were converted back to cysteine. Based on the coordinates of the deoxyuracil octamer found in the complex, the oligonucleotides (dT)6 and U6 served as the DNA and RNA substrates, respectively. The first and second residues were deleted because their electron densities were not detected in 1aiv and their positions are not critical for interaction with the enzyme.
We modeled four substrate-enzyme complexes: RNA-W501, RNA-W501A, DNA-W501, and DNA-W501A. The models were neutralized with Na+ counterions and then solvated in a TIP3 water box with edges at least 10 Å from any atoms in the complex by using the Xleap module in Amber 7 (3). Minimization was performed using the Sander module in Amber 7 with parm98 all-atom parameters (3) by the following procedure: 2000 steps steepest-descent (SD) minimization with a 5,000-kcal/mol harmonic restraint on all solute atoms and all bonds to hydrogen constrained using SHAKE (tolerance = 0.000001); 1,000 steps SD with a 50-kcal/mol restraint on all atoms; 1,000 steps SD with a 10-kcal/mol restraint on all atoms; and 10,000 steps SD with no restraint. Root mean square deviations (in ångstroms) of protein backbone and phosphoribose heavy atoms were as follows: DNA-W501A, 0.49 and 0.82; DNA-W501, 0.49 and 0.80; RNA-W501A, 0.49 and 0.87; and RNA-W501, 0.50 and 0.92. SYBYL was used to visualize the final structure and to identify hydrogen bonds.
RESULTS
Expression and purification of the HCV NS3 helicase domain.
To study the role played by amino acids V432 and W501 on the HCV NS3 helicase function, we produced a total of seven mutants. These were V432A, V432D, V432R, W501A, W501E, W501R, and W501F. Construction of a pET-based expression vector encoding the HCV NS3 helicase domain as a fusion protein with a histidine tag was described previously (12). Expression of the recombinant wild-type and mutant proteins was induced by the addition of IPTG, and the recombinant proteins in the E. coli lysate (Fig. 1, lane 2) were purified by Ni-agarose column and DEAE column chromatography. Normalized amounts of each protein were analyzed by SDS-PAGE and Coomassie blue staining to ensure that similar quantities of mutant enzymes were used in the comparative study. The purified proteins were homogeneous as determined by SDS-PAGE (lanes 4 to 11).
FIG. 1.
Expression and purification of the wild-type and HCV NS3 helicases from E. coli. The recombinant proteins were analyzed by SDS-PAGE (10% polyacrylamide) followed by Coomassie brilliant blue R-250 staining. Lanes: 1, molecular size maker; 2, soluble fraction of total E. coli lysate; 3, peak fraction of 100 mM imidazole eluate from the nickel-agarose column; 4 to 11, peak high-performance liquid chromatography fraction containing the wild-type (lane 4) and the mutant HCV helicases (lanes 5 to 11) eluted from the Protein-Pak 5PW anion-exchange column. The mutant proteins were V432A, V432D, V432R, W501A, W502E, W502R, and W501F (lanes 5 to 11, respectively).
RNA and DNA helicase substrate.
In our experiment, we employed the native HCV-1a sequence as the helicase substrate (Fig. 2). The substrate sequence contains a variable region and a portion of the poly(U) tract within the 3′ UTR of the HCV-1a genome (14, 38). Although the replication initiation site within the HCV genome in vivo is unknown, we speculated that a poly(U) tract near the 3′ end of the HCV genome could constitute an entry site for the replication complex, which should contain host and viral factors including the NS3 and the NS5B proteins. This speculation was based on the fact that HCV helicase strongly prefers poly(U) for the stimulation of ATPase activity (8, 34, 37). It also has high binding affinity to poly(U), a property which is being widely exploited to purify the recombinant HCV helicase by using a poly(U) affinity column. The partially dsRNA substrate (05R/06R) and dsDNA substrate (05D/06D) shown in Fig. 2 were made by annealing synthetic oligonucleotides. These substrates obviously differ in the sugar moiety but are identical in nucleotide sequence except for U's in the RNA and the corresponding T's in the DNA.
RNA and DNA helicase activities in wild-type HCV NS3 helicase.
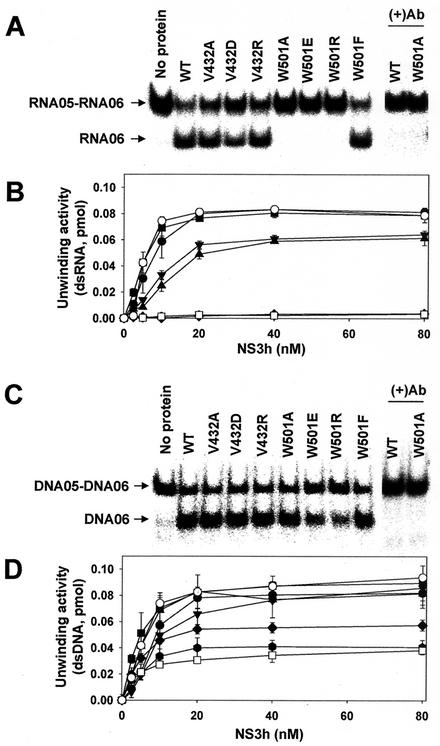
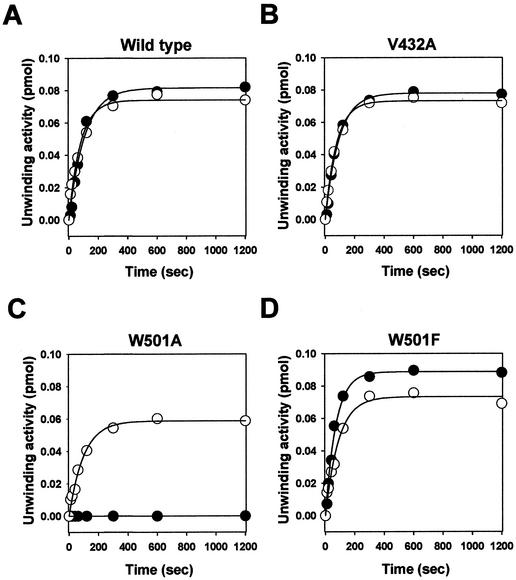
The RNA and DNA helicase activities of the NS3 enzyme were measured by using the dsRNA (05R/06R) or dsDNA substrate (05D/06D). Titration of the wild-type NS3 helicase under standard reaction conditions showed that the unwinding activity was linear up to 10 nM, reaching a plateau around 20 nM at about a 90% strand displacement (Fig. 3A and B). Similar results were obtained with the corresponding dsDNA substrate (Fig. 3C and D). The addition of NS3-specific polyclonal antibody at a 1/500 dilution completely abolished the strand displacement activity (Fig. 3A and C), demonstrating that no contaminating E. coli proteins in the enzyme preparations contributed to the unwinding activities shown in our experiments. Our data with the wild-type enzyme agree with other published data (18, 25, 36). The time courses of the wild-type enzyme reactions and unwinding kinetics are shown in Fig. 4A and Table 1. As indicated, the unwinding rates were similar for the two helicase activities but the unwinding amplitude of RNA helicase activity was slightly higher (∼10%) than that of the DNA helicase activity in the wild-type enzyme.
FIG. 3.
Unwinding activity of the wild-type and mutant HCV NS3 helicases on dsRNA or dsDNA substrate. (A) PAGE analysis of unwinding activity on dsRNA. A standard helicase reaction product containing 10 nM each indicated enzyme was analyzed on a native 15% polyacrylamide gel. In lanes (+)Ab, monoclonal antibody against wild-type HCV NS3 helicase was preincubated with the indicated enzyme before the enzyme reaction. (B) The dependence of the dsRNA unwinding activities on enzyme concentrations. The enzyme activity was measured for different concentration (0 to 80 nM) in the presence of dsRNA. (C) PAGE analysis of unwinding activity on dsDNA. (D) The dependence of the dsDNA unwinding activity on enzyme concentrations. The unwinding activities were quantified by determining the ratio between the amount of radioactivity associated with the release strand (RNA05 or DNA05) and the total radioactivity. The error bars represent the standard errors for two or more measurements. Symbols: •, wild type; ▪, V432A; ▴, V432D; ▾, V432R; ⧫, W501A; , W501E; □, W501R; ○, W501F.
FIG. 4.
The time course of the unwinding reaction. The unwinding reaction was performed with dsRNA (•) or dsDNA (○). The reaction was started by adding 20 nM HCV NS3 helicase and stopped by adding a termination buffer. The reaction product was resolved on a native 15% polyacrylamide gel immediately after incubation for the indicated time and then quantified using a phosphorimager. The solid lines represent nonlinear least-squares fits of the data to the exponential equation.
TABLE 1.
Kinetic parameters of the time course unwinding assaya
| NS3 helicase | RNA05-RNA06 |
DNA05-DNA06 |
||
|---|---|---|---|---|
| 10−2A (pmol)b | 10−2kobs (s−1) | 10−2A (pmol) | 10−2kobs (s−1) | |
| Wild type | 8.2 ± 0.2 | 0.94 ± 0.09 | 7.4 ± 0.2 | 1.28 ± 0.13 |
| V432A | 7.8 ± 0.2 | 1.08 ± 0.09 | 7.3 ± 0.1 | 1.32 ± 0.06 |
| W501A | NDb | ND | 5.9 ± 0.2 | 1.02 ± 0.09 |
| W501F | 8.9 ± 0.2 | 1.40 ± 0.09 | 7.3 ± 0.2 | 1.12 ± 0.11 |
The data were fit to the following equation to obtain the kinetic parameters from Fig. 3: f(t) = A [1 − exp(−kobs t)], where f(t) is the amplitude at time t, A is the amplitude of unwinding, and kobs is rate constant.
ND, nondetectable.
RNA and DNA helicase activities in mutant HCV NS3 helicases.
Mutant enzymes with amino acid substitutions at V432 and W501 showed various degrees of unwinding activities (Fig. 3). With the RNA substrate, all three W501 mutants (W501A, W501E, and W501R) showed no detectable unwinding activity over the background (<3% of that of the wild-type enzyme) throughout the enzyme concentrations tested (Fig. 3A and B). However, replacement of W501 by Phe (W501F) led to 120% of wild-type activity. These data were consistent with other published results (18, 25, 36) and with what was predicted from the three-dimensional structure by Kim et al. (13). These results suggest that W501 is critical for the RNA helicase activity of the HCV NS3 protein. In contrast to the prediction made by Kim et al. (13), all three V432 mutants were active. The unwinding activity of V432A was almost indistinguishable from that of the wild-type enzyme. Even in the V432D and V432R mutants, in which a negatively or a positively charged amino acid was introduced, the unwinding activity remained at about 70% of that of the wild-type enzyme (Fig. 3A and B). Taken together, these results suggest that V432 is not required for the RNA helicase activity of the HCV NS3 enzyme. The same result for the V432A mutant has been published by Tai et al. (36).
With the DNA substrate, all three W501 mutants (W501A, W501E, and W501R) showed significant unwinding activities ranging from 50 to 80% of that of the wild-type enzyme (Fig. 3C and D). Kinetic analysis showed that both the amplitude and the rate of unwinding of the W501A mutant were about 80% of those of the wild-type enzyme (Table 1). As was the case with the RNA substrate, all three V432 mutants showed strand displacement activities similar to that of the wild-type enzyme in the DNA substrate. The results with the V432 mutants were not consistent with the results of a biochemical analysis reported by Preugschat et al. (25), where strand displacement activity with a DNA substrate was measured by a fluorescence-based unwinding assay. The source of this discrepancy is not clear but may be associated with the assay system used. Taken together, these results suggested that W501 is required for RNA helicase activity but is not essential for the DNA helicase activity of HCV NS3 protein. The results also suggest that V432 is not essential for either the RNA or DNA helicase activity of the NS3 protein.
ATPase activity of the HCV NS3 helicase.
The ATPase activity of the purified HCV NS3 helicase was examined using thin-layer chromatography. As shown Table 2, the wild-type and mutant HCV NS3 helicases had similar basal-level ATPase activities, which were stimulated to various degrees by the presence of different ssRNA and ssDNA sequences. In the V432 mutants (V432A, V432D, and V432R), the stimulation of ATPase activity by template strands (05R, 05D) was more efficient than that by release strands (06R, 06D), and this effect appears to be correlated with the presence of U15 and (dT)15-tracts on the template strands. Poly(U)100 and poly(dT)45 stimulated ATPase activity more efficiently than did the other template strands, with poly(U)100 being the most efficient. The presence of a negatively charged Asp residue in the mutant enzyme (V432D) lowered the ATPase stimulation in all nucleic acids tested, whereas a positively charged Arg residue in the mutant enzyme (V432R) increased stimulation. In the W501 mutants (W501A, W501E, and W501R), overall patterns of enzyme stimulation by RNAs and DNAs were similar to those in the V432 mutants. Apparently, there is little correlation between the ATPase activity and the unwinding activity of mutant enzymes. These results suggest that a threshold ATPase activity was present in all enzymes tested and was sufficient for the unwinding activity detected in this study. Our data show that the loss of unwinding activity in W501 mutants was not due to altered ATPase activity in the mutant enzymes.
TABLE 2.
ATPase activities of wild-type and mutant HCV NS3 helicases in the presence or absence of selected ssRNA or ssDNA
| NS3 helicase | ATPase activity (mM s−1 mM−1)a |
||||||
|---|---|---|---|---|---|---|---|
| Basal | RNA06 | DNA06 | RNA05 | DNA05 | Poly(U) | (dT)45 | |
| Wild type | 0.8 | 1.5 | 1.9 | 5.0 | 2.4 | 11.7 | 3.7 |
| V432A | 0.6 | 1.8 | 3.7 | 12.4 | 3.9 | 22.7 | 5.0 |
| V432D | 0.4 | 0.7 | 1.1 | 2.2 | 1.6 | 2.7 | 1.7 |
| V432R | 0.6 | 8.3 | 7.0 | 16.0 | 5.4 | 24.6 | 9.0 |
| W501A | 0.6 | 1.6 | 3.6 | 6.0 | 8.0 | 8.2 | 8.3 |
| W501E | 0.7 | 0.8 | 1.4 | 3.5 | 3.3 | 4.1 | 3.8 |
| W501R | 0.6 | 6.6 | 5.5 | 14.3 | 10.2 | 18.1 | 14.9 |
| W501F | 1.0 | 3.5 | 12.2 | 18.8 | 13.3 | 23.9 | 17.1 |
Each value represents the average of two or more independent experiments. The standard error was less than 10%.
Single-stranded nucleic acid binding of the HCV NS3 helicase protein.
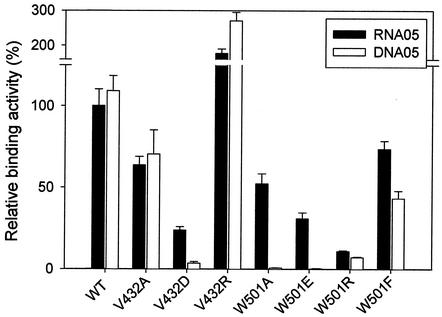
We confirmed the published results that the HCV helicase forms a stable complex with nucleic acid substrate in the absence of ATP but does not form a detectable complex in the presence of ATP (24, 26). We first measured the profile of single-stranded nucleic acid (RNA05 and DNA05) binding in the absence of ATP. Each substrate bound to the wild-type enzyme at a similar level, and binding was linear up to 200 nM protein (data not shown). We measured the binding activities of wild-type and mutant NS3 enzymes at a concentration within the linear range; the results are shown in Fig. 5 and Table 3. The binding of both substrates to the protein was reduced in all the V432 mutants except the V432R mutant, in which the activity sharply increased. This suggests that the residue at position 432 is exposed on the surface of the mutant enzyme and that an ionic interaction was probably involved in substrate binding. Profiles of binding activity in the W501 mutants were similar to that in the V432 mutants in most cases; however, the W501R mutant showed no increased binding activity. This result suggests that the W501 residue is not implicated in substrate binding. In the absence of ATP, the binding experiments did not show parallel titration of unwinding activities in mutant enzymes. These data appear to indicate that the gel or filter assay, which is routinely used as a helicase binding assay, measures stoichiometric binding of nucleic acid to the NS3 helicase, not catalytic binding. During the unwinding reaction in the presence of ATP, binding of substrate to enzyme should occur within the catalytic pocket. Our attempt to measure the catalytic binding by UV cross-linking probably did not work because the catalytic binding is transient in nature.
FIG. 5.
Binding activities of the wild-type and mutant HCV NS3 helicases in the absence of ATP. A gel retardation reaction was performed as described in Materials and Methods. The enzyme concentrations used for the binding assay were 25 to 200 nM for wild type, V432A, V432R, and W501F and 75 to 600 nM for V432D, W501A, W501E, and W501R. The calculated binding activity of each enzyme was expressed as a percentage of that of the wild-type enzyme on the RNA05 template (100%). Each bar represents the standard error from four independent experiments.
TABLE 3.
Binding activities of the wild-type and mutant HCV NS3 helicases
| NS3 helicase | % Binding toa: |
|
|---|---|---|
| RNA05 | DNA05 | |
| Wild type | 100 ± 10 | 109 ± 9 |
| V432A | 64 ± 5 | 70 ± 15 |
| V432D | 24 ± 2 | 4 ± 1 |
| V432R | 177 ± 25 | 271 ± 24 |
| W501A | 52 ± 12 | <1 (0.8 ± 0.1) |
| W501E | 31 ± 7 | <1 (0.3 ± 0.1) |
| W501R | 11 ± 1 | 7 ± 0.4 |
| W501F | 74 ± 10 | 43 ± 5 |
The values represent the average of at least four different independent experimental data at the points showing linear binding activity to the enzyme concentration. Each value was converted to a percentage relative to the RNA05 binding activity of the wild-type enzyme (taken as 100%).
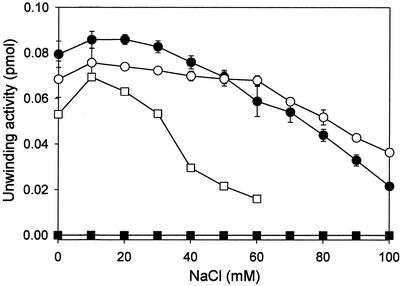
Effect of salt on RNA and DNA helicase activity.
It is known that HCV NS3 helicase activity is inhibited in the presence of monovalent cations (7, 8, 37). Lee et al. (16) reported that the DNA helicase activity of the Drosophila maleless protein was more salt sensitive than was the RNA helicase activity. In this regard, we examined the influence of salt on the RNA and DNA unwinding activities of the wild-type NS3 helicase and the W501A mutant. As shown Fig. 6, both RNA and DNA helicase activities were inhibited by increasing concentrations of NaCl. In the wild-type enzyme, the salt sensitivity between the RNA and DNA helicase activities was distinguishable. This suggests that the RNA and DNA helicase activities in the wild-type enzyme are not controlled by the same enzymatic reaction. The DNA helicase activity was more severely inhibited by salt in the W501A mutant than in the wild-type enzyme.
FIG. 6.
Effect of salt on the RNA or DNA unwinding activity of HCV NS3 helicase. The unwinding reactions were performed in the presence of the indicated concentrations of NaCl. The enzyme concentration in each reaction was 20 nM. Symbols: •, wild-type enzyme on a dsRNA substrate; ○, wild-type enzyme on a dsDNA substrate; ▪, W501A on a dsRNA substrate; □, W501A on a dsDNA substrate.
Computer-aided three-dimensional analysis of the enzyme-substrate complex.
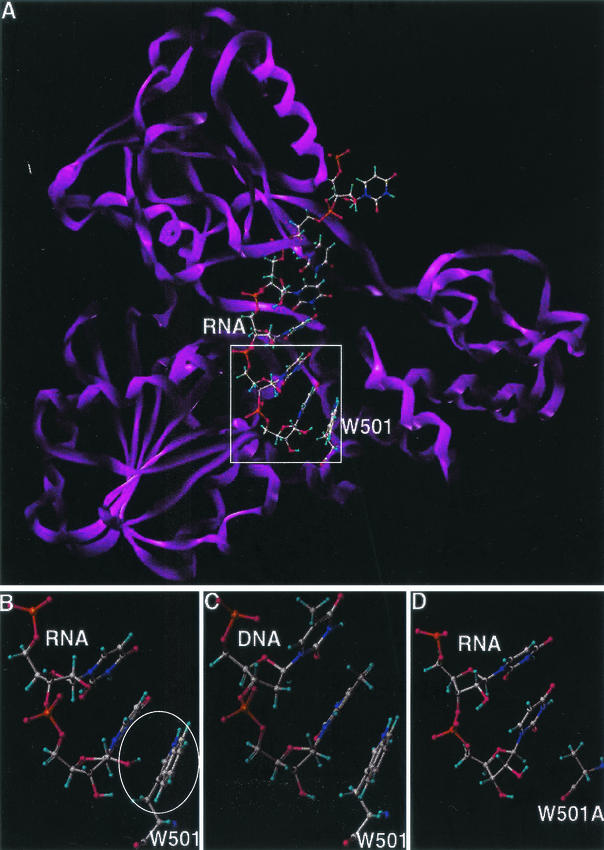
Similar protein contacts and an identical number of U6-protein and U6-solvent hydrogen bonds were observed in the minimized structures of the U6/W501 wild-type and the U6/W501A mutant enzyme. The ring-to-ring (π-π) stacking patterns observed among the bases of U6 and the indole ring of W501 were comparable to those among the (dT)6 bases and W501. The backbone of U6 retained a conformation similar to that of the nucleic acid in 1aiv. One structural difference between U6/W501 (Fig. 7B) and U6/W501A (Fig. 7D) appears to be near the 2′-OH sugar group at the 3′ end of the oligonucleotide. In the wild-type model, the hydroxyl group is approximately 2 to 3 Å from the centroid of the six-membered ring of W501, indicative of a possible π-facial interaction. However, this hydroxyl in the mutant model is not compensated by any hydrogen bonds with the enzyme, substrate, or solvent. In the (dT)6/W501 (Fig. 7C) and (dT)6/W501A models (data not shown), such an interaction was apparently missing and the two structures showed similar protein contacts and patterns of hydrogen bonding.
FIG. 7.
Molecular modeling of wild-type and mutant HCV helicases. Carbon is represented in grey, oxygen in red, hydrogen in light blue, nitrogen in indigo, and phosphorus in orange. (A) Wild-type HCV helicase domain complexed with ssRNA U8. The protein is depicted as a ribbon structure (violet); the RNA and W501 residue of the helicase are depicted as a ball-and-stick model. (B) Interaction between the 2′OH of U8 base in ssRNA (top) and the aromatic ring of W501 in the wild-type helicase (bottom). The putative π-facial hydrogen bond is encircled. (C) Lack of the π-facial hydrogen bond interaction between the ssDNA (top) and W501 in the wild-type helicase. (D) Lack of the π-facial hydrogen bond interaction between the ssRNA (top) and Ala side residue in the W501A mutant helicase (bottom).
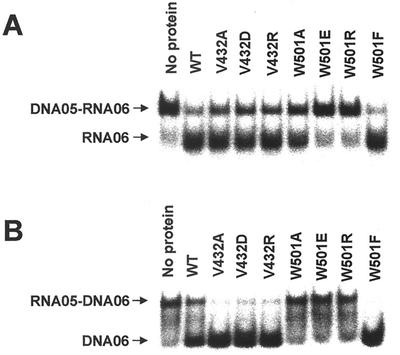
HCV NS3 helicase activity on heteroduplex substrates.
The HCV NS3 helicase has been proven capable of unwinding heteroduplex substrates (12, 18). Using this property, we attempted to test whether the differential substrate specificities seen in the V432 and W501 mutants were reproducible in the heteroduplex substrate. We prepared substrates in two different combinations: an RNA template strand with a DNA release strand (05R/06D) and a DNA template strand with an RNA release strand (05D/06R). These substrates differ in that the nucleic acid strand facing W501 of the enzyme is RNA in 05R/06D but DNA in 05D/06R. In this design, the W501 mutants except for W501F were expected to unwind 05D/06R but not 05R/06D due to the lack of aromatic residue. However, all V432 mutants were expected to unwind either substrate due to the nonessential nature of V432 in the enzyme activity. As shown, the overall pattern of strand displacement in the wild-type and mutant enzymes in the 05R/06D substrate (Fig. 8B) was similar to that in the duplex RNA substrate (Fig. 3A), and the pattern in 05D/06R (Fig. 8A) was similar to that in the duplex DNA substrate (Fig. 3C). This result confirms our prediction and thus highlights the requirement and the possible role of the aromatic side chain of W501 in the RNA unwinding activity of HCV helicase.
FIG. 8.
Unwinding of heteroduplex substrates by the wild-type and mutant HCV NS3 helicases. (A) Unwinding reaction on the DNA05-RNA06 substrate. (B) Unwinding reaction on the RNA05-DNA06 substrate. The reaction was performed with 10 nM enzyme under standard conditions, and the reaction product was analyzed on a native 15% polyacrylamide gel.
DISCUSSION
HCV and BVDV are the two most closely related RNA viruses within the family Flaviviridae and have significant nucleotide and amino acid sequence homologies at the 5′ UTR and the NS3 helicase region (9, 22). Nevertheless, the helicases of these two viruses differ with respect to substrate specificity. Both HCV and BVDV replicate in the cytoplasm, and the replication cycle does not involve a DNA intermediate; therefore, the role of DNA helicase activity in the HCV enzyme has been under scrutiny. We have been interested in identifying the region(s) responsible for this substrate specificity within HCV helicase. Based on the structure of HCV helicase complexed with d(U)8, Kim et al. proposed a ratchet model as a working mechanism of the HCV NS3 helicase (13). In this monomeric model, residues W501 and V432 are in direct contact with substrate bases within the catalytic pocket and were predicted to play a key role in HCV NS3 helicase function. It has been hypothesized that substrate specificity might be determined by the specific interaction between the enzyme and substrate at the catalytic pocket (41). It has also been demonstrated that unwinding of nucleic acids by the HCV NS3 helicase is sensitive to the structure of the duplex (35). Based on these findings, we suspected that W501 and/or V432 might also be involved in the substrate specificity of HCV helicase. We therefore mutagenized W501 and V432 residues individually, produced a total of seven mutant enzymes, and analyzed their abilities to unwind dsRNA and dsDNA HCV substrates.
In our results, replacement of W501 with nonaromatic hydrophobic or charged residues resulted in a complete loss of RNA unwinding activity. However, the same mutation to Ala resulted in very little change in DNA unwinding activity and the mutation to Glu or Arg resulted in a reduced but still significant DNA unwinding activity. In contrast, similar mutations at V432 resulted in RNA and DNA unwinding activities that were virtually indistinguishable. These results demonstrate that W501 is essential for RNA helicase activity but not for HCV NS3 DNA helicase activity. This supports the old hypothesis that a specific binding interaction between the enzyme and the substrate at the catalytic pocket is important for substrate specificity. It also shows that RNA helicase and DNA helicase activities are related but that each activity is separable in the HCV NS3 protein. This view is further supported by the fact that RNA and DNA unwinding activities of the HCV helicase have different salt sensitivities.
The specific loss of the RNA helicase activity, not the DNA helicase activity, in W501 mutants was unexpected. To see whether an altered ATPase activity or substrate binding activity in the mutant enzymes was responsible for the observed phenotype, we analyzed these parameters in the mutant enzymes. Our data ruled out the possibility that a change in the ATPase activity of the mutant enzymes was directly responsible for the phenotype. However, the precise role of substrate binding in the W501 mutants on the RNA unwinding activity is still obscure in this study. It is worth noting that the HCV NS3 helicase has at least two reported RNA binding sites, one at the catalytic pocket as described previously (5, 13) and the other near the Arg-rich motif (460QRRGRTGR467) in domain 2 (4, 12). Since the latter site has been identified by binding assays only under nonphysiological conditions without ATP, it probably represents a substrate holding site as suggested (41). Due to the limitation of our current RNA binding assay, we probably have measured the stoichiometric RNA binding at the holding site. We suspect that substrate binding at the catalytic site would be transient and rate limiting. It is possible that the W501 mutant enzymes had a defect in this catalytic site and that the resultant nonproductive RNA binding was responsible for the loss of RNA helicase activity in the mutant enzymes.
The fact that mutant enzymes bearing amino acid changes with nonaromatic side chains were inactive whereas the mutant enzyme bearing Phe at W501 restored dsRNA unwinding activity illustrates the requirement of an aromatic side chain. This side chain requirement was first explained by Kim et al. (13) in terms of ring-to-ring stacking between W501 and the bases of the substrate. While this explanation may be applied to account for the dsRNA unwinding activities seen in our experiment, it fails to account for our results showing dsDNA unwinding activity which did not require an aromatic side chain. Therefore, to find another logical explanation, we studied further the detailed structure of wild-type and mutant HCV helicase complexed with an ssRNA or ssDNA substrate by using molecular modeling techniques. In this analysis, we identified a putative π-facial interaction between the 2′ OH of the RNA substrate and the W501 aromatic ring. The OH hydrogen is approximately 2.5 Å from the centroid of the six-member ring and is in a favorable orientation to interact with the face of the ring. Putative π-facial interactions have been identified in a number of protein (30) and small-molecule structures (19) and might make important contributions to specificity and enzyme catalysis (10, 33). High-level ab initio calculations estimate the interaction energy of these bonds to be approximately 1 to 3 kcal/mol (19, 40). In the wild-type complex, the π-facial interaction can compensate for the free energy cost of desolvating the 2′-OH of the RNA nucleotide, whereas the W501A mutation leaves the 2′-OH unsatisfied. The loss of this interaction might explain, in part, the decreased activity of the W501A helicase mutant with respect to the wild-type helicase for the RNA substrate. In the structures of DNA-W501 and DNA-W501A, there were no clear differences in protein interactions and hydrogen bonding patterns that would suggest that one complex is more stable than the other. This is supported by the similar levels of activity of both wild-type and W501A helicase with a DNA substrate in biochemical analysis. Based on these structural and biochemical evidences, we hypothesize that an aromatic side chain at amino acid 501 is required and that the π-facial interaction is involved in RNA helicase function of HCV NS3 protein.
One critical experiment which tests our hypothesis is to measure the unwinding of the RNA-DNA or DNA-RNA heteroduplex substrate by the mutant HCV NS3 helicase. Our data show that the RNA-DNA hybrid was unwound by the W501A mutant only if the nucleic acid strand facing W501 was DNA (DNA05/RNA06), not RNA (RNA05/DNA06). In contrast, either heteroduplex substrate was unwound by V432A, regardless of the strand combination. This is consistent with our hypothesis and further confirms the fact that the V432 residue is nonessential for HCV helicase function.
The double stranded region of RNA (as well as RNA-DNA hybrids) is known to adopt the A-form duplex as opposed to the B-form found in most natural dsDNA. In the A-form duplex, base pairings are tilted 19° away from the normal helix axis because of what is known as C3′-endo puckering caused by the 2′ OH of ribose present in RNA. Under the circumstance, the minor groove nearly vanishes, making the duplex shorter and wider and thus making the phosphates in the A-form duplex bind fewer water molecules than do the phosphates in the B-form duplex. The degree of structural distortion in dsRNA is also affected by the nucleotide sequence (32). Because the structural difference between dsRNA and dsDNA is rather large, it is possible that the mode of their binding to HCV NS3 helicase might differ significantly, resulting in different interactions with W501. To date, the structure of the NS3 helicase complexed with dsRNA or dsDNA has not been solved; therefore, we cannot rule out the possibility that the HCV enzyme interacts with the dsRNA or dsDNA substrate in different ways other than the 2′OH of RNA. In addition, there is a large difference in the energetics involved in the unwinding of dsRNA and dsDNA. However, it is not known whether HCV helicase unwinds dsRNA by an active or passive mechanism. Therefore, the extent of the possible energy contribution of the π-facial H bond to dsRNA unwinding is unknown. In these aspects, our hypothesis about the π-facial hydrogen bond based on the molecular modeling approach is still limited. Elucidation of precise role played by W501 on substrate specificity phenotype of HCV helicase requires further work.
The monomeric ratchet model by Kim et al. requires two capping residues, W501 and V432, which intercalate the base of the substrate at the 3′ and 5′ end, respectively. This model also requires two major ATP binding sites, one at Walker motifs A and B in domain 1 and the other at Arg-rich motifs in domain 2 (R464 and R467). Binding of ATP to helicase triggers the close of the intercleft domain between domains 1 and 2 and thereby translocates domain 1 in the 3′-to-5′ direction along with a bound substrate strand. Hydrolysis of ATP then facilitates reopening of the cleft and releases ADP, with a concomitant movement of domain 2 in the same direction. Thus, W501 and V432 alternatively disrupt base stacking at either end of the single-stranded region, which is critical to a net translocation of the helicase as a monomer. This model therefore predicts that an amino acid substitution at V432 would affect the unwinding activity of the HCV helicase to a similar extent to that seen with an amino acid substitution at W501. In our study, however, replacement of V432 had little effect on the RNA and DNA unwinding activity of HCV helicase, indicating that V432 was not critically required. Initial structural analysis by Kim et al. (13) indicated that most of the enzyme-substrate interactions within the catalytic pocket involve hydrogen bonds between amino acid residues and the phosphate backbone. Subsequent mutagenesis work confirmed the importance of some of these residues in the unwinding activity (18, 23, 25, 29). Whether one of these residues is needed for the 5′-capping function in place of V432 to account for the monomeric model is not clear. Recent evidence shows that HCV helicase works as a dimer or an oligomer (11, 17). If this is the case, W501 of the HCV helicase may carry out a capping function, as seen in F182 of E. coli Rep replicase as proposed by Korolev et al. (15). In addition, Chang et al. showed that R464 does not bind ATP but is important to RNA binding (4). Moreover, Rho et al. showed that R467 is a methylation site by arginine methyl transferase 1 (29). Further studies are required to refine the working mechanism of the unwinding function of HCV NS3 helicase.
Our findings in this study provide an explanation for the conservation of the W501 residue in HCV NS3 helicase and for the requirement of an aromatic ring at the W501 site for the dsRNA unwinding activity. These findings provide further insight into the mechanism of the HCV helicase function and may have important ramification for the design of antiviral drugs.
Acknowledgments
We thank Chi-Gun Lee for helpful discussions.
This work was supported by a grant from the Pohang University of Science and Technology. J. W. Kim was supported by a postdoctoral training program at the Department of Life Science, Pohang University of Science and Technology. A. Shelat was supported by an NDSEG fellowship from the U.S. Department of Defense. D. T. Moustakas was partially supported by a Sandler Award from the UCSF Foundation.
REFERENCES
- 1.Alter, M. J. 1997. Epidemiology of hepatitis C. Hepatology 3(Suppl. 1):62S-65S. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 1998. Recommendations for prevention and control of hepatitis C virus (HCV) infection and HCV-related chronic disease. Morb. Mortal. Wkly. Rep. 47:1-39. [Google Scholar]
- 3.Case, D. A., D. A. Pearlman, J. W. Caldwell, T. E. Cheatham III, J. Wang, W. S. Ross, C. L. Simmerling, T. A. Darden, K. M. Merz, R. V. Stanton, A. L. Cheng, J. J. Vincent, M. Crowley, V. Tsui, H. Gohlke, R. J. Radmer, Y. Duan, J. Pitera, I. Massova, G. L. Seibel, U. C. Singh, P. K. Weiner, and P. A. Kollman. 2002. Amber 7. University of California, San Francisco.
- 4.Chang, S. C., J. C. Cheng, Y. H. Kou, C. H. Kao, C. H. Chiu, H. Y. Wu, and M. F. Chang. 2000. Roles of the AX4GKS and arginine-rich motifs of hepatitis C virus RNA helicase in ATP- and viral RNA-binding activity. J. Virol. 74:9732-9737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis, G. L., R. Esteban-Mur, V. Rustgi, J. Hoefs, S. C. Gordon, C. Trepo, M. L. Shiffman, S. Zeuzem, A. Craxi, M. H. Ling, and J. Albrecht. 1998. Interferon alfa-2b alone or in combination with ribavirin for the treatment of relapse of chronic hepatitis C. The International Hepatitis Interventional Therapy Group. N. Engl. J. Med. 339:1493-1499. [DOI] [PubMed] [Google Scholar]
- 6.Gorbalenya, A. E., and E. V. Koonin. 1993. Helicase: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 3:419-429. [Google Scholar]
- 7.Gwack, Y., D. W. Kim, J. H. Han, and J. Choe. 1997. DNA helicase activity of the hepatitis C virus nonstructural protein 3. Eur. J. Biochem. 250:47-54. [DOI] [PubMed] [Google Scholar]
- 8.Gwack, Y., D. W. Kim, J. H. Han, and J. Choe. 1996. Characterization of RNA binding activity of the hepatitis C virus NS3 protein. Biochem. Biophys. Res. Commun. 225:654-659. [DOI] [PubMed] [Google Scholar]
- 9.Han, J. H., V. Shyamala, K. H. Richman, M. J. Brauer, B. Irvine, M. S. Urdea, P. Tekamp-Olson, G. Kuo, Q. L. Choo, and M. Houghton. 1991. Characterization of the terminal regions of hepatitis C viral RNA: identification of conserved sequences in the 5′ untranslated region and poly (A) tails at the 3′ end. Proc. Natl. Acad. Sci. USA 88:1711-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jedrzejas, J. M., S. Singh, W. J. Brouilette, W. G. Laver, M. Air, and M. Luo. 1995. Structures of aromatic inhibitors of influenza virus neuraminidase. Biochemistry 34:3144-3151. [DOI] [PubMed] [Google Scholar]
- 11.Khu, Y. L., E. Koh, S. P. Lim, Y. H. Tan, S. Brenner, S. G. Lim, W. J. Hong, and P. Y. Goh. 2001. Mutations that affect dimer formation and helicase activity of hepatitis C virus helicase. J. Virol. 75:205-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim, D. W., J. Kim, Y. Gwack, J. H. Han, and J. Choe. 1997. Mutational analysis of the hepatitis C virus RNA helicase. J. Virol. 71:9400-9409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim, J. L., K. A. Morgenstern, J. P. Griffith, M. D. Dwyer, J. A. Thomson, M. A. Murcko, C. Lin, and P. R. Caron. 1998. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure 6:89-100. [DOI] [PubMed] [Google Scholar]
- 14.Kolykhalov, A. A., S. M. Feinstone, and C. M. Rice. 1996. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus. J. Virol. 70:3363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korolev, S., N. Yao, T. M. Lohman, P. C. Weber, and G. Waksman. 1998. Comparisons between the structures of HCV and Rep helicases reveal structural similarities between SF1 and SF2 super-families of helicases. Protein Sci. 7:605-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, C. G., K. A. Chang, M. I. Kuroda, and J. Hurwitz. 1997. The NTPase/helicase activities of Drosophila maleless, an essential factor in dosage compensation. EMBO J. 16:2671-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levin, M. K., and S. S. Patal. 1999. The helicase from hepatitis C virus is active as an oligomer. J. Biol. Chem. 274:31839-31846. [DOI] [PubMed] [Google Scholar]
- 18.Lin, C., and J. L. Kim. 1999. Structure-based mutagenesis study of hepatitis C virus NS3 helicase. J. Virol. 73:8798-8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malone, J. F., C. M. Murray, M. H. Charlton, R. Docherty, and A. J. Lavery. 1997. X-H:π (phenyl) interactions. Theoretical and crystallographic observations. J. Chem. Soc. Faraday Trans. 93:3429-3436. [Google Scholar]
- 20.Matson, S. W., and K. A. Kaiser-Rogers. 1990. DNA helicases. Annu. Rev. Biochem. 59:289-329. [DOI] [PubMed] [Google Scholar]
- 21.McHutchison, J. G., S. C. Gordon, E. R. Schiff, M. L. Shiffman, W. M. Lee, V. K. Rustgi, Z. D. Goodman, M.-H. Ling, S. Cort, and J. K. Albrecht. 1998. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. Hepatitis Interventional Therapy Group. N. Engl. J. Med. 339:1485-1492. [DOI] [PubMed] [Google Scholar]
- 22.Miller, R. H., and R. H. Purcell. 1990. Hepatitis C virus shares amino acid sequence similarity with pestiviruses and flaviviruses as well as members of two plant virus supergroups. Proc. Natl. Acad. Sci. USA 87:2057-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paolini, C., A. Lahm, R. De Francesco, and P. Gallinari. 2000. Mutational analysis of hepatitis C virus NS3-associated helicase. J. Gen. Virol. 81:1649-1658. [DOI] [PubMed] [Google Scholar]
- 24.Porter, D. J. T., S. A. Short, M. H. Hanlon, F. Preugschat, J. E. Wilson, D. H. Willard, Jr., and T. G. Consler. 1998. Product release is the major contributor to kcat for the hepatitis C virus helicase catalyzed strand separation of short duplex DNA. J. Biol. Chem. 273:18906-18914. [DOI] [PubMed] [Google Scholar]
- 25.Preugschat, F., D. P. Danger, L. H. Carter III, R. G. Davis, and D. J. T. Porter. 2000. Kinetic analysis of the effects of mutagenesis of W501 and V432 of the hepatitis C virus NS3 helicase domain on ATPase and strand-separating activity. Biochemistry 39:5174-5183. [DOI] [PubMed] [Google Scholar]
- 26.Preugschat, F., D. R. Averett, B. E. Clarke, and D. J. T. Porter. 1996. A steady-state and pre-steady-state kinetic analysis of the NTPase activity associated with the Hepatitis C virus NS3 helicase domain. J. Biol. Chem. 271:24449-24457. [DOI] [PubMed] [Google Scholar]
- 27.Reed, K. E., and C. M. Rice. 2000. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr. Top. Microbiol. Immunol. 242:55-84. [DOI] [PubMed] [Google Scholar]
- 28.Rice, C. M. 1996. Flaviviridae: the viruses and their replication, p. 931-960. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 29.Rho, J. S. Choi, Y. R. Seong, J. Choi, and D. S. Im. 2001. The arginine-1493 residue in QRRGRTGR1493G motif IV of the hepatitis C virus NS3 helicase domain is essential for NS3 protein methylation by the protein arginine methyltransferase 1. J. Virol. 75:8031-8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Samanta, U., D. Pal, and P. Chakrabarti. 1999. Packing of aromatic rings against tryptophan residues in proteins. Acta Crystallogr. D55:1421-1427. [DOI] [PubMed] [Google Scholar]
- 31.Simmonds, P. J. 1999. Viral heterogeneity of the hepatitis C virus. Hepatology 31(Suppl. 1):54-60. [DOI] [PubMed] [Google Scholar]
- 32.Stryer, L. 1995. Biochemistry 4th ed., p. 787-818. W. H. Freeman & Co. New York, NY.
- 33.Sulpizi, M., and P. Carloni. 2000. Cation-π versus OH-π interactions in proteins: a density functional study. J. Phys. Chem. Ser. B 104:10087-10091. [Google Scholar]
- 34.Suzich, J. A., J. K. Tamura, F. Palmer-Hill, P. Warrener, A. Grakoui, C. M. Rice, S. M. Feinstone, and M. S. Collett. 1993. Hepatitis C virus NS3 protein polynucleotide-stimulated nucleoside triphosphatase and comparison with the related pestivirus and flavivirus enzymes. J. Virol. 67:6152-6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tackett, A. J, L. Wei, C. E. Cameron, and K. D. Raney. 2001. Unwinding of nucleic acids by HCV NS3 helicase is sensitive to the structure of the duplex. Nucleic Acids Res. 29:565-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tai, C. L., W. C. Pan, S. H. Liaw, U. C. Yang, L. H. Hwang, and D. S. Chen. 2001. Structure-based mutational analysis of the hepatitis C virus NS3 helicase. J. Virol. 75:8289-8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tai, C. L., W. K. Chi, D. S. Chen, and L. H. Hwang. 1996. The helicase activity associated with hepatitis C virus nonstructural protein 3 (NS3). J. Virol. 70:8477-8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanaka, T., N. Kato, M. J. Cho, K. Sugiyama, and K. Shimotohno. 1996. Structure of the 3′ terminus of the hepatitis C virus genome. J. Virol. 70:3307-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsukiyama-Kohara, K., N. Iizuka, M. Kohara, and A. Nomoto. 1992. Internal ribosome entry sites within hepatitis C virus RNA. J. Virol. 66:1476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsuzuki, S., K. Honda, T. Uchimaru, M. Mikami, and K. Tanabe. 2000. Origin of the attraction and directionality of the NH/π interaction: comparison with OH/π and CH/π interactions. J. Am. Chem. Soc. 122:11450-11458. [DOI] [PubMed] [Google Scholar]
- 41.von Hippel, P. H., and E. Delagoutte. 2001. A general model for nucleic acid helicases and their “coupling” within macromolecular machines. Cell 104:177-190. [DOI] [PubMed] [Google Scholar]
- 42.World Health Organization. 1999. Global surveillance and control of hepatitis C. Report of a WHO consultation organized with the Viral Hepatitis Prevention Board. J. Viral Hepatitis 6:35-47. [PubMed] [Google Scholar]
- 43.Yao, N., T. Hesson, M. Cable, Z. Hong, A. D. Kwong, H. V. Le, and P. C. Weber. 1997. Structure of the hepatitis C virus RNA helicase domain. Nat. Struct. Biol. 4:463-467. [DOI] [PubMed] [Google Scholar]