Abstract
Two CD209 family genes identified in humans, CD209 (DC-SIGN) and CD209L (DC-SIGNR/L-SIGN), encode C-type lectins that serve as adhesion receptors for ICAM-2 and ICAM-3 and participate in the transmission of human and simian immunodeficiency viruses (HIV and SIV, respectively) to target cells in vitro. Here we characterize the CD209 gene family in nonhuman primates and show that recent evolutionary alterations have occurred in this family across primate species. All of the primate species tested, specifically, Old World monkeys (OWM) and apes, have orthologues of human CD209. In contrast, CD209L is missing in OWM but present in apes. A third family member, that we have named CD209L2, was cloned from rhesus monkey cDNA and subsequently identified in OWM and apes but not in humans. Rhesus CD209L2 mRNA was prominently expressed in the liver and axillary lymph nodes, although preliminary data suggest that levels of expression may vary among individuals. Despite a high level of sequence similarity to both human and rhesus CD209, rhesus CD209L2 was substantially less effective at binding ICAM-3 and poorly transmitted HIV type 1 and SIV to target cells relative to CD209. Our data suggest that the CD209 gene family has undergone recent evolutionary processes involving duplications and deletions, the latter of which may be tolerated because of potentially redundant functional activities of the molecules encoded by these genes.
CD209 (DC-SIGN) and CD209L (DC-SIGNR/L-SIGN) are homologous type II membrane-associated C-type lectins (2, 9, 12, 20). CD209 was first discovered as a human placental protein capable of binding human immunodeficiency virus type 1 (HIV-1) gp120 (9) and was later identified on the surface of human dendritic cells (DCs) derived from monocytes that had been treated with granulocyte-macrophage colony-stimulating factor and interleukin-4 in vitro (monocyte-derived DCs [MDDCs]) (12). CD209 binds ICAM-2 and ICAM-3 and mediates HIV-1 capture and transmission by MDDCs to target T cells in vitro (11, 12). In vivo, CD209 has been detected on the surface of cells with DC morphology in lymphoid tissues and the lamina propria of mucosal tissues (12, 14) and on placental macrophages (24), locations where it may participate in HIV transmission. Although CD209L is 85% identical to CD209 and has functional activity very similar to that of CD209, their expression patterns are quite distinct in that CD209L is expressed mainly on endothelial cells of the lymph nodes (LN), liver, and placenta (2, 20).
The in vitro functional characterization of CD209 has led to speculation regarding a potential role for the molecule in HIV-1 capture and transmission by DCs at mucosal sites and in vertical transmission of the virus through the placenta (11, 24). Alternatively, Langerhans cells, which are potentially critical for virus transport from the mucosal epithelium to LN (5), are CD209 negative (12, 23), supporting the possibility that other receptors are involved in this process independently of CD209 (25).
The genes encoding CD209 and CD209L are located on human chromosome 19p13.2-3, within a 30-kb segment, and have similar exon-intron structures, suggesting that they were derived from the duplication of an ancestral precursor gene (2, 22). Recently, five mouse homologues of human CD209 that have 65 to 70% sequence similarity to human CD209 in the carbohydrate recognition domain (CRD) and have diverged from one another substantially on the basis of sequence similarity were described (17). Only one of these genes appears to be expressed in DCs (17). Rhesus monkey (Macaca mulatta), pigtailed macaque (Macaca nemestrina), and chimpanzee (Pan troglodytes) CD209 orthologues have also been characterized (1, 10, 27), all of which show a high level of sequence similarity to human CD209 and ligand specificity overlapping that of human CD209. Rhesus monkey CD209 and pigtailed macaque CD209 were shown to transmit HIV-1, HIV-2, and SIV to T cells in vitro (1, 27). Curiously, no detectable level of CD209 in rhesus MDDCs was observed even though these cells perform efficiently in HIV transmission assays (27), again implicating novel molecules in HIV transmission. Immunohistochemical staining with antibodies against CD209 do, however, indicate its expression in rhesus monkey lymphoid and mucosal tissues (10, 14), similar to expression patterns observed in human tissues. On the other hand, attempts to clone rhesus CD209L were unsuccessful, as was detection of a CD209L homologue in rhesus tissues by using human CD209L-specific antibodies (14). Immunohistochemical staining with antibody AZN-D3, which recognizes both CD209 and CD209L (2), indicated the expression of one or both of these molecules in chimpanzee LN (10).
The multigenic nature of the CD209 family in humans and mice raises the possibility that other novel homologues exist in primates. In an attempt to map the recent evolutionary history of the CD209 gene family, distinct members of this family in various primate species were identified in the present study. OWM and apes have retained orthologues of the CD209 gene, perhaps because of some particularly useful function conferred by its protein product. On the other hand, genetic mechanisms such as duplication, deletion, and mutation have led to the differentiation of other CD209 homologues among primate species to some extent. A novel member of the family, named CD209L2, was identified in rhesus monkeys and subsequently in other OWM and nonhuman apes. Although the rhesus CD209L2 gene sequence is very similar to that of CD209 (at least as similar as CD209L), their sequence divergence has resulted in differential functional behavior.
MATERIALS AND METHODS
Primate DNA sources.
Nonhuman primate DNAs from a collection of samples described previously (3, 4) were used to test for the presence of CD209 homologues: galago (Galago crassicaudatus), squirrel monkey (Saimiri sciureus), three baboon species (Papio cynocephalus, Papio anubis, and Papio papio), patas (Erythrocebus patas), grivet (Cercopithecus sabaeus), sooty mangabey (Cercocebus atys), gelada (Theropithecus gelada), colobus (Colobus guereza), two species of langur (Presbytis obscurus and Presbytis senex), two species of gibbon (Hylobates lar and Hylobates concolor), and gorilla (Gorilla gorilla).
Some of the DNA samples were extracted from cultured cells with the QIAGEN Genomic-tip kit. An owl monkey (Aotus trivirgatus) fibroblast cell line and Epstein-Barr virus-transformed stump-tailed macaque (Macaca arctoides), gibbon (Hylobates lar), orangutan (Pongo pygmaeus), and chimpanzee (Pan troglodytes) lymphoblastoid cell lines were provided by Roscoe Stanyon. Human DNA was isolated from freshly isolated peripheral blood mononuclear cells and the THP-1 cell line. Rhesus monkey (Macaca mulatta) DNA was isolated from the B-cell line rh-B116 (J. D. Lifson laboratory).
The QIAamp DNA Blood Mini kit was used for isolation of DNA from rhesus monkey and chimpanzee whole blood or peripheral blood mononuclear cells (provided by Genoveffa Franchini and Barbara Rehermann, respectively). DNA samples from several orangutans were provided by Stephen O'Brien.
Animal care was provided in accordance with the procedures outlined in reference 16a.
Isolation of Mm-CD209L2 cDNA.
A rhesus monkey CD209L2 (Mm-CD209L2) cDNA fragment was amplified from total rhesus monkey MDDC RNA. Culture conditions for MDDCs, RNA extraction, and reverse transcriptase PCR (RT-PCR) conditions were similar to those described for mac-DC-SIGN (Mm-CD209) (27). The primers used for amplification were s15 (ATGAGTGACTCCAAGGAACCAAG) and s18 (GGCTTAAAAGTGGCGAAGTGCCA). The primer sequences were complementary to genomic regions that were conserved in four rhesus monkeys. The amplified cDNA fragment was cloned into the expression vector pcDNA3.1/V5-His/TOPO (Invitrogen). Five clones were sequenced to eliminate the possibility that a PCR error had occurred.
Southern blot analysis.
Genomic DNA (2 to 5 μg) was digested overnight with 50 U of EcoRI, HindIII, BamHI, or XbaI (New England Biolabs) under the conditions specified by the manufacturer. Digested DNA was loaded onto a 0.8% agarose gel and run overnight at low voltage. After electrophoresis, DNA was visualized by ethidium bromide staining and transferred to Hybond-XL (Amersham Pharmacia Biotech) as previously described (7). The Southern blots were hybridized with the PCR product (i) exon 5 probe (nucleotides [nt] 2321 to 2463; GenBank accession no. AF209479); (ii) exon 6 probe (nt 3185 to 3337; GenBank accession no. AF209479); or (iii) exon 7 probe, which was an equimolar combination of three DNA fragments corresponding to the human genes CD209 (nt 4177 to 4377; GenBank accession no. AF209479) and CD209L1 (nt 1044 to 1217; GenBank accession no. AF290887) and the Mm-CD209L2 gene (nt 580 to 794; GenBank accession no. AY074781). The probes were radioactively labeled with Megaprime labeling systems (Amersham Pharmacia Biotech). Hybridization was performed in the ExpressHyb Solution (Clontech Laboratories, Inc.) under conditions described by the manufacturer.
Phylogenetic analysis.
Phylogenetic trees were constructed by the neighbor-joining method (21). The phylogeny of primate genes was based on the number of nucleotide substitutions per site in exon 7 fragments (∼250 bp; see the following section), as this was the only region of the CD209 gene family that was available for a large number of species. Additional phylogenetic trees were based on the proportion of amino acid differences in the complete coding sequences and in the CRDs of primate and rodent CD209 family members.
Sequencing of primate CD209 genes.
Partial sequences of primate CD209 and CD209L1 were obtained by sequencing PCR products generated with primers specific for the corresponding human sequences. Primate genomic fragments of 4.5 to 6.5 kb were amplified by using Platinum Taq High Fidelity DNA polymerase (Invitrogen) at a 60°C annealing temperature with primers 5ut5 (CATCCCACTGCTCAGCCATC) and 3dc2 (ACATAGCAGCTACACATGGC) for CD209 and primers sim1 (CTGGGGACAGCGGGAAA) and sim4 (GCAGTTACAACATTTACCACTTTATTATAAAGGC) for CD209L1. The single exception was the use of primer s40 (GTCCCTAGGAGCCCTGAACAT), instead of sim1, for gibbon CD209L1. The fragments were purified from agarose gels after electrophoresis, diluted 50 times, and reamplified with intronic primers (supplemental Tables 1 and 2 at URL address http:/home.ncifcrf.gov/ccr/lgd/cd209/sup_data.htm) at a 55°C annealing temperature in order to obtain sequences for each exon. (Note that for species other than rhesus monkeys and chimpanzees, putative exons are provided as they were not confirmed by cDNA sequencing.) The chimpanzee cDNA fragments for CD209 and CD209L1 containing putative full coding sequences (on the basis of similarity to the human genes) were cloned from total RNA extracted from a liver biopsy (provided by Barbara Rehermann).
TABLE 1.
The CD209 gene family in primatesa
| Species | CD209 | CD209L1 | CD209L2 |
|---|---|---|---|
| OWM | + | − | + |
| Nonhuman apes | + | + | + |
| Humans | + | + | − |
+, present; −, not present.
TABLE 2.
Polymorphism of the repeat region in nonhuman primates
| Species | No. of chromosomes | No. of CD209 alleles (repeat range) | No. of CD209L1 alleles (repeat range) |
|---|---|---|---|
| Baboon | 8 | 2 (4-5) | |
| Patas | 2 | 1 (6) | |
| Grivet | 2 | 1 (6) | |
| Mangabey | 4 | 1 (6) | |
| Gelada | 4 | 1 (4) | |
| Rhesus | 72 | 1 (6) | |
| Stump-tailed macaque | 2 | 1 (6) | |
| Colobus | 4 | 1 (6) | |
| Gibbon | 8 | 4 (6-10) | 2 (7-8) |
| Orangutan | 76 | 4 (5-8) | 1 (3) |
| Gorilla | 16 | 2 (8-9) | 2 (6-7) |
| Chimpanzee | 42 | 2 (8-9) | 5 (5-12) |
Exon 7 fragments of CD209L2 used in phylogenetic analysis were amplified with primers 17P (TATTGGAACAGAGGAGAGCC) and s36/37 (ACCAGGGGANCTTGGAGGCAT [N = C/A]) at a 55°C annealing temperature. The primers may anneal to all three genes, but because of the insertion of an Alu repeat that is located downstream from the sequence used for the phylogenetic analysis, the CD209L2 fragment is ∼300 bp larger than the corresponding fragments in the other genes. The CD209L2 fragments (∼650 bp) were separated on an agarose gel, extracted, and sequenced.
Repeat region polymorphism was typed by PCR at a 55°C annealing temperature with the following pairs of primers: (i) s5 (GCTCCATAAGTCAGGAACAATCCA) and i4r (CCCCGTGTTCTCATTTCACAG) for CD209 and (ii) sim18 (GCTCCCTAAGTCAGGAACAATCCGA) and sim19 (AAATCGGTCAGTTCTTGATAGATTTG) for CD209L1. To evaluate the size of CD209L2 exon 4 in rhesus monkeys and chimpanzees, large genomic fragments (∼5 to 7 kb) were amplified by using Platinum Taq High Fidelity DNA polymerase at a 60°C annealing temperature with primers s16 (AAGAGGAAGAGCTGATAACTAGCA) and s37 (ACCAGGGGACCTTGGAGGCAT), diluted 50-fold, and reamplified with primers sim27 (TGTCCAAGGTCCCCAGCTCC) and i4r at a 55°C annealing temperature.
Northern blot and RT-PCR analyses.
Total RNAs from the liver of rhesus monkey rh-A01-42 (provided by Andrew Lackner), a number of tissues from rhesus monkey rh-94C009, an axillary LN (Ax-LN) from rhesus monkey rh-96D551, and the B-cell line rh-B116 were isolated by using Trizol (Invitrogen). RNA (3 μg) was electrophoresed on a 1% agarose gel, transferred to Hybond-XL (Amersham Pharmacia Biotech) as previously described (7), and hybridized to a CD209L2 probe (nt 1 to 794; GenBank accession no. AY074781) in ExpressHyb Solution (Clontech Laboratories, Inc.). An actin control probe (Clontech Laboratories, Inc.) was used to evaluate the quantity of RNA loaded in each lane.
Total RNA (1 μg) was used as the template for reverse transcription with the SuperScript First Strand Synthesis System (Invitrogen). First-strand cDNA was amplified at a 60°C annealing temperature for 35 cycles with primers 15p (AACTTCCTACAGCTGCAGTC) and s-9m (27) for Mm-CD209 and primers 15p and s18 for Mm-CD209L2. The human G3PDH amplimer set (Clontech Laboratories, Inc.) was used as a positive control (60°C annealing temperature, 25 cycles).
Antibodies.
Monoclonal antibodies (MAbs) against CD209/DC-SIGN were obtained from R&D Systems (Minneapolis, Minn.) and have been previously described (27). CD209 MAb 120526 is abbreviated as MAb 526 in the remainder of the text. All other antibodies were purchased from B-D/PharMingen unless otherwise stated.
Cell culture.
The THP-1, THP-1/Hs-CD209 (human DC-SIGN), THP-1/Hs-CD209L1 (human L-SIGN), and THP-1/Mm-CD209 (mac-DC-SIGN) cell lines have been previously described (2, 15, 27). THP-1/Mm-CD209L2 cells were generated by electroporation of THP-1 cells with the pcDNA3.1-Mm-CD209L2 expression construct. Cells showing resistance to G418 (Invitrogen) were positively sorted for Mm-CD209L2 cell surface expression.
Hut/CC chemokine receptor CCR5 (Hut/CCR5) cells are the transformed human T-cell line Hut78 stably transduced with CCR5 (27). HEK 293T cells are human embryonic kidney cells containing a single temperature-sensitive allele of the simian virus 40 large-T antigen. GHOST/X4/R5 cells are HIV indicator cells derived from human osteosarcoma cells (6).
THP-1, THP-1/Hs-CD209, THP-1/Hs-CD209L1, THP-1/Mm-CD209, THP-1/Mm-CD209L2, and Hut/CCR5 cells were maintained in RPMI 1640 medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS; HyClone Laboratories). HEK 293T and GHOST/X4/R5 cells were grown in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% FBS.
Flow cytometry.
To assess expression of different CD209 family molecules, THP-1, THP-1/Hs-CD209, THP-1/Hs-CD209L1, THP-1/Mm-CD209, and THP-1/Mm-CD209L2 cells were treated with MAb 526 and assayed by fluorescence-activated cell sorter (FACS) as previously described (27). Briefly, 2 × 105 cells were incubated in ice-cold phosphate-buffered saline (PBS) containing 2% FBS, 0.02% sodium azide (FACS buffer), and 1 μg of MAb per ml in a total volume of 100 μl. After 30 min at 4°C, the cells were washed with the FACS buffer and resuspended in 100 μl of FACS buffer containing 1 μg of phycoerythrin-conjugated goat anti-mouse immunoglobulin G (IgG) antibody (Caltag) per ml. Cells were incubated for 30 min at 4°C, washed with PBS containing 2% FBS, and analyzed with a FACScalibur apparatus (Becton Dickinson).
ICAM-3 adhesion assay.
Carboxylate-modified TransFluorSpheres (1.0 μm; 488-nm excitation wavelength and 645-nm emission wavelength; Molecular Probes) were coated with soluble, recombinant ICAM-3 (R&D Systems) as described previously (13). The fluorescent bead adhesion assay was performed as described previously (27). Briefly, THP-1, THP-1/Hs-CD209, THP-1/Hs-CD209L1, THP-1/Mm-CD209, and THP-1/Mm-CD209L2 cells (1.5 × 105) were resuspended in 20 mM Tris-HCl (pH 7.4)-150 mM NaCl-1 mM CaCl2-2 mM MgCl2-0.5% BSA-20 nM sodium azide (adhesion buffer) and preincubated with MAb 526 or a mouse IgG isotype control (10 μg/ml) for 10 min at room temperature. Adhesion of ICAM-3 to the CD209 proteins was determined by measuring the detectable percentage of cells that bound fluorescent beads by using flow cytometry on a FACScalibur (Becton Dickinson).
Virus stocks.
Single-round infectious, pseudotyped HIV-1 stocks were generated by calcium phosphate cotransfections of HEK 293T cells with proviral vector plasmid NL-Luc-E−R− (HIV-Luc) containing a firefly luciferase reporter gene (8) and an expression plasmid for either the R5-tropic HIV-1ADA (HIV-Luc/ADA) or the SIV-1MAC1A11 (HIV-Luc/SIVMAC1A11) envelope glycoprotein. Viral stocks were evaluated by limiting dilution on GHOST/X4/R5 cells.
HIV-1 infection assays.
HIV-1 capture and transmission assays were performed as described previously (27). In brief, THP-1, THP-1/Hs-CD209, THP-1/Mm-CD209, and THP-1/Mm-CD209L2 cells (2.5 × 105) were preincubated with cross-reactive MAb 526 or the mouse IgG isotype control (10 μg/ml) for 30 min at 37°C and then the cells were incubated with pseudotyped HIV-1 (multiplicity of infection, ∼0.1) in a total volume of 400 μl for 3 h to allow cellular adsorption of the virus. After 3 h, cells were washed with 1 ml of PBS and cocultured with Hut/CCR5 target cells (105) in the presence of 10 μg of Polybrene in 1 ml of cell culture medium. Cell lysates were obtained 2 days after infection and analyzed for luciferase activity with a commercially available kit (Promega).
Nucleotide sequence accession numbers.
The accession number for Mm-CD209L2 cDNA in GenBank is AY074781. The GenBank accession numbers of the primate genomic sequences used for phylogenetic analyses are AF480022 to AF480057. Exon sequences for baboons, gibbons, orangutans, gorillas, and chimpanzees, as well as two cDNA sequences for chimpanzees, CD209 and CD209L1, have been submitted to the GenBank database and assigned accession no. AY078807 to AY078920.
RESULTS
CD209L2.
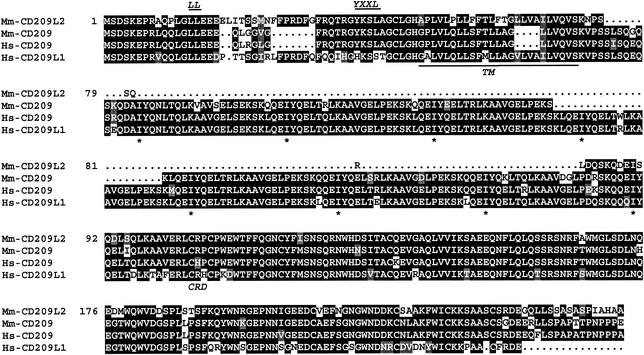
The identification of CD209 in rhesus monkeys (M. mulatta) and chimpanzees (P. troglodytes) (1, 10, 27) indicated that an orthologue of human CD209L might also be present in other primate species. Although CD209L sequences were readily detected by PCR with human-specific primers to amplify chimpanzee, gorilla, gibbon, and orangutan DNAs, we failed to amplify CD209L in rhesus monkey and other OWM DNAs (data not shown). Rather, a novel member of the CD209 family was discovered, which we have named CD209L2. As a result of this finding, we suggest that CD209L be renamed CD209L1, and this terminology is employed throughout the remainder of this report. On the basis of comparisons with other members of the gene family, the cDNA fragment cloned from a rhesus monkey is likely to be the full coding sequence for CD209L2. PCR analysis and partial sequencing of genomic DNA indicated that the exon-intron structure of the rhesus CD209L2 (Mm-CD209L2) gene is similar to that of human CD209 and CD209L1, consisting of seven exons. The putative Mm-CD209L2 amino acid sequence has 81% similarity to Mm-CD209 (Fig. 1), the primary difference being the length of the membrane-proximal neck domain, which is shorter in Mm-CD209L2 than in human and rhesus CD209, as well as in human CD209L1. The neck domain of CD209 and CD209L1 contains tandem repeats of 23 amino acids, whereas the Mm-CD209L2 neck region contains only a fraction of a single repeat element.
FIG. 1.
Amino acid alignment of putative Mm-CD209, Mm-CD209L2, human CD209 (Hs-CD209), and Hs-CD209L1. The potential internalization motifs, LL and YXXL, are annotated in the cytoplasmic domain, and the transmembrane domain (TM) is underlined. The starts of the repeats in the neck region (*) and the beginning of the CRD are indicated below the sequences.
With primers specific for Mm-CD209L2, at least one CD209L2 genomic fragment was successfully amplified in various OWM and nonhuman apes (exon 7 fragment; see below). However, PCR amplification of human genomic DNA did not reveal any sequences for this gene, corresponding to the lack of evidence of CD209 family members apart from CD209 and CD209L1 in the human genome databases.
Southern analysis of DNAs from various primates with CD209 family-specific probes.
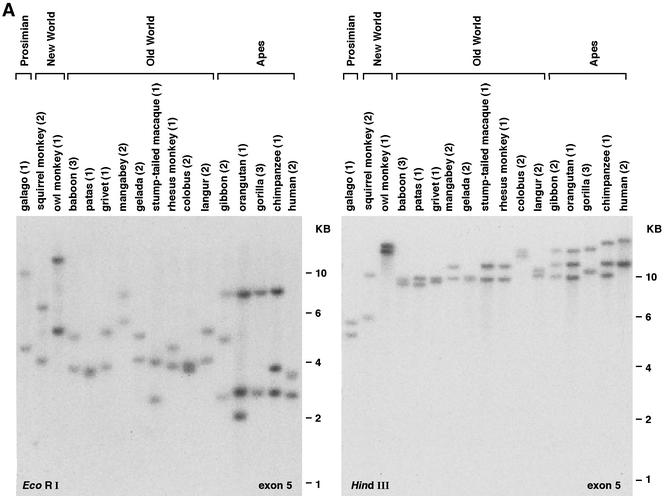
PCR amplification with a number of gene-specific primer pairs suggested that CD209L1 is absent in OWM, whereas CD209L2 is absent in humans. However, the success of PCR amplification depends on sequence conservation at primer sites and these genes could have been missed inadvertently because of variation among genes at these sites. Since hybridization with a fairly long probe (i.e., >100 bp) is less sensitive to nucleotide variations than is oligonucleotide annealing and extension during PCR, Southern blot hybridization was used to confirm the presence or absence of CD209 gene family members. Blots of genomic DNAs from 17 primate genera, including a prosimian, New World monkeys (NWM), OWM, and apes, were prepared, and probes designed to recognize all three genes (CD209, CD209L1, and CD209L2) were used to determine the number of homologous restriction fragments in each species (Fig. 2).
FIG. 2.
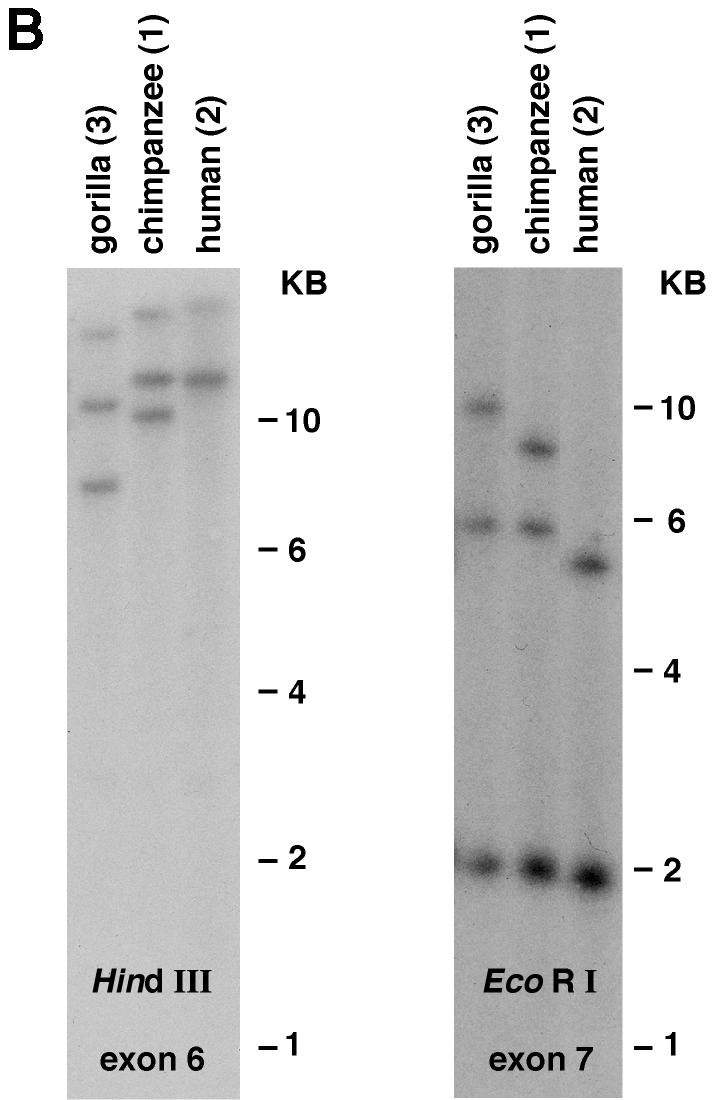
Southern blot analysis of primate genomic DNA. Hybridization patterns suggest the presence of two or three CD209 family genes in various primate species. The number of animals of each genus tested is indicated in parentheses. Individuals from the same genus either showed the same banding pattern (most cases) or revealed restriction fragment length polymorphisms (see supplemental Fig. 1 at URL address http:/home.ncifcrf.gov/ccr/lgd/cd209/sup_data.htm). (A) Hybridization with the human CD209 exon 5 probe. (B) Hybridization with the exon 6 and exon 7 probes showing three bands in the gorilla sample.
Hybridization of primate DNA using a probe corresponding to human CD209 exon 5, a sequence that encodes part of the highly conserved CRD, revealed two or three fragments in DNA samples digested with EcoRI or HindIII (Fig. 2A). The indistinct bands observed in lanes containing patas and colobus DNAs (EcoRI digest), as well as baboon, grivet, and gelada DNAs (HindIII digest), are each likely to represent two restriction fragments that are close in size. In each of these cases, two distinct bands were obtained with the second restriction enzyme. The doublet observed in the human EcoRI digest (upper band of about 3.8 kb) represents two allelic variants corresponding to polymorphism in the CD209L1 neck region, as genotyping of this sample revealed two alleles at the CD209L1 locus distinguished by seven and five full repeats in the neck region.
Although the exon 5 probe and an exon 4 probe (data not shown for exon 4) hybridized to two restriction bands in gorilla DNA, an exon 6 probe and an exon 7 probe revealed three distinct fragments (Fig. 2B). These data suggest that the gene corresponding to this inconsistent banding pattern is either truncated or highly divergent in the region upstream of exon 6. Additional PCR and sequencing analysis revealed that this gene is CD209L2.
The assumption that the number of restriction fragments is equal to the number of homologous genes could be inaccurate under some circumstances. Fewer fragments than expected on the basis of the gene copy number could be observed if (i) hybridizing fragments very close in size are not resolved (as discussed above), (ii) two or more genes map to one restriction fragment, or (iii) hybridization is inhibited because of major diversity in the sequence corresponding to the probe being used. Alternatively, restriction fragment length polymorphism could lead to the appearance of more bands than expected on the basis of the gene copy number (Fig. 2A, upper EcoRI doublet band in human DNA, and supplemental Fig. 1 at URL address http:/home.ncifcrf.gov/ccr/lgd/cd209/sup_data.htm). Therefore, a thorough analysis with four restriction enzymes (EcoRI, Hind III, BamHI, and XbaI), four different probes (corresponding to exons 4, 5, 6, and 7), and DNA samples obtained from several animals for each species (when possible) was performed. Consistent patterns derived from the Southern analyses indicated the presence of two CD209 family genes in prosimians, NWM, OWM, and humans and three genes in nonhuman apes. On the basis of these results and the PCR assay and sequence data described in the previous and following sections, we conclude that nonhuman apes have retained CD209, CD209L1, and CD209L2; humans have retained CD209 and CD209L1; and OWM have retained CD209 and CD209L2 (Table 1). Unfortunately, because of sequence divergence, we were not able to amplify CD209 family genes in prosimians and NWM. Thus, the two CD209 family genes present in these primate groups according to the Southern analysis remain unknown.
Sequence and phylogenetic analyses of CD209 genes in primates.
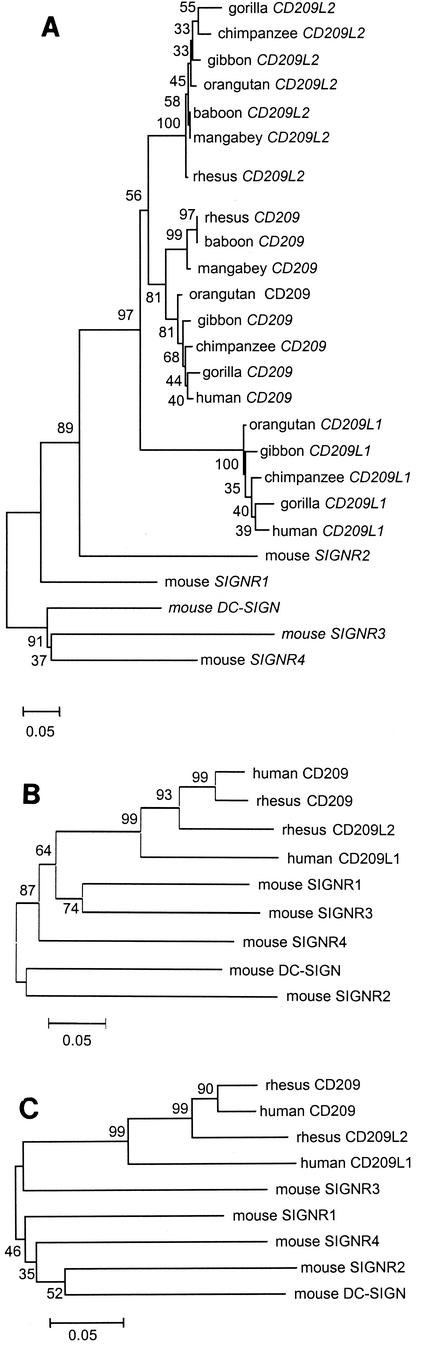
Having established the existence of a third member of the CD209 gene family that appears to be present in nonhuman primates, sequence comparisons of the genes in various species was performed in order to determine the level of divergence among orthologous and paralogous genes. Fragments homologous to a 249-bp fragment of human CD209 exon 7 (nt 1060 to 1308; GenBank accession no. AF290886) were amplified and sequenced in four ape genera for CD209, CD209L1, and CD209L2; three OWM genera for CD209 and CD209L2; and humans for CD209 and CD209L1. Five mouse homologues that were reported previously (17) were also included in the analysis. The 249-bp fragment consists of the 3′ coding terminus of human CD209 (nt 1 to 165) and part of the 3′ untranslated region (nt 166 to 249). A neighbor-joining tree was constructed (Fig. 3A) in which three separate clades supported by bootstrap values of 81, 100, and 100% were formed for CD209, CD209L1, and CD209L2, respectively. The mouse homologues have very long branch lengths, suggesting a fair degree of gene divergence within this species, and there is no detectable orthologous relationship between any of the three primate CD209 family genes and mouse family members.
FIG. 3.
Phylogenetic analysis of the CD209 gene family. (A) Neighbor-joining tree of nucleotide sequences homologous to a 249-bp fragment of human CD209 exon 7. Two to four individuals from each genus were sequenced. (B) Neighbor-joining tree of putative full-length amino acid sequences of the CD209 family proteins. (C) Neighbor-joining tree of amino acid sequences of the CRDs in the CD209 family proteins.
Phylogenetic analysis of complete amino acid sequences representing the CD209 family in rhesus monkeys and humans, as well as the mouse homologues, indicated that CD209 and CD209L2 are more closely related to one another than they are to CD209L1 (Fig. 3B). In order to optimize sequence alignments with Mm-CD209L2 in this analysis, repeat sequences in the neck regions of CD209 and CD209L1 were deleted, leaving only the partial repeat that is present in CD209L2.
The CRD is the most conserved region in the CD209 family. Phylogenetic analysis of the CRD (Fig. 3C) suggested relationships among the molecules that are quite similar to those observed in Fig. 3A and B, where CD209 is more closely related to CD209L2 than either is to CD209L1. Also common to the previous analyses, rhesus monkey and human CD209 proteins form a clade separate from all of the mouse homologues with strong bootstrap support.
We have generated amino acid sequences of putative gibbon, orangutan, and gorilla CD209 and CD209L1 proteins by assembling potential exon sequences that were obtained by sequencing genomic DNA (supplemental Fig. 2 at URL address http:/home.ncifcrf.gov/ccr/lgd/cd209/sup_data.htm), whereas the chimpanzee protein sequences are based on cloned cDNA fragments. Apart from repeat length, minor sequence variations among the compiled protein sequences were observed, the most interesting of which was a stop codon (TAG) in exon 5 of the orangutan orthologue of human CD209L1. All 14 of the orangutans tested were homozygous for the stop codon. Termination of transcription in exon 5 would result in disruption of the CRD, suggesting the absence of a functional CD209L1 protein in this species. We also observed an insertion mutation that would lead to a frameshift in one of six alleles of orangutan CD209L1 exon 4 (GenBank accession no. AY078831).
The repeat region of CD209L1 is polymorphic in humans, ranging from three to nine repeats, whereas this region is conserved with seven repeats in human CD209 (2). Analysis of CD209 and CD209L1 exon 4, which encodes the neck repeats, in various nonhuman primates indicated that both genes are polymorphic in this region (Table 2). However, only the single CD209L2 partial repeat was observed in all of the rhesus monkeys and chimpanzees tested (n = 10 for each species), suggesting that this region of the gene is conserved within and across species.
Analysis of Mm-CD209L2 expression.
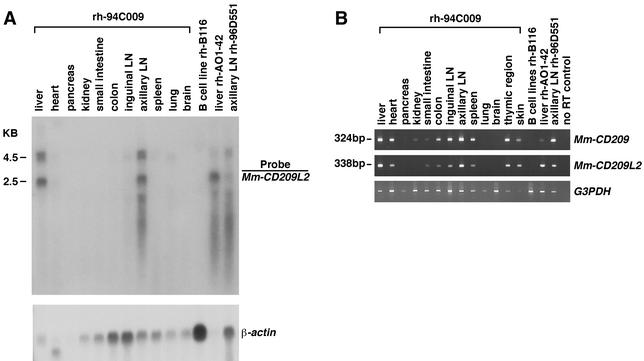
Differential expression patterns of CD209 family members have been observed within species (e.g., human CD209 and CD209L1 are expressed on different cell types) (2, 20) and across species (human MDDCs express CD209, whereas rhesus monkey MDDCs do not) (27). Thus, we examined the expression pattern of Mm-CD209L2 in a number of rhesus monkey tissues by Northern blot hybridization with a 794-bp probe containing the putative full-length coding sequence of Mm-CD209L2. Because of the sequence similarity between Mm-CD209L2 and Mm-CD209, the probe was expected to detect both genes. Tissues and cells from four unrelated animals were used as sources of mRNA: multiple tissues from monkey rh-94C009, liver tissue from monkey rh-AO1-42, Ax-LN from monkey rh-96D551, and a B-cell line generated from monkey rh-B116 (Fig. 4A). mRNA extracted from tissue samples was partially degraded, which was evident in banding patterns from rRNA (data not shown) and an actin control (Fig. 4A, compare actin signals in tissues with that in the B-cell line). Degradation was inevitable, as the tissues were collected about an hour postmortem. Nevertheless, a doublet at ∼4.5 kb and a single band at ∼2.5 kb were observed in the liver and Ax-LN of monkey rh-94C009, indicating substantial expression of the CD209 homologues in these tissues. Expression levels in Ax-LN tissue of monkey rh-96D551 was substantially lower than that observed in Ax-LN of monkey rh-94C009, suggesting that these genes are expressed in an inducible manner or are constitutively expressed but at different levels in different individuals. The intensity of the 2.5- and 4.5-kb bands observed in liver tissue from monkey rh-94C009 were fairly evenly distributed, whereas the rh-AO1-42 liver RNA revealed an intense 2.5-kb band and a nearly undetectable 4.5-kb band. Additional bands masked by degraded material may also be present in Ax-LN samples of monkeys rh-94C009 and rh-96D551, as well as in the rh-AO1-42 liver samples.
FIG. 4.
Expression of the Mm-CD209L2 gene in rhesus monkey tissues. (A) Northern analysis of the Mm-CD209L2 gene. Total RNAs isolated from various tissues and a B-cell line were hybridized with a 794-bp Mm-CD209L2 cDNA probe. The 2.5-kb band is presumably Mm-CD209L2 mRNA, and the 4.5-kb doublet most likely corresponds to Mm-CD209. A human β-actin probe was used as a control for RNA loading. (B) RT-PCR analysis of Mm-CD209 and Mm-CD209L2 expression. Gene-specific primers were used to analyze expression of the two genes. In addition to the RNA samples represented in panel A, skin and thymic region RNAs were also included. Human G3PDH primers were used as a positive control. Four individual rhesus monkeys (rh-94C009, rh-A01-42, rh-95D551, and rh-B116) are represented.
RT-PCR of the RNA panel was performed by using gene-specific primers in order to examine the expression patterns of the two CD209 family genes independently (Fig. 4B). The highest level of expression of both genes in monkey rh-94C009 was observed in the liver and Ax-LN, supporting data obtained by Northern analysis (Fig. 4A). Coexpression of the genes was also observed in several other tissues but was not detected in pancreas, lung, or brain tissue or in the B-cell line. However, absence of expression in pancreas and lung tissues may be due to greater RNA degradation, as the G3PDH control band is relatively faint in these tissues.
Transcription of the Mm-CD209 and Mm-CD209L2 genes in monkey rh-94C009 may be regulated by the same mechanism, as similar patterns of expression of these genes in tissue were observed by RT-PCR analysis. On the other hand, expression of CD209 in the liver was low in rhesus monkey rh-AO1-42 relative to that in rh-94C009, whereas comparable expression of CD209L2 in the liver was observed in these animals on the basis of RT-PCR. These observations, along with banding patterns obtained by Northern analysis, led to the conclusion that the lower 2.5-kb hybridizing band in Fig. 4A represents Mm-CD209L2 and the upper 4.5-kb bands represent alternative transcripts of Mm-CD209. Furthermore, the 2.5-kb fragment was more resistant to stringent washing conditions than were the upper bands, presumably because of more precise complementarity with the Mm-CD209L2 probe.
Functional characterization of Mm-CD209L2.
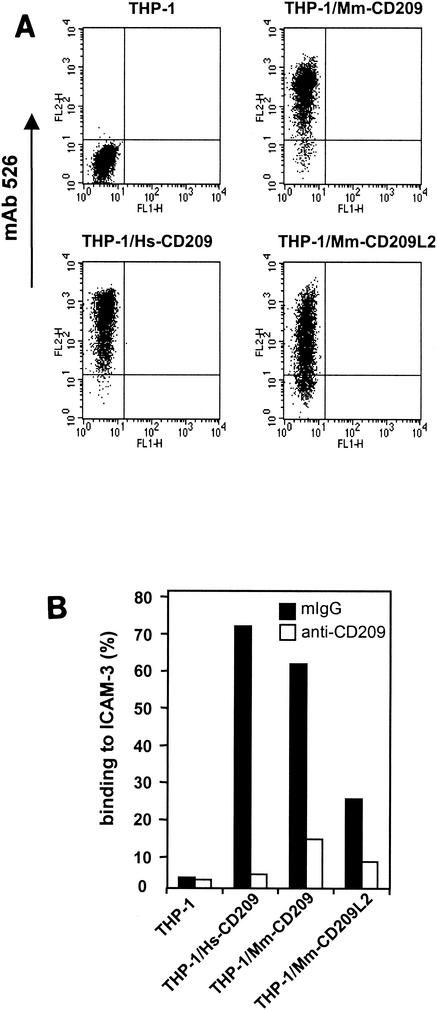
Human CD209 and CD209L1 and rhesus CD209 are all capable of binding HIV gp120 and enhancing HIV-1 infection of T-cell targets in vitro, although the efficiency with which they do so varies to some extent (1, 2, 11, 20, 27). To characterize the putative protein encoded by the novel rhesus monkey gene Mm-CD209L2, we generated a stable THP-1 cell line expressing the gene (THP-1/Mm-CD209L2) and tested whether MAbs generated against human CD209 and CD209L1 recognize Mm-CD209L2. MAb 526 was strongly cross-reactive with Mm-CD209L2 (Fig. 5A). With a fluorescent bead adhesion assay (13), THP-1/Mm-CD209L2 cells were shown to bind human ICAM-3 (Fig. 5B), although 2.5 times less efficiently than THP1/Mm-CD209 cells. Binding was inhibited by the addition of cross-reactive MAb 526. Thus, while rhesus CD209L2 binds human ICAM-3 poorly relative to both human CD209 and rhesus CD209, specific, low-level binding was clearly observed. Rhesus ICAM-3, which has not been cloned to date, would be a more appropriate ligand for this analysis, and the weaker binding of human ICAM-3 to rhesus CD209L2 may be due to ICAM-3 orthologue differences. However, it is very likely that the significant structural differences between CD209 and CD209L2 account for most or all of the observed effect (Fig. 5B).
FIG. 5.
Cell lines expressing CD209 family molecules bind ICAM-3. (A) THP-1 monocytes stably transduced with human and macaque CD209 family molecules were assessed for protein expression with cross-reactive MAb 526. Positive expression is indicated on the FL-2 axis. Isotypic antibody control stainings of the different cell lines were uniformly negative. (B) Adhesion of ICAM-3 to THP-1/Hs-CD209, THP-1/Hs-CD209L, THP-1/Mm-CD209, and THP-1/Mm-CD209L2 cells was measured by FACS analysis with a fluorescent bead adhesion assay as previously described (27). Control mouse IgG or CD209 MAb 526 (10 μg/ml) was preincubated with cells. Adhesion of ICAM-3 to THP-1 parental cells was less than 5%. The results of one experiment representative of two are shown.
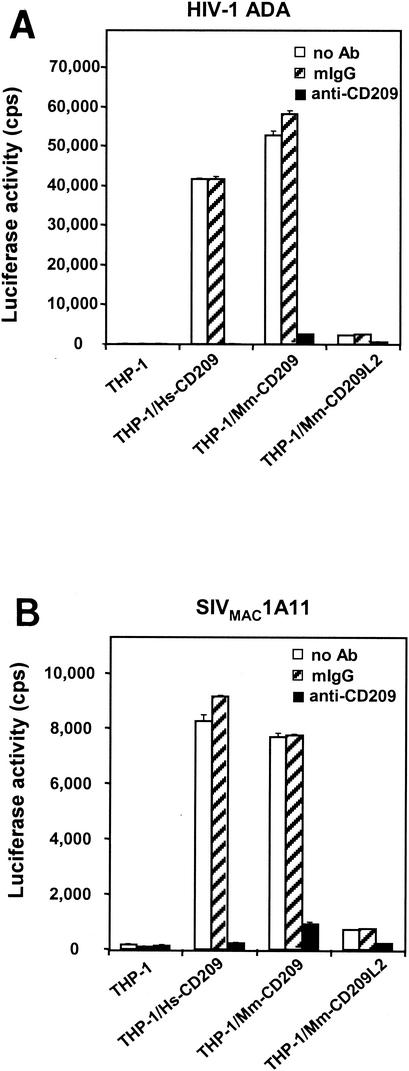
Mm-CD209L2 was then tested for the ability to capture HIV-1 or SIV and transmit it to target T cells in a virus capture-transmission assay (2, 11, 27). Stable THP-1 cells expressing Mm-CD209, human CD209, and Mm-CD209L2 were pulsed with an HIV-Luc vector pseudotyped with either HIV-1 envelope HIV-1ADA (Fig. 6A) or SIV envelope SIVMAC1A11 (Fig. 6B), unbound virus was removed, and the cells were incubated with human T cells (Hut/CCR5) for 2 days. As expected, human and rhesus monkey CD209-transfected THP-1 cells efficiently transmitted the virus to the target T cells and cross-reactive MAb 526 significantly inhibited transmission. Mm-CD209L2 transfectants, on the other hand, were 90 to 95% less effective at viral transmission than were cells transfected with either rhesus monkey or human CD209. Nevertheless, Mm-CD209L did transmit the virus at a low level on the basis of complete inhibition of this activity in the presence of MAb 526. Thus, sequence divergence observed between rhesus CD209L2 and CD209 may have resulted in discrimination of their physiological activities, particularly with regard to HIV-SIV interactions.
FIG. 6.
Impaired virus transmission by Mm-CD209L2. Relative virus capture and transmission by CD209 family molecules was assayed with HIV-Luc pseudotyped with HIV-1 ADA Env (A) or SIVMAC1A11 Env (B). THP-1, THP-1/Hs-CD209, THP-1/Mm-CD209, and THP-1/Mm-CD209L2 donor cells were preincubated with the MAbs (10 μg/ml) for 30 min at 37°C, after which HIV-Luc pseudotypes were added and the mixture was incubated for an additional 3 h at 37°C. The cells were then washed and cocultured with Hut/CCR5 target cells in the presence of Polybrene. HIV-1 infection was determined after 2 days by measuring the luciferase activity. Treatment with MAb 526 was used to evaluate the necessity of CD209 molecules for transmission, and mouse IgG (mIgG) was used as a nonspecific antibody control. Each set of data represents the mean of three separate wells of infected cells. The results of one experiment representative of three are shown. cps, counts per second.
DISCUSSION
At least three CD209 family members have appeared during primate evolution, including the novel CD209L2 gene identified in OWM and nonhuman apes. Tandem arrangement of CD209 family members has been observed consistently across species, as human CD209 and CD209L1 and the five mouse CD209 homologues are located adjacent to one another on human chromosome 19 and mouse chromosome 8, respectively (2, 17, 22). Further, we have established that rhesus monkey CD209 and CD209L2 map to a single BAC clone, indicating their close physical proximity (data not shown). Given the relative positions of CD209 and CD209L1 in humans, as well as CD209 and CD209L2 in rhesus monkeys, it is likely that all three genes present in nonhuman apes are also adjacent to one another.
The combination of CD209 and CD209L2 in OWM and apes except for humans suggests that these two genes were present in a common primate ancestor at least 20 million years ago and that through some genetic mechanism, such as unequal crossing over, CD209L2 was lost in the human lineage. CD209L1, which is missing in OWM, may have arisen by gene duplication in a common ancestor of the apes approximately 20 million years ago, but if so, comparisons of the gene sequences do not conclusively indicate whether CD209L1 was most likely derived by duplication of CD209 or CD209L2. On the other hand, CD209L1 may be more ancient than CD209 or CD209L2 if mutation rates are assumed to be constant for all three genes, since phylogenetic analysis suggests that CD209L1 is more distantly related to CD209 and CD209L2 than CD209 and CD209L2 are to each other. In this case, an ancestor of OWM would have to have lost the CD209L1 gene. Sequence data from the CD209 gene family members in prosimians and NWM, which appear to be somewhat divergent on the basis of our inability to amplify these genes with the primers described herein, may provide further insight into the evolutionary history of this gene family in primates.
The 69-bp repeats encoding the neck region of the CD209 family molecules have also been subject to genetic mechanisms resulting in insertions or deletions. The wide range of repeats observed for CD209L1 in both humans and chimpanzees implies that if its gene product is essential, then the physiologic function of CD209L1 is not very sensitive to the length of its neck region. The absence of CD209L1 in OWM, as well as its possible pseudogene status in orangutans, questions the necessity of this gene in the ape species that actually express the gene. If, indeed, this molecule is highly beneficial in species that express it, then such functions may have developed in recent evolutionary history. Alternatively, CD209L1 and CD209L2 may share functional characteristics and the absence of one may be compensated for by the presence of the other. The conserved nature of the region encoding the CD209L2 neck is most reasonably explained by the occurrence of a severe deletion that became fixed. Because of its nonrepetitive nature (it contains only a fraction of the 69-bp repeat), it may subsequently have become resistant to processes resulting in insertions or deletions. Like CD209L1, the functional necessity of CD209L2 is uncertain given its absence in humans and the possibility that it is a pseudogene in gorillas. Finally, the conserved nature of the human CD209 repeat region is curious in light of its somewhat polymorphic nature in other ape species. If, indeed, this region of the CD209 gene family is prone to expansion and contraction in general, then the specific neck length of human CD209 must confer a particularly favorable function, or unique structural characteristics of the DNA in the region of the human CD209 gene have evolved that inadvertently inhibit repeat insertions or deletions. The presence of this gene in OWM and apes, as well as the ability of its protein product to bind ICAM-3 at relatively high levels, may suggest that it has maintained a function in cell-cell interaction over millions of years.
Several studies suggest the importance of neck region length for efficient virus transmission to target T cells. The mouse CD209 molecule, referred to as DC-SIGN (1) or SIGNR1 (17), which contains a short neck region of roughly 2.5 repeat lengths, has been shown to bind HIV-1, but it is inefficient in virus transmission to T cells (1). Transfection studies with a human CD209 construct that lacks the entire coding sequence for the neck region indicated that this segment is essential for both binding of HIV-1 and efficient virus transmission by CD209 (18). Thus, the inefficient virus transmission observed when cells expressing Mm-CD209L2 were used could be explained in part by its truncated neck domain, although other parts, such as the cytoplasmic tail, may also influence the process. Whether the poor virus transmission is caused by a weak interaction between the viral envelope and CD209L2 remains to be determined. However, virus binding and transmission by CD209 are dissociable functions (19), and it is possible that the structural differences between CD209 and CD209L2 specifically affect the process of virus transmission by CD209L2 while maintaining strong binding of the virus. Further investigation of the mechanistic basis of the poor transmission of HIV-1 or SIV by CD209L2 should involve detailed analysis of both virus binding and transmission by using various chimeric and mutant constructs. Given that CD209 and CD209L1 can form tetramers via their neck domains (16) and Mm-CD209 and Mm-CD209L2 may be coexpressed, it will also be interesting to examine whether CD209L2 expression has dominant negative or regulatory effects on CD209-mediated virus transmission.
Patterns of Mm-CD209 and Mm-CD209L2 expression are similar to those of human CD209L1 in that it is expressed most notably in the liver and LN (Fig. 4A), the only tissues containing mRNA for CD209 homologues detectable by Northern analysis. The more sensitive RT-PCR method indicated that a number of additional tissues also express the molecule but evidently at lower levels. Insufficient amounts of mRNA from skin prohibited its use in Northern analysis, but strong signals representing CD209 and CD209L2 were observed by RT-PCR in spite of nearly undetectable levels of the G3PDH control (Fig. 4B). Thus, skin may also express significant amounts of the CD209 homologues. Previous studies have indicated the expression of Mm-CD209 by DCs and macrophages in lymphoid and mucosal tissues, as well as by sinusoidal endothelial cells in the liver (10, 14); however, given the possibility that antibodies raised against Mm-CD209 cross-react with CD209L2, it is necessary to determine which molecule(s) is responsible for the observed staining patterns.
Expression of the rhesus genes in inguinal LN was detectable by RT-PCR but not by Northern analysis, whereas their expression in Ax-LN was relatively pronounced and readily detectable by Northern analysis. This observation may reflect unique properties specific to secondary lymphoid tissues differentially located in the body. For example, during an early response to SIV infection, rhesus LN in the upper body, such as Ax-LN, are activated relative to those located in the lower body, such as inguinal LN (26).
The level of Mm-CD209 and Mm-CD209L2 expression may vary among individuals (Fig. 4A), although a more thorough analysis involving larger sample sizes is required in order to define the factors that may influence this heterogeneity (e.g., age, disease status, polymorphisms, etc.). Rhesus MDDCs have been shown to lack expression of CD209 (27), and we now infer that these cells also failed to express CD209L2 since the probes and MAbs used in the previous analyses recognize both family members. Given the possibility of heterogeneity in CD209 family gene expression among different individuals or under different conditions within a single individual, reassessment of MDDCs may be warranted.
Insight regarding the differential biological functions of the CD209 family of molecules may explain the fluctuation in the number and type of CD209 family genes observed across primate species. If CD209 is truly important in enhancing HIV pathogenesis by facilitating infection of T cells in vivo, selection for deleterious mutations in CD209 may occur (particularly in regions of the world where HIV infection is highly endemic), potentially leading to the demise of what is currently the most stable CD209 gene family member.
Acknowledgments
We thank G. K. Pei for sequence data and A. Lackner, R. Stanyon, G. Franchini, B. Rehermann, and S. O'Brien for providing DNA, tissue, and cell samples from nonhuman primates.
This study was funded in whole or in part by federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400, as well as by Division of Research Resources (National Institutes of Health) grant RR00168.
The content of this report does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government.
REFERENCES
- 1.Baribaud, F., S. Pohlmann, T. Sparwasser, M. T. Kimata, Y. K. Choi, B. S. Haggarty, N. Ahmad, T. Macfarlan, T. G. Edwards, G. J. Leslie, J. Arnason, T. A. Reinhart, J. T. Kimata, D. R. Littman, J. A. Hoxie, and R. W. Doms. 2001. Functional and antigenic characterization of human, rhesus macaque, pigtailed macaque, and murine DC-SIGN. J. Virol. 75:10281-10289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bashirova, A. A., T. B. Geijtenbeek, G. C. van Duijnhoven, S. J. van Vliet, J. B. Eilering, M. P. Martin, L. Wu, T. D. Martin, N. Viebig, P. A. Knolle, V. N. KewalRamani, Y. van Kooyk, and M. Carrington. 2001. A dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin (DC-SIGN)-related protein is highly expressed on human liver sinusoidal endothelial cells and promotes HIV-1 infection. J. Exp. Med. 193:671-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benveniste, R. E., R. Heinemann, G. L. Wilson, R. Callahan, and G. J. Todaro. 1974. Detection of baboon type C viral sequences in various primate tissues by molecular hybridization. J. Virol. 14:56-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benveniste, R. E., L. Kuller, S. T. Roodman, S. L. Hu, and W. R. Morton. 1993. Long-term protection of macaques against high-dose type D retrovirus challenge after immunization with recombinant vaccinia virus expressing envelope glycoproteins. J. Med. Primatol. 22:74-79. [PubMed] [Google Scholar]
- 5.Blauvelt, A., S. Glushakova, and L. B. Margolis. 2000. HIV-infected human Langerhans cells transmit infection to human lymphoid tissue ex vivo. AIDS 14:647-651. [DOI] [PubMed] [Google Scholar]
- 6.Cecilia, D., V. N. KewalRamani, J. O'Leary, B. Volsky, P. Nyambi, S. Burda, S. Xu, D. R. Littman, and S. Zolla-Pazner. 1998. Neutralization profiles of primary human immunodeficiency virus type 1 isolates in the context of coreceptor usage. J. Virol. 72:6988-6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chomczynski, P. 1992. One-hour downward alkaline capillary transfer for blotting of DNA and RNA. Anal. Biochem. 201:134-139. [DOI] [PubMed] [Google Scholar]
- 8.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 9.Curtis, B. M., S. Scharnowske, and A. J. Watson. 1992. Sequence and expression of a membrane-associated C-type lectin that exhibits CD4-independent binding of human immunodeficiency virus envelope glycoprotein gp120. Proc. Natl. Acad. Sci. USA 89:8356-8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geijtenbeek, T. B., G. Koopman, G. C. van Duijnhoven, S. J. van Vliet, A. C. van Schijndel, A. Engering, J. L. Heeney, and Y. van Kooyk. 2001. Rhesus macaque and chimpanzee DC-SIGN act as HIV/SIV gp120 trans-receptors, similar to human DC-SIGN. Immunol. Lett. 79:101-107. [DOI] [PubMed] [Google Scholar]
- 11.Geijtenbeek, T. B., D. S. Kwon, R. Torensma, S. J. van Vliet, G. C. van Duijnhoven, J. Middel, I. L. Cornelissen, H. S. Nottet, V. N. KewalRamani, D. R. Littman, C. G. Figdor, and Y. van Kooyk. 2000. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell 100:587-597. [DOI] [PubMed] [Google Scholar]
- 12.Geijtenbeek, T. B., R. Torensma, S. J. van Vliet, G. C. van Duijnhoven, G. J. Adema, Y. van Kooyk, and C. G. Figdor. 2000. Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3 receptor that supports primary immune responses. Cell 100:575-585. [DOI] [PubMed] [Google Scholar]
- 13.Geijtenbeek, T. B., Y. van Kooyk, S. J. van Vliet, M. H. Renes, R. A. Raymakers, and C. G. Figdor. 1999. High frequency of adhesion defects in B-lineage acute lymphoblastic leukemia. Blood 94:754-764. [PubMed] [Google Scholar]
- 14.Jameson, B., F. Baribaud, S. Pohlmann, D. Ghavimi, F. Mortari, R. W. Doms, and A. Iwasaki. 2002. Expression of DC-SIGN by dendritic cells of intestinal and genital mucosae in humans and rhesus macaques. J. Virol. 76:1866-1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwon, D. S., G. Gregorio, N. Bitton, W. A. Hendrickson, and D. R. Littman. 2002. DC-SIGN-mediated internalization of HIV is required for trans-enhancement of T cell infection. Immunity 16:135-144. [DOI] [PubMed] [Google Scholar]
- 16.Mitchell, D. A., A. J. Fadden, and K. Drickamer. 2001. A novel mechanism of carbohydrate recognition by the C-type lectins DC-SIGN and DC-SIGNR. Subunit organization and binding to multivalent ligands. J. Biol. Chem. 276:28939-28945. [DOI] [PubMed] [Google Scholar]
- 16a.National Institutes of Health. 1985. Guide for the care and use of laboratory animals. NIH publication 86-23. National Institutes of Health, Bethesda, Md.
- 17.Park, C. G., K. Takahara, E. Umemoto, Y. Yashima, K. Matsubara, Y. Matsuda, B. E. Clausen, K. Inaba, and R. M. Steinman. 2001. Five mouse homologues of the human dendritic cell C-type lectin, DC-SIGN. Int. Immunol. 13:1283-1290. [DOI] [PubMed] [Google Scholar]
- 18.Pohlmann, S., F. Baribaud, B. Lee, G. J. Leslie, M. D. Sanchez, K. Hiebenthal-Millow, J. Munch, F. Kirchhoff, and R. W. Doms. 2001. DC-SIGN interactions with human immunodeficiency virus type 1 and 2 and simian immunodeficiency virus. J. Virol. 75:4664-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pohlmann, S., G. J. Leslie, T. G. Edwards, T. Macfarlan, J. D. Reeves, K. Hiebenthal-Millow, F. Kirchhoff, F. Baribaud, and R. W. Doms. 2001. DC-SIGN interactions with human immunodeficiency virus: virus binding and transfer are dissociable functions. J. Virol. 75:10523-10526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pohlmann, S., E. J. Soilleux, F. Baribaud, G. J. Leslie, L. S. Morris, J. Trowsdale, B. Lee, N. Coleman, and R. W. Doms. 2001. DC-SIGNR, a DC-SIGN homologue expressed in endothelial cells, binds to human and simian immunodeficiency viruses and activates infection in trans. Proc. Natl. Acad. Sci. USA 98:2670-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 22.Soilleux, E. J., R. Barten, and J. Trowsdale. 2000. DC-SIGN; a related gene, DC-SIGNR; and CD23 form a cluster on 19p13. J. Immunol. 165:2937-2942. [DOI] [PubMed] [Google Scholar]
- 23.Soilleux, E. J., and N. Coleman. 2001. Langerhans cells and the cells of Langerhans cell histiocytosis do not express DC-SIGN. Blood 98:1987-1988. [DOI] [PubMed] [Google Scholar]
- 24.Soilleux, E. J., L. S. Morris, B. Lee, S. Pohlmann, J. Trowsdale, R. W. Doms, and N. Coleman. 2001. Placental expression of DC-SIGN may mediate intrauterine vertical transmission of HIV. J. Pathol. 195:586-592. [DOI] [PubMed] [Google Scholar]
- 25.Turville, S. G., J. Arthos, K. M. Donald, G. Lynch, H. Naif, G. Clark, D. Hart, and A. L. Cunningham. 2001. HIV gp120 receptors on human dendritic cells. Blood 98:2482-2488. [DOI] [PubMed] [Google Scholar]
- 26.Wallace, M., R. Pyzalski, D. Horejsh, C. Brown, M. Djavani, Y. Lu, J. M. Hanson, J. L. Mitchen, S. B. Perlman, and C. D. Pauza. 2000. Whole body positron emission tomography imaging of activated lymphoid tissues during acute simian-human immunodeficiency virus 89.6PD infection in rhesus macaques. Virology 274:255-261. [DOI] [PubMed] [Google Scholar]
- 27.Wu, L., A. A. Bashirova, T. D. Martin, L. Villamide, E. Mehlhop, A. O. Chertov, D. Unutmaz, M. Pope, M. Carrington, and V. N. KewalRamani. 2002. Rhesus macaque dendritic cells efficiently transmit primate lentiviruses independently of DC-SIGN. Proc. Natl. Acad. Sci. USA 99:1568-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]