Abstract
Lens epithelium-derived growth factor p75 (LEDGF/p75) is a DNA-binding, transcriptional co-activator that participates in HIV-1 integration site targeting. Using complementary approaches, we determined the mechanisms of LEDGF/p75 DNA-binding in vitro and chromatin-association in living cells. The binding of highly-purified, recombinant protein was assayed by surface plasmon resonance (SPR) and electrophoretic mobility gel shift. Neither assay revealed evidence for sequence-specific DNA-binding. Residues 146–197 spanning the nuclear localization signal (NLS) and two AT-hook motifs mediated non-specific DNA-binding, and DNA-binding deficient mutants retained the ability to efficiently stimulate HIV-1 integrase activity in vitro. Chromatin-association was assessed by visualizing the localization of EGFP fusion proteins in interphase and mitotic cells. Although a conserved N-terminal PWWP domain was not required for binding to condensed mitotic chromosomes, its deletion subtly affected the nucleoplasmic distribution of the protein during interphase. A dual AT-hook mutant associated normally with chromatin, yet when the mutations were combined with NLS changes or deletion of the PWWP domain, chromatin-binding function was lost. As the PWWP domain did not readily bind free DNA in vitro, our results indicate that chromatin-association is primarily affected through DNA-binding, with the PWWP domain likely contributing a protein interaction to the overall affinity of LEDGF/p75 for human chromatin.
INTRODUCTION
Transcriptional co-activators play important roles in regulating gene expression in higher eukaryotes. Some, like acetyltransferases, are enzymes that directly modify chromatin. Other co-activators lack enzyme function and effect transcription by interacting with the general transcription apparatus [reviewed in ref. (1)]. Transcriptional co-activator p75, also known as lens epithelium-derived growth factor (LEDGF), fits in this latter category. A member of the hepatoma-derived growth factor (HDGF) family of proteins, LEDGF/p75 and its smaller splice-variant LEDGF/p52 were identified as activities that co-purified with the PC4 general transcriptional co-activator (2).
LEDGF/p75 can affect cellular or organismal physiology. Its expression can protect cells from stress-induced apoptosis (3,4). Elevated levels of the protein are observed in some prostate cancers (5) and relatively rare cases of acute and chronic myeloid leukemia harbor NUP98-PSIP1 translocations that predictably express NUP98–LEDGF/p75 fusion proteins (6–8). LEDGF/p75 is a nuclear protein that intimately associates with chromatin (9–11) and like other chromatin-binding proteins, antibodies to LEDGF/p75 occur in some autoimmune diseases [reviewed in ref. (12)]. However the regions and amino acid residues of LEDGF/p75 that mediate chromatin-association are unknown. In this study, we identify the LEDGF/p75 determinants that effect its interaction with chromatin in live cells.
Partial proteolysis and sequence analyses previously revealed two ∼10 kDa structured domains within the 530-residue human protein: an N-terminal PWWP domain (residues 1–93) that is present in a variety of chromatin-associated proteins (13,14) and an integrase-binding domain (IBD, residues 347–429) that occurs in one other HDGF family member, HDGF-related protein 2 (15,16). LEDGF/p75 binds to DNA in vitro (17,18) and as a prelude to cell-based experiments, the regions and amino acid residues that mediate DNA-binding were mapped using surface plasmon resonance (SPR) and electrophoretic mobility shift assay (EMSA). Unlike previous reports (17), the preferential binding of purified LEDGF/p75 protein to specific DNA sequences was not detected. The nuclear localization signal (NLS) (16,19) and a nearby dual copy of the AT-hook DNA-binding motif (20) mediate LEDGF/p75 non-specific DNA activity in vitro. The PWWP domain is dispensable for binding to naked DNA, but functions together with the NLS and AT-hooks to effect the interaction with chromatin in cells.
LEDGF/p75 is a dominant cellular binding partner of HIV-1 integrase (10,11,19,21–24) and potently stimulates the in vitro activities of the viral enzyme (10,15,25). More recently, LEDGF/p75 was shown to play a role in targeting HIV-1 to active gene regions and A/T-rich sequences during integration (26). LEDGF/p75 mutant proteins defective for DNA-binding, integrase-binding or both activities were analyzed for stimulation of integrase activity using a novel real-time PCR-based integration assay. As the results of these assays discount an important role for DNA-binding in stimulating integrase function, we infer that the ability for LEDGF/p75 to target HIV-1 to A/T-rich sequences and active genes is separable from its integrase-stimulatory activity.
MATERIALS AND METHODS
Plasmid constructs
Deletion mutants were expressed in bacteria as fusions to glutathione S-transferase (GST) using pGEX-6P3-based vectors (Amersham Biosciences). Sense primers for truncated LEDGF coding sequences were tagged at their 5′ ends with a BamHI site, followed by an initiating ATG codon. DNA fragments amplified by PCR from pCP-Nat75 (11) using PfuUltra DNA polymerase (Stratagene) were digested with BamHI and ligated with BamHI/SmaI-digested vector DNA. Full-length LEDGF/p75 and missense variants were expressed from pFT-1-LEDGF (27) as fusions to an N-terminal hexahistidine (His6) tag. Mutations were introduced into pFT-1-LEDGF by QuikChange mutagenesis (Stratagene).
For mammalian cell-expression, sequences were amplified from corresponding pFT-1-LEDGF constructs using PfuUltra polymerase and either 5′-GCGCAGATCTCGATGACTCGCGATTTCAAAC (sense primer for full-length constructs) or 5′-GCGCAGATCTCGTCAAGTGAACAGGCAGCAAC (sense primer for PWWP deletion constructs) and 5′-CGCGAATTCCTAGTTATCTAGTGTAGAATCC (antisense primer for all constructs). The underlined sequences are BglII and EcoRI restriction sites in sense and antisense primers, respectively. BglII/EcoRI-digested fragments were ligated with BglII/EcoRI-digested pEGFP-C2 (Clontech). The initial 116 codons of LEDGF/p75 were fused to enhanced green fluorescent protein (EGFP) to create EGFP-PWWP. The sequences of all plasmids that were derived by PCR in this study were verified by DNA sequencing.
Expression and purification of recombinant proteins
GST-LEDGF proteins expressed in BL21 cells were purified by a two-step procedure as described previously (15) with slight modifications. Bacteria were grown at 30°C to OD600 0.8–0.9, protein expression was induced by adding isopropyl-thio-β-d-galactopyranoside (IPTG) to 0.5 mM, followed by growth at 37°C for 2 h. Harvested bacterial pellets were frozen at −80°C, and thawed cells were resuspended in buffer containing 500 mM NaCl, 1 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 2 mM DTT, 50 mM Tris–HCl (pH 7.2). Sonicated lysates clarified by centrifugation were injected on to a GSTrap HP column (Amersham Biosciences), and bound proteins were eluted with 500 mM NaCl, 25 mM reduced glutathione, 50 mM Tris–HCl (pH 7.7). Peak fractions were pooled and the GST-tag was removed by overnight digestion at 4°C with 4 U of PreScission Protease per mg of protein. Cleaved protein diluted 5-fold with 50 mM NaH2PO4 (pH 7.2) to reduce NaCl concentration was injected on to a HiTrap SP Sepharose column (Amersham Biosciences), followed by elution with a linear gradient of NaCl from 0.2 to 1 M in 50 mM NaH2PO4. If necessary, pooled LEDGF fractions were further purified by gel filtration chromatography using a Superdex 200 column (Amersham Biosciences) in 100 mM NaCl, 50 mM NaH2PO4 (pH 7.2). Fractions containing purified protein were pooled, concentrated by ultrafiltration using Centricon-10 (Millipore Corp.), supplemented with 10% (w/v) glycerol, flash frozen in liquid N2 and stored at −80°C.
His6-tagged LEDGF/p75 expressed in BL21(DE3)pLysS cells (Novagen, Inc.) was purified using three chromatography steps. Cells grown at 32°C to OD600 0.8–0.9 were induced by adding IPTG to 0.5 mM, followed by growth at 28°C for 4 h. Bacterial pellets were frozen, and thawed cells were lysed in buffer containing 1 M NaCl, 0.5 mM PMSF, 15 mM imidazole, 25 mM Tris–HCl (pH 7.4) in the presence of EDTA-free Complete protease inhibitor (Roche Molecular Biochemicals). Sonicated lysates clarified by centrifugation were incubated in batch with Ni-nitrilotriacetate agarose beads (QIAGEN) for 1 h at 4°C. Bound proteins were eluted with 1 M NaCl, 200 mM imidazole, 25 mM Tris–HCl (pH 7.4). Peak fractions were pooled and the His6-tag was removed by overnight digestion with PreScission Protease as described above. LEDGF was further purified by cation-exchange chromatography as described above for deletion mutant proteins. Pooled LEDGF-containing fractions were then purified by gel filtration chromatography (Superdex 200) in 120 mM NaCl, 25 mM Tris–HCl (pH 7.4). Fractions containing purified protein were pooled, concentrated, frozen and stored as above. Protein concentrations were determined by the Bio-Rad protein assay using BSA as a standard. Protein purity was analyzed using Coomassie-stained images and Quantify One version 4.1.1 software (Bio-Rad Laboratories). Non-tagged LEDGF/p75 and GST were purified as described (15); RNase A was purchased from QIAGEN.
SPR
Double-stranded oligonucleotides were constructed from high performance liquid chromatography (HPLC)-purified single-strands (Integrated DNA Technologies). AE2019 (5′-biotin-GATTTAACGAGAGAAGGTTCCAGA), AE2042 (5′-biotin-GATTTAACTAAATAAATTTTAAAA), AE2043 (5′-biotin GATTTAACGAGATAATTTTACAGA), AE1198 (5′-TCTGGAACCTTCTCTCGTTAA), AE2000 (5′-TTTTAAAATTTATTTAGTTAA) and AE2002 (5′-TCTGTAAAATTATCTCGTTAA) were used for SPR. AE1198 annealed to AE2019 yielded the 21 bp consensus heat shock element (HSE) sequence that supported LEDGF/p75 binding, whereas AE2000/AE2042 and AE2002/AE2043 yielded Mut1 and Mut2 derivatives, respectively, which reportedly failed to bind LEDGF/p75 (17). Biotinylated strands were 24 bases to incorporate GAT single-strand linkers.
SPR was performed on a BIAcore 3000 instrument (Biacore International AB) at 25°C. A streptavidin-coated SA sensor chip was washed with running buffer [140 mM NaCl, 0.005% (v/v) surfactant P20, 10 mM HEPES (pH 7.4)] for 30 min, then exposed to three consecutive, 1 min injections of conditioning buffer (1 M NaCl, 50 mM NaOH) at a flow rate of 10 µl/min. Biotinylated oligonucleotides (25 nM) were injected at 5 µl/min for 1 min, which reproducibly yielded ∼150 response units (RU). Complementary strands (25 nM) were injected until the observed response equaled that of the biotinylated strand, giving rise to a homogeneous surface of double-stranded DNA. Typically, 3 of the 4 chip channels were primed with DNA, with the other serving as a reference to determine aspecific levels of LEDGF/p75 binding in the absence of DNA.
Wild-type or mutant LEDGF/p75 (25 nM) was injected using the KINJECT command with an association phase of 60 s or 120 s (for kinetics measurements) and a dissociation phase ranging from 5 to 30 min. A flow rate of 30 µl/min was used to minimize mass transport and rebinding effects. Following each protein injection, the chip surface was regenerated with three injections of conditioning buffer, followed by reannealing of non-biotinylated complementary strands. Similar levels of double-stranded DNA immobilization were reproducibly achieved after each cycle of annealing/protein injection/chip stripping. Data were analyzed using BIAevaluation software version 4.0.1 (Biacore International AB).
EMSA
The 5′ ends of AE1197 (5′-TTAACGAGAGAAGGTTCCAGA), AE1199 (5′-TTAACTAAATAAATTTTAAAA) and AE2001 (5′-TTAACGAGATAATTTTACAGA) were labeled with T4 polynucleotide kinase (Amersham Biosciences) and complementary strands (see above) were annealed as described (28) to yield 21 bp blunt-ended DNAs.
DNA-binding reactions (15 µl) contained 120 mM NaCl, 5 mM MgCl2, 2 mM DTT, 10% glycerol, 50 µg/ml BSA, 100 nM DNA, 25 mM Tris–HCl (pH 7.4) and 80 ng/ml poly dIdC (Amersham Biosciences) as non-specific competitor. LEDGF proteins diluted in 120 mM NaCl, 25 mM Tris–HCl (pH 7.4) were added last, and binding proceeded for 30 min at room temperature. Reactions (without the addition of sample buffer) were fractionated on 4% Metaphor agarose gels (Cambrex Bio Science) supplemented to contain 0.1 µg/ml heparin (29). Electrophoresis was for 2.5 h at 4°C in 0.5× TBE (44.5 mM Tris base, 44.5 mM borate, 1 mM EDTA) supplemented with 0.1 µg/ml heparin. Dried gels were visualized and quantified by PhosphorImager using ImageQuant v1.11 (Amersham Biosciences).
Supershift experiments were conducted by preincubating LEDGF/p75 (2 µM) with 100 nM DNA at room temperature for 5 min. Incubation proceeded for 30 min after adding 2.25 µg of mouse anti-LEGDF/p75 monoclonal antibody (BD Biosciences) or purified IgG1 isotype control (Sigma–Aldrich Corp.). Electrophoresis was for 7.5 h.
Cell transfection, western blot and confocal microscopy
HeLa cells were maintained in DMEM supplemented with 10% fetal calf serum (FCS), 100 IU/ml penicillin and 100 µg/ml streptomycin. For blotting, cells plated in 6 well dishes to reach confluency were transfected with 4 µg plasmid using Lipofectamine 2000 (Invitrogen Corp.). Twenty-four hours thereafter, cells were harvested, washed twice with phosphate-buffered saline and lysed in buffer containing 25 mM Tris–HCl (pH 7.4), 400 mM NaCl, 0.01% (v/v) NP-40, 0.1% (w/v) SDS, 0.1 mM PMSF and EDTA-free Complete protease inhibitor (Roche Molecular Biochemicals). Protein concentration was determined using the BCA protein assay (Pierce Biotechnology Inc.) and 15 µg fractionated on 4–20% Tris-Glycine Gels (Invitrogen Corp.) were electroblotted on to polyvinylidene difluoride membranes. Anti-EGFP antibody (BD Biosciences) was used at 1:5000 and secondary horseradish peroxidase (HRP)-conjugated goat anti-rabbit antibody (Dakocytomation California Inc.) at 1:10 000. Blots were developed with the ECL + chemiluminescent HRP substrate (Amersham Biosciences).
For confocal microscopy, cells were plated at 2 × 105 cells/well in a 24 well plate 24 h prior to transfection. Cells were transfected as above with 800 ng plasmid and 24 h later, cells were trypsinized and seeded into 8-well LabTek chambered cover glass cuvettes (Nalge Nunc International, NY) at ∼60% confluency. Live images were obtained using a Nikon C1 Laser Confocal Microscope 24 h thereafter. The cell permeable DNA stain DRAQ5™ (Alexis Biochemicals) (1 µM) was added immediately before data acquisition. All images were acquired at 1024 × 1024 resolution using a 60× water immersion objective. EGFP was excited at 488 nm and DRAQ5™ at 633 nm.
In vitro integration assays
Recombinant HIV-1 integrase was purified as described previously (15). Integration reactions (40 µl) contained 300 ng linearized mini-HIV substrate DNA (30), 0.6 µM integrase, 110 mM NaCl, 5 mM MgCl2, 10 mM DTT, 4 µM ZnCl2, 10 mM HEPES (pH 7.5). Wild-type or mutant LEDGF/p75 was used at 0.2 µM. For quantitative analyses, 300 ng supercoiled pTZ18U/PL (31) was added as target DNA. After 90 min at 37°C, reactions stopped by addition of 25 mM EDTA and 0.5% SDS were deproteinized using 10 µg of proteinase K at 37°C for 30 min. Reaction products recovered by precipitation with ethanol were resuspended in 20 µl TE buffer [10 mM Tris–HCl (pH 8), 1 mM EDTA]. DNA (10 µl) fractionated through 0.8% agarose gels in TAE buffer (40 mM Tris base, 20 mM acetate, 1 mM EDTA) was detected by staining with ethidium bromide.
Integration into pTZ18U/PL was quantified by real-time PCR using HIV-1 U5 primer AE2397 (5′-GGTTAGACCAGATCTGAGCCTGGGAG) and target DNA primers AE2315 (5′-CTACTTACTCTAGCTTCCCGGCAAC) and AE2316 (5′-TTCGCCAGTTAATAGTTTGCGCAAC). Reactions (30 µl) containing 1 µl of a 1:100 dilution of DNA, 0.3 µM of each primer and QuantiTect SYBR Green PCR Kit (QIAGEN) components were cycled in a DNA Engine Opticon thermal cycler (MJ Research Inc.) using 20 s of denaturation at 94°C, 20 s of annealing at 58°C and 45 s of extension at 72°C. A standard curve was generated for each set of integration reactions by serially diluting the wild-type LEDGF/p75-containing sample prior to PCR. Results were analyzed using OpticonMONITOR™ Analysis Software version 2.01 supplied by the manufacturer.
RESULTS
Characterization of the in vitro DNA-binding activity of LEDGF/p75
LEDGF/p75 was reported to bind consensus HSE (nGAAn) and stress-related element [STRE; (T/A)GGGG(A/T)] DNA sequences in the promoter regions of stress-related genes (3,17,32). To characterize the region(s) of LEDGF/p75 responsible for DNA-binding, a set of three previously described 21 bp oligonucleotide substrates were utilized. LEDGF/p75 reportedly bound to the HSE substrate (which contains two GAA motifs) in a polyacrylamide gel-based EMSA, whereas Mut1 and Mut2, containing either TAA or AAA in place of GAA (Figure 1A), failed to support detectable binding (17).
Figure 1.
LEDGF/p75 binds to various DNA sequences in vitro. (A) Sequences of 21 bp substrates. The HSE sequence, containing two 5′-GAA motifs (underlined), has a G/C content of 47%. Mut1 (5% G/C rich) and Mut2 (24% G/C rich) were derived from HSE by mutating G or C residues to A or T (17). Altered residues are in bold type. (B) Binding of LEDGF/p75 to DNA as determined by SPR. The response for each substrate is plotted as a function of time. Values obtained from a blank streptavidin-coated surface were subtracted from DNA-dependent signals. Similar results were obtained over multiple (more than a dozen) independent experiments. (C) A subset of the LEDGF proteins used in this study. Approximately 2 µg of the following purified proteins were detected by staining with Coomassie blue following polyacrylamide gel electrophoresis: lane 1, non-tagged wild-type LEDGF/p75; lane 2, PreScission Protease-treated wild-type LEDGF/p75; lane 3, 347–530; lane 4, 1–116; lane 5, 93–226; lane 6, MutL1; lane 7, MutL3; lane 8, MutL4; lane 9, MutL5. The migration positions of molecular mass standards in kDa are shown next to the gel. (D) LEDGF/p75 DNA-binding as determined by EMSA. The reactions in lanes 2, 9 and 16 contained 125 nM LEDGF/p75. Lanes 3, 10 and 17 contained 250 nM LEDGF/p75; lanes 4, 11 and 18, 500 nM; lanes 5, 12 and 19, 1 µM; lanes 6, 13 and 20, 2 µM; lanes 7, 14 and 21, 4 µM. LEDGF/p75 was omitted from the reactions in lanes 1, 8 and 15. The migration positions of starting substrates, shifted complexes and slowly-migrating high molecular weight (HMW) complexes are indicated. Similar results were obtained following multiple independent experiments. (E) Supershift experiment. LEDGF/p75 was omitted from the reaction in lane 1. Lane 2 contained 2 µM LEDGF/p75 and 100 nM Mut1 DNA. Lane 3, anti-LEDGF/p75 antibody was added 5 min after LEDGF/p75 and DNA. Lane 4, same as lane 3 except control IgG1 was substituted for the specific antibody. Results are representative of five independent supershift experiments.
SPR was initially used to study DNA-binding. Unlike gel-based EMSAs, SPR can monitor molecular interactions in real-time under native conditions like isotonic salt, and can also detect transient interactions (33,34). Biotinylated substrates were immobilized on to a streptavidin-coated sensor chip, LEDGF/p75 was injected for 1 min at the sub-saturating concentration of 25 nM, and protein dissociation was monitored for 30 min by flowing running buffer over the chip surface. Unexpectedly, LEDGF/p75 displayed near equal efficiencies of binding to all three DNA substrates (Figure 1B). DNA-binding was characterized by a relatively fast association, followed by biphasic dissociation. LEDGF/p75 dissociated from DNA relatively quickly for ∼2 min, and then more slowly, giving rise to stable binding after ∼15 min. Based on kinetic measurements, LEDGF/p75 DNA interactions could fit, though with significant residuals, to a simple 1:1 Langmuir binding model. We were unable to observe any significant difference between association/dissociation curves or apparent dissociation constants for LEDGF/p75 binding to the different substrates (Figure 1B and data not shown). Similar binding characteristics were observed using a biotinylated 38 bp oligonucleotide that modeled the U5 end of HIV-1 DNA. We note that these results were independent of the LEDGF/p75 purification procedure, as protein recovered via the use of a His6 affinity tag (three total chromatographic steps; Figure 1C, lane 2) as well as non-tagged LEDGF/p75 (four chromatographic steps; Figure 1C, lane 1) displayed indistinguishable DNA-binding profiles to the HSE, Mut1 and Mut2 substrates.
Due to the discrepancy observed here with SPR and previous results that primarily used EMSA, we next assayed LEDGF/p75 DNA-binding by gel-based methods. Protein was incubated with non-biotinylated DNA substrates, and reactions were fractionated through 4% MetaPhor agarose gels (Figure 1D). As observed with SPR, each of the three previously described substrates supported LEDGF/p75 DNA complex formation. Faster-migrating (presumably lower molecular weight) complexes were first observed with 250 ng LEDGF/p75 (Figure 1D, lanes 3, 10 and 17) and better-defined, more slowly-migrating complexes were observed at 2 and 4 µM protein (lanes 6, 7, 13, 14, 20 and 21). As was the case with SPR, the different preparations of LEDGF/p75 protein yielded indistinguishable patterns of DNA-binding (data not shown). Similar overall patterns of DNA-binding were observed in 4.5% polyacrylamide gels, though we note the formation of better-defined nucleoprotein complexes in MetaPhor agarose.
The different preparations of wild-type LEDGF/p75 protein used in these assays were ∼95 and 80% pure (Figure 1C, lanes 1 and 2). Considering the use of different purification procedures, we felt reasonably confident that the observed DNA-binding was due to LEDGF/p75 and not a minor co-purifying contaminant(s). To demonstrate specificity, a supershift experiment was performed (Figure 1E). To enhance the separation of potential high molecular weight complexes, gel running times were increased from 2.5 to 7.5 h. At these conditions, LEDGF/p75 yielded a heterogeneous population of shifted complexes (compare lane 2 to lane 1). Importantly, complex mobilities were retarded by an anti-LEDGF/p75 monoclonal antibody under conditions where migration positions were unaffected by an isotype control antibody (compare lanes 2–4). Based on these observations as well as the identical behavior of different protein preparations, we conclude that SPR (Figure 1B) and EMSA (Figure 1D) detected LEDGF/p75 DNA-binding activity.
DNA-binding activity maps to residues 93–226
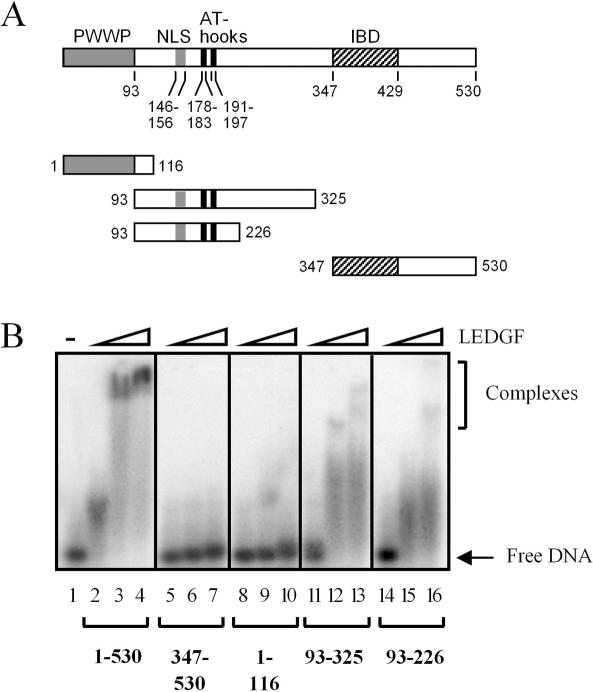
Deletion analysis was utilized to locate the region(s) of LEDGF/p75 responsible for DNA-binding. Choices of protein boundaries were based on previously defined conserved sequences and functional domains (10,13–15). The PWWP domain, encompassing the N-terminal 93 residues of LEDGF/p75, is the most conserved part of the protein (15). The other conserved domain, comprised of residues 347–429, mediates binding to integrase (15,16,25,35) (Figure 2A). Shorter stretches of conserved residues were also targeted. Residues 146RRGRKRK152 comprise the NLS (16,19,36), whereas residues 178–197 resemble two AT-hook DNA-binding motifs (10,15) (see below). Four deletion mutant proteins were constructed and purified to address the roles of these elements in DNA-binding. The 1–116 fragment contains the PWWP domain, and the C-terminal 347–530 fragment harbors the IBD (Figures 1C and 2A). Two internal fragments, 93–325 and 93–226, contain the NLS and AT-hooks (Figure 2A). Proteins were produced in bacteria as fusions to GST, and the tags were removed prior to DNA-binding assays (Figure 1C).
Figure 2.
LEDGF/p75 residues 93–226 harbor DNA-binding activity. (A) Schematic representation of wild-type LEDGF/p75 and deletion mutant proteins. Conserved sequence motifs/functional domains are indicated above the protein, with boundaries indicated below. NLS, nuclear localization signal; IBD, integrase-binding domain. (B) DNA-binding activities of deletion mutant proteins. DNA (100 nM Mut1) was reacted with the following protein concentrations: 250 nM (lanes 2, 5, 8, 11 and 14), 2 µM (lanes 3, 6, 9, 12 and 15) or 5 µM (lanes 4, 7, 10, 13 and 16). LEDGF was omitted from the reaction in lane 1. The results are representative of four independent gel shift experiments. The migration positions of free DNA and nucleoprotein complexes are indicated.
Neither the PWWP domain (Figure 2B, lanes 8–10) nor the larger C-terminal 347–530 fragment (lanes 5–7) readily bound to DNA, even at high protein concentrations. On the contrary, internal fragments 93–325 and 93–226 shifted the DNA substrate, though we note initial complex formation required more deletion mutant protein than wild-type (compare lanes 11 and 14 to lane 2) and high molecular weight complexes formed less efficiently than with the wild-type. Akin to the full-length protein, 93–226 bound similarly to the HSE, Mut1 and Mut2 substrates (data not shown). SPR analyses with this set of mutant proteins yielded similar results (data not shown). We conclude that the DNA-binding activity of LEDGF/p75 resides within residues 93–226, and by extension, that neither the PWWP domain nor residues 347–530 contribute significantly to DNA-binding function in vitro.
The NLS and both copies of the AT-hook mediate DNA-binding
Based on the previous results, the NLS and putative AT-hook motifs were mutagenized. The core AT-hook consensus sequence is Pro-Arg-Gly-Arg-Pro (with RGRP being invariant) (37). Figure 3A aligns this sequence to the putative LEDGF/p75 AT-hook sequences; as the NLS was observed to contain three of the four invariant residues, it was included in the alignment. The second AT-hook (hereafter referred to as AT2) contains 193PRGRP197, which precisely matches the consensus sequence. AT1, though mismatched at the first Pro position, contains 180RGRP183, which matches the four invariant residues. The NLS, which harbors 147RGR149, substitutes Lys-150 for the invariant Pro (Figure 3A, asterisk). We note that Lys-150 plays a key role in NLS function (19).
Figure 3.
The NLS and both AT-hooks mediate DNA-binding. (A) Alignment of invariant amino acids within human LEDGF/p75 AT2, AT1 and NLS sequences to the AT-hook consensus motif. Of the five residues that comprise the motif, four (RGRP) are invariant (Inv) (37). Residues in LEDGF/p75 that match the consensus sequence are underlined. Lys-150 within the NLS (marked by an asterisk) is essential for nuclear import (19). (B) Residues altered in MutL1–MutL5. Asterisks indicate amino acid residues that are identical among human, chicken and frog LEDGF/p75 orthologs (15). Invariant NLS and AT-hook residues are underlined with continuous and dotted lines, respectively. Mutated residues are in bold type. (C) MutL1–MutL5 activities, expressed as percent of wild-type DNA-binding. Error bars represent the variation obtained from duplicate measurements.
Based on these observations, the following full-length LEDGF/p75 mutant proteins were constructed (Figure 3B and Table 1). Each of the six basic residues within the NLS was changed to Ser or Ala in MutL2. MutL4 and MutL5 contained Ala-Ala-Ala in place of Arg-Gly-Arg in the AT1 and AT2 consensus sequences, respectively. MutL3 combined the MutL4 and MutL5 changes, whereas MutL1 contained a total of 12 amino acid substitutions (MutL3 plus MutL2). Proteins were expressed in bacteria as His6-tag fusions, and tags were removed prior to DNA-binding assays (Figure 1C). Each mutant protein was expressed and displayed purification properties similar to wild-type LEDGF/p75, indicating that the noted amino acid substitutions did not significantly affect protein expression, stability or folding.
Table 1.
LEDGF/p75 mutants
| Mutant name | Targeted region(s)a | Mutations |
|---|---|---|
| MutL1 | NLS, AT1, AT2 | R146S, R147A, R149A, K150A, R151S, K152A, R180A, G181A, R182A, R194A, G195A, R196A |
| MutL2 | NLS | R146S, R147A, R149A, K150A, R151S, K152A |
| MutL3 | AT1, AT2 | R180A, G181A, R182A, R194A, G195A, R196A |
| MutL4 | AT1 | R180A, G181A, R182A |
| MutL5 | AT2 | R194A, G195A, R196A |
| MutL6 | NLS, AT1, AT2, IBD | R146S, R147A, R149A, K150A, R151S, K152A, R180A, G181A, R182A, R194A, G195A, R196A, D366N |
| MutL7 | PWWP | Δ1-92 |
| MutL8 | PWWP, NLS, AT1, AT2 | Δ1-92, R146S, R147A, R149A, K150A, R151S, K152A, R180A, G181A, R182A, R194A, G195A, R196A |
| MutL9 | PWWP, AT1, AT2 | Δ1-92, R180A, G181A, R182A, R194A, G195A, R196A |
aNLS, nuclear localization signal (residues 146–152); AT1, AT-hook motif 1 (residues 178–183); AT2, AT-hook motif 2 (residues 191–197); PWWP, PWWP domain (residues 1–93); IBD, integrase-binding domain (residues 347–429) (15).
The level of mutant protein that remained stably bound to Mut1 DNA after 20 min of dissociation was quantified by SPR, and results were expressed as percentage of wild-type DNA-binding activity (Figure 3C). Each of the five mutant proteins dissociated from DNA significantly faster than wild-type, indicating that the NLS and each AT-hook contributed to DNA-binding. However, the respective contribution of each of these elements to DNA-binding differed. MutL4 retained the most function (∼45% of wild-type), revealing AT1 180RGR182 as the least important of the three invariant sequences under these conditions. MutL2 displayed about 20% of wild-type function, whereas MutL5 retained about 13% residual DNA-binding activity (Figure 3C). Based on these results, the contribution of the three different elements to DNA-binding was ranked AT2>NLS>AT1. The NLS-defective K150A point mutant protein (19) displayed wild-type DNA-binding activity, indicating that Lys-150 on its own does not supply critical DNA-binding function (data not shown). Mutation of four additional lysine residues surrounding the core AT2 sequence (positions 179, 189, 198 and 201; Figure 3B) did not further reduce the residual activity of MutL5, indicating that invariant 194RGR196 residues are primarily responsible for AT2-mediated DNA recognition.
The dual AT-hook mutant MutL3 (Figure 1C, lane 7) displayed about 7% of wild-type DNA-binding activity, revealing an additive effect of combining separate AT1 and AT2 mutations. The NLS and dual AT-hook mutant MutL1 (Figure 1C, lane 6) harbored just 3% residual DNA-binding function (Figure 3C). Similar results were obtained for the five full-length LEDGF/p75 mutant proteins using HSE or Mut2 substrate DNA in place of Mut1 (data not shown). Based on this, we conclude that a tripartite motif comprised of the NLS and both copies of the AT-hook mediates the binding of LEDGF/p75 to DNA in vitro.
The PWWP domain, though dispensable for binding to naked DNA in vitro, contributes to chromatin-association in vivo
Results of in vitro assays discounted a major role for the PWWP domain in binding to DNA (Figures 2B and 3C). The PWWP domain is found in more than 60 eukaryotic proteins and where analyzed, appears to play a central role in mediating interactions with chromatin (38,39). To analyze the role of the PWWP domain as well as the NLS and AT-hook motifs in the association of LEDGF/p75 with human chromatin, EGFP fusion proteins were visualized in interphase and mitotic cells using laser-scanning microscopy. The following mutants were constructed for these analyses. MutL7 lacked the PWWP domain. MutL9 contained this change along with the dual AT-hook changes. The NLS was additionally altered in MutL8 (Table 1).
LEDGF/p75 displays a characteristic irregular intranuclear distribution during interphase where it is in large part excluded from nucleoli, and associates intimately with chromosomes during mitosis (9–11,16) (Figures 4A and 5A). We reported that transiently-expressed NLS mutants of LEDGF/p75 were excluded from nuclei at 24 h post-transfection, with a small number of cells displaying the wild-type nuclear distribution pattern (19). Herein, where measurements were obtained 48 h post-transfection, EGFP-MutL2 (NLS-deficient) mainly localized to cell nuclei, displaying the wild-type intranuclear distribution pattern (data not shown). In accordance with Vanegas et al. (16), the NLS mutant also concentrated on condensed chromosomes, indicating that nuclear accumulation occurred through chromosomal capture during open mitoses. Because of this design, the inability for a mutant fusion protein to interact normally with chromatin was primarily due to defects in chromatin-association as compared to LEDGF/p75 nuclear import. EGFP-MutL3, with both AT-hooks mutated, behaved as the wild-type fusion protein in interphasic and mitotic cells (data not shown). In contrast, EGFP-MutL1, with the NLS and both AT-hooks mutated, distributed diffusively throughout interphase cells (Figure 4B) and failed to concentrate onto condensed chromosomes (Figure 5B). As the results of western blotting revealed stable expression of full-length EGFP-MutL1 protein in these cells (Supplementary Figure 1), we infer that DNA-binding plays a central role in chromatin-binding in vivo.
Figure 4.
Distribution of wild-type and mutant LEDGF/p75 fusion proteins in live cells. (A) Wild-type (WT) LEDGF/p75. Note the characteristic irregular nucleoplasmic distribution and exclusion from nucleoli (9,11). These two cells, not adjacent in the original image, were brought together to highlight the wild-type phenotype. (B–F) Intracellular/nuclear distribution of the indicated LEDGF mutant proteins. Two cells expressing relatively low levels of EGFP-MutL7 were highlighted in panel C; part of a cell expressing higher levels of mutant protein is seen at the upper edge in the first and third frames. The other panels depict two to three cells that are representative of the total population of transfected cells.
Figure 5.
The PWWP domain, NLS and AT-hooks contribute to chromatin interactions in vivo. (A–F) Cells undergoing mitosis were analyzed for expression of the indicated EGFP fusion protein and DNA content.
Proteins lacking the PWWP domain were expressed at significantly higher levels than wild-type LEDGF/p75 or other EGFP fusion proteins (Supplementary Figure 1). Because of this, detailed information was gleaned by lowering the intensity of the scanning laser or by visualizing cells expressing relatively low levels of mutant protein (Figure 4C; similar results obtained using either method). Similar to the wild-type, nuclear EGFP-MutL7 primarily co-localized with DNA (Figure 4A and C). Yet, the PWWP deletion mutant signal appeared more diffuse, and nucleolar exclusion was somewhat less pronounced, as compared to the wild-type (Figure 4A and C; data not shown). Because EGFP-MutL7 efficiently interacted with mitotic chromosomes (Figure 5C), we conclude that the PWWP domain by itself is not a dominant determinant of chromatin-association. Additional mutant analyses, however, revealed an important contribution to this activity. EGFP-MutL9 (PWWP and dual AT-hook mutations) distributed diffusively throughout interphasic nuclei (Figure 4D) and did not effectively engage mitotic chromosomes (Figure 5D). Thus, although separate disruption of the PWWP domain or AT-hooks did not detectably alter binding to condensed chromosomes (Figure 5C and data not shown), the combination of these changes did. To directly assess the chromatin-binding potential of the PWWP domain, EGFP-PWWP was analyzed. This protein, which was predominantly nuclear during interphase (Figure 4F), interacted with mitotic chromosomes (Figure 5F). Yet as a fraction of EGFP-PWWP remained cytosolic during mitosis (Figure 5F), we conclude that the PWWP domain interacts with condensed chromosomes less efficiently than does either wild-type LEDGF/p75 or, more importantly, the MutL7 mutant lacking the domain (Figure 5, compare panel F to panels A and C). EGFP-MutL8, altered at the NLS in addition to the PWWP domain and AT-hooks, localized exclusively to the cytoplasm of interphasic cells (Figure 4E) and was predictably deficient for binding to condensed chromosomes (Figure 5E). The somewhat unexpected cytoplasmic localization of EGFP-MutL8 was not due to loss of LEDGF/p75 epitopes, as only full-length protein was detected by blotting with the anti-EGFP antibody (Supplementary Figure 1). As leptomycin B did not affect the intracellular distribution of EGFP-MutL8 (data not shown), the phenotype was independent of CRM1-mediated nuclear export (40).
A minor role for DNA-binding in stimulating HIV-1 integrase activity
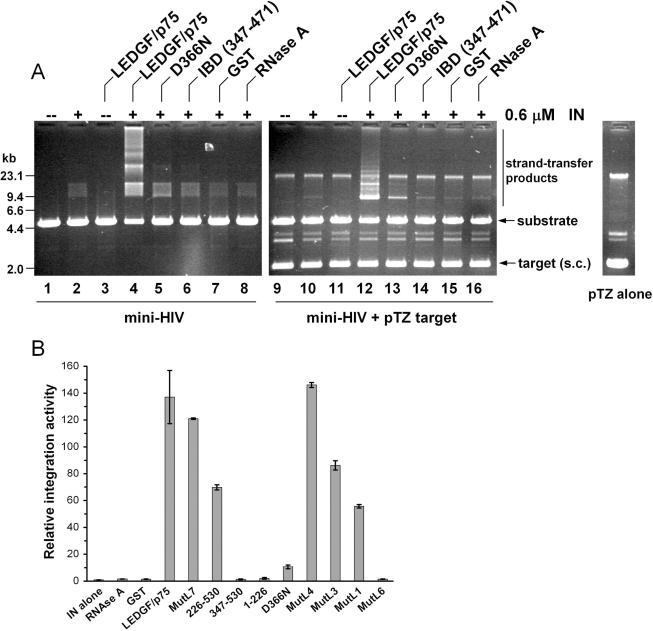
LEDGF/p75 potently stimulates the in vitro catalytic activities of HIV-1 integrase (10). The interaction with integrase, mediated through the IBD, is necessary though not sufficient for activity (15,25). To investigate the potential contribution of DNA-binding to integration co-factor function, purified mutant proteins were tested in an in vitro integration assay alongside wild-type LEDGF/p75. To facilitate quantitative read-out, a previously described mini-HIV integration assay (30) was adapted for real-time PCR. The published assay monitors the integration of substrate DNA into a second mini-HIV molecule, readily revealing the stimulatory effect of LEDGF/p75 (Figure 6A, compare lane 4 to lane 2). For real-time PCR, a supercoiled plasmid lacking viral sequences was included in the reaction as an integration target. Preliminary experiments revealed efficient use of pTZ18U/PL (Figure 6A, compare lane 12 to lanes 9–11) and parallel behavior of previously-assessed LEDGF/p75 loss-of-function mutants (compare lanes 12–14 to lanes 4–6).
Figure 6.
The role of LEDGF/p75 DNA-binding in stimulating HIV-1 integration in vitro. (A) Efficient use of pTZ18U/PL as an integration target. Reactions in lanes 2, 4–8, 10 and 12–16 contained 0.6 µM integrase, whereas reactions in lanes 3–8 and 11–16 contained 0.2 µM of the indicated protein. Reactions in lanes 9–16 additionally contained pTZ18U/PL. The migration positions of mini-HIV substrate DNA, supercoiled (s.c.) pTZ18U/PL and strand transfer reaction products are indicated. The migration pattern of the plasmid on its own is shown on the right; molecular mass standards are to the left. (B) Integrase (IN) alone, integration reaction conducted with integrase alone. Other reactions additionally contained 0.2 µM of the indicated protein. Relative levels of integration activity were quantified by real-time PCR; IN alone activity was arbitrarily set to 1.0. Error bars are variation obtained from duplicate real-time assays. Similar levels of wild-type and mutant LEDGF/p75 activities were observed following independent sets of integration reactions.
Real-time PCR results were quantified by comparing control or mutant LEDGF/p75 activities to a standard curve generated by serially diluting wild-type LEDGF/p75-dependent integration products. With this design, wild-type LEDGF/p75 was determined to stimulate HIV-1 integrase activity ∼137-fold (Figure 6B). GST and RNase A, each of which failed to stimulate integration based on gel analyses (Figure 6A, lanes 7, 8, 15 and 16), revealed about 1.1 and 1.2% of wild-type LEDGF/p75 function, respectively (panel B). In accordance with our previous report (25), the D366N missense mutant, which failed to interact with HIV-1 integrase, was greatly impaired in its ability to stimulate integration, revealing about 8% of wild-type function in the quantitative assay (Figure 6).
Because the N-terminal deletion mutant MutL7 displayed about 88% of wild-type function (Figure 6B), we conclude that the PWWP domain does not play an important role in integrase co-factor function in vitro. As the AT1 mutant MutL4 (about 45% of wild-type DNA-binding; Figure 3C) stimulated integrase as efficiently as wild-type (Figure 6B), we conclude that wild-type levels of LEDGF/p75 DNA-binding are not required for maximal activity. This conclusion was further bourn out by analyzing MutL3 (AT1/2 mutant) and MutL1 (NLS + AT1/2): whereas MutL3 and MutL1 retained but 7 and 3% of wild-type DNA-binding activity (Figure 3C), ∼63 and 41% of wild-type LEDGF/p75 stimulatory function, respectively, was observed (Figure 6B). Combining the D366N integrase knockout change with MutL1 DNA-binding mutations effectively abrogated co-factor function (Figure 6B, MutL6; ∼1.1% of wild-type activity). An N-terminal deletion mutant lacking the PWWP domain, NLS and AT-hooks interestingly supported about 51% of wild-type function (Figure 6B, 226–530). In agreement with previous findings (15), further truncation from the N-terminus of this fragment to residue 347 abolished stimulation (Figure 6B; 1.0% of wild-type function), indicating that sequences that lie between LEDGF/p75 residues 226 and 347 play a role in HIV-1 co-factor function in vitro.
DISCUSSION
LEDGF/p75 and its smaller splice-variant LEDGF/p52 are transcriptional co-activators that appear to function by interacting with general transcriptional machinery (2). Like other proteins involved in transcriptional regulation (41), LEDGF/p75 contains extensive unstructured regions (15). The protein harbors two ∼10 kDa conserved domains, the PWWP domain and IBD (15). Shorter stretches of invariant residues also play critical roles in LEDGF/p75 function: 146RRGRKRK152 define the NLS (16,19,36) and here we demonstrate a role for two AT-hook motifs in binding to DNA and to chromatin.
Identification and characterization of a novel non-specific DNA-binding motif in LEDGF/p75
Using a combination of deletion and missense mutagenesis, we determined that a tripartite motif comprised of the NLS and two AT-hooks mediates the binding of LEDGF/p75 to DNA in vitro (Figures 2B and 3C) and ranked the individual elements AT2>NLS>AT1 in terms of their relative contributions to overall DNA-binding function (Figure 3C). Consistent with these findings, the vast majority of NLS sequences enriched in basic amino acids contribute to nucleic acid-binding function (42,43).
The NLS and AT-hooks account for fully one-half of all invariant residues between the PWWP domain (residue 93) and residue 226 in LEDGF/p75 (Figure 3B) (15). From this and sequence predictions programs (15), we infer that the conserved tripartite DNA-binding element sits in a relatively unstructured part of the protein. The brunt of LEDGF/p75 is efficiently digested after short exposure to a limited amount of trypsin, as the IBD and PWWP domain resist proteolysis and accumulate over time (15). Including DNA did not detectably alter the kinetics or extent of proteolysis (data not shown), indicating that DNA-binding does not induce significant structural rearrangements within this or other regions of the protein. We suppose that unstructured AT-hooks might assume crescent shapes upon engaging minor grooves in DNA, as demonstrated for AT-hooks 2 and 3 in high mobility group protein A1 (HMGA1) (44). Although we demonstrate the importance of invariant RGR motifs in both AT1 and AT2 function, our results did not reveal significant affinity for AT-rich DNA over other sequences (Figure 1A, B and D). SPR revealed faster dissociation rates and less stable binding of LEDGF/p75 to different DNA substrates when the concentration of salt was incrementally increased from 140 to 300 mM (data not shown), suggesting that LEDGF/p75 might primarily interact with DNA in vitro though electrostatic interactions. Consistent with this interpretation, binding to single-stranded DNA and RNA templates was detected by EMSA (data not shown).
It is somewhat difficult to reconcile our observation of sequence non-specific binding with results of previous studies. Shinohara and colleagues (17) reported that LEDGF/p75 specifically bound to nGAAn and (T/A)GGGG(A/T) sequences in the promoter regions of Hsp27 and αB-crystallin genes, respectively. Our substrates were based on findings that changing GAA to TAA or AAA within the consensus HSE prevented LEDGF/p75 binding (17), a result we failed to recapitulate (Figure 1B and D). Although we failed to detect sequence-specific binding utilizing independent preparations of LEDGF/p75 protein, we cannot rule-out that LEDGF/p75 might display specificity for particular DNA sequences under certain experimental conditions.
More recently, Singh et al. (36) reported that the PWWP domain and two HTH-like motifs in the C-terminal region of LEDGF/p75 mediate binding to STRE and HSE sequences, respectively. Yet, our results indicate that highly-purified PWWP domain (Figure 1C, lane 4) and C-terminal domain (Figure 1C, lane 3) protein failed to significantly bind DNA under conditions where purified 93–226 protein (Figure 1C, lane 5), harboring the NLS and AT-hooks, readily shifted DNA substrates (Figure 2B). As Singh et al. (36) utilized single-step purification procedures and detected the migration positions of recombinant deletion mutant proteins by western blotting, we infer that the deletion variants manufactured herein were of greater biochemical purity. Consistent with our findings, it was recently reported that the AT-hooks participate in the interaction of LEDGF/p75 with chromatin in vivo (26). Additionally, our previous sequence and structure-based analyses failed to reveal evidence for HTH-like motifs within C-terminal regions 421–442 and 471–492. Residues 412–428 form the C-terminal α helix of the pseudo HEAT repeat analogous topology (PHAT) IBD both in solution (25) and as part of a co-crystallized complex with HIV-1 integrase (35). HEAT (named after Huntingtin, Elongation factor 3, A subunit of phosphatase 2A, yeast PI3 kinase TOR) repeats mediate protein–protein interactions (45), and NMR spectroscopy failed to detect any stable secondary structure within residues 431–471 (25). Furthermore, we were unable to confirm evidence for a HTH-like motif within residues 471–492 using either the PROF (46) or SMART (47) prediction program. As internal fragments 93–226 and 93–325 bound DNA less efficiently than the full-length protein (Figure 2B), it seems the N- and/or C-terminal regions could contribute to the overall affinity of LEDGF/p75 for nucleic acid. If so, we conclude these affects are distinct from direct DNA-binding.
The PWWP domain cooperates with the NLS and AT-hooks to mediate chromatin-binding in vivo
The PWWP domain, present in more than 60 eukaryotic proteins, is the most conserved region of LEDGF/p75 (13,15). The Dnmt3a and Dnmt3b methyltransferase enzymes have been most thoroughly studied in terms of PWWP domain contribution(s) to DNA and chromatin-binding activities. We found that the LEDGF/p75 PWWP domain behaved similarly to the Dnmt3a domain, as neither effectively bound to free DNA in vitro [Figure 2B and ref. (38)]. In contrast, the Dnmt3b domain displayed DNA-binding activity (38,48). Despite this difference, the Dnmt3a and Dnmt3b domains each played a central role in pericentric heterochromatin localization (38) and mutations within the Dnmt3b domain prevented the binding of full-length enzyme to condensed chromosomes (39).
Based on the phenotypes of EGFP fusion proteins in transfected cells, we propose that the PWWP domain works in concert with the NLS and AT-hook motifs to mediate the association of LEDGF/p75 with chromatin in vivo. The PWWP deletion mutant MutL7 interacted normally with condensed chromosomes (Figure 5C), though we noted a subtle difference in nucleoplasmic localization (Figure 4C). The dual AT-hook mutant MutL3 displayed the wild-type chromatin-interaction pattern, whereas the tripartite PWWP/AT-hook MutL9 (Table 1) mutant failed to effectively engage mitotic chromosomes (Figure 5D). As EGFP-PWWP remained partially cytosolic under conditions wherein the PWWP deletion MutL7 bound to mitotic chromosomes indistinguishably from the wild-type (Figure 5A, C and F), we conclude that DNA-binding likely imparts the dominant chromatin-association function in vivo. It is clear that the PWWP domain contributes to this activity but unlike Dnmt3b, it is not the dominant determinant [Figure 5C and ref. (39)]. Our results agree with and extend previous suggestions (14,38) that the PWWP domain is likely to mediate protein interaction(s) with chromatin.
DNA-binding activity plays a minor role in HIV-1 co-factor function in vitro
A variety of cellular factors have been reported to stimulate HIV-1 integration, and the proteins can be divided into two broad classes based on their propensity to bind integrase. For example, integrase-interactor 1 (49) and LEDGF/p75 bind to integrase and, at least in the latter case, integrase-binding is essential for co-factor function in vitro (15,25) (Figure 6). Other potential integration co-factors such as the barrier-to-autointegration factor (50) and HMGA1 (51) appear to lack affinity for retroviral integrases (52,53) and instead stimulate integration mainly through their ability to bind DNA [(28,54); for review see ref. (22,55)]. The IBD is necessary but not sufficient to stimulate integrase activity in vitro (15,25), so an outstanding question was the contribution of LEDGF/p75 DNA-binding activity to this process.
The tripartite DNA-binding mutant protein MutL1 was about 2.5-fold reduced for integrase stimulation under conditions where the D366N interaction-impaired mutant was about 12.5-fold defective (Figure 6B). From these data we conclude that DNA-binding plays a modest role in stimulating integrase activity and, by extension, that integrase-binding is ∼5-fold more important than DNA-binding under these assay conditions. Furthermore, the 226–530 fragment lacking the PWWP domain, NLS and AT-hooks supported about 51% of wild-type LEDGF/p75 function in vitro (Figure 6B). The isolated IBD (residues 347–471; Figure 6A) and somewhat larger 347–530 fragment (Figure 6B) did not appreciably stimulate integrase activity, indicating that residues between 226 and 347 play a role in integrase co-factor function in vitro. What function might this region contribute? The region is fairly well-conserved across LEDGF/p75 orthologs, harboring 50 invariant residues that mainly cluster into small groupings (15). Although nearly half the invariant residues are lysine (15,36), fragments encompassing residues 226–347 or 226–283 did not detectably bind to DNA in vitro. Additional experiments will be required to address the role of this region in stimulating integrase activity in vitro as well as in other LEDGF/p75 functions. As the AT-hooks were recently implicated in targeting HIV-1 integration to active gene regions and A/T-rich DNA sequences in vivo (26), our results indicate that viral integration targeting is in large part independent of LEDGF/p75's ability to stimulate integrase catalytic function in vitro.
CONCLUSIONS
Though established to bind to DNA in vitro and interact with chromatin in cells, the regions and residues of LEDGF/p75 that mediate chromatin-association were unknown. In this work, we define a novel tripartite DNA-binding element comprised of the NLS and two copies of the AT-hook motif that mediates the binding of LEDGF/p75 to DNA, which, together with our previous sequence and structure-based analyses, casts doubt on recent claims for the PWWP domain and two C-terminal HTH-like motifs in directing sequence-specific DNA-binding activity. We also showed that DNA-binding contributes a dominant function to chromatin-binding in cells, with a likely PWWP domain-mediated protein interaction playing more of a modulatory role than observed in previously analyzed systems. In contrast, DNA-binding plays only a modest role in stimulating HIV-1 integrase activity in vitro. Our results uncovered essential elements of LEDGF/p75 function, a transcriptional co-activator implicated in a variety of human diseases including autoimmunity, cancer and AIDS.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
Funding for this work and to cover Open Access publication charges for this article were provided by US NIH grants AI39394 and AI52014. Core facilities were supported by a Center for AIDS Research grant (AI60354) and the Dana-Farber Cancer Institute/Harvard Cancer Center.
Conflict of interest statement. None declared.
REFERENCES
- 1.Spiegelman B.M., Heinrich R. Biological control through regulated transcriptional coactivators. Cell. 2004;119:157–167. doi: 10.1016/j.cell.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 2.Ge H., Si Y., Roeder R.G. Isolation of cDNAs encoding novel transcription coactivators p52 and p75 reveals an alternate regulatory mechanism of transcriptional activation. EMBO J. 1998;17:6723–6729. doi: 10.1093/emboj/17.22.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shinohara T., Singh D.P., Fatma N. LEDGF, a survival factor, activates stress-related genes. Prog. Retin. Eye Res. 2002;21:341–358. doi: 10.1016/s1350-9462(02)00007-1. [DOI] [PubMed] [Google Scholar]
- 4.Wu X., Daniels T., Molinaro C., Lilly M.B., Casiano C.A. Caspase cleavage of the nuclear autoantigen LEDGF/p75 abrogates its pro-survival function: implications for autoimmunity in atopic disorders. Cell Death Differ. 2002;9:915–925. doi: 10.1038/sj.cdd.4401063. [DOI] [PubMed] [Google Scholar]
- 5.Daniels T., Zhang J., Gutierrez I., Elliot M.L., Yamada B., Heeb M.J., Sheets S.M., Wu X., Casiano C.A. Antinuclear autoantibodies in prostate cancer: immunity to LEDGF/p75, a survival protein highly expressed in prostate tumors and cleaved during apoptosis. Prostate. 2005;62:14–26. doi: 10.1002/pros.20112. [DOI] [PubMed] [Google Scholar]
- 6.Ahuja H.G., Hong J., Aplan P.D., Tcheurekdjian L., Forman S.J., Slovak M.L. t(9;11)(p22;p15) in acute myeloid leukemia results in a fusion between NUP98 and the gene encoding transcriptional coactivators p52 and p75-lens epithelium-derived growth factor (LEDGF) Cancer Res. 2000;60:6227–6229. [PubMed] [Google Scholar]
- 7.Hussey D.J., Moore S., Nicola M., Dobrovic A. Fusion of the NUP98 gene with the LEDGF/p52 gene defines a recurrent acute myeloid leukemia translocation. BMC Genet. 2001;2:20. doi: 10.1186/1471-2156-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grand F.H., Koduru P., Cross N.C., Allen S.L. NUP98-LEDGF fusion and t(9;11) in transformed chronic myeloid leukemia. Leuk. Res. 2005;29:1469–1472. doi: 10.1016/j.leukres.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 9.Nishizawa Y., Usukura J., Singh D.P., Chylack L.T.J., Shinohara T. Spatial and temporal dynamics of two alternatively spliced regulatory factors, lens epithelium-derived growth factor (ledgf/p75) and p52, in the nucleus. Cell Tissue Res. 2001;305:107–114. doi: 10.1007/s004410100398. [DOI] [PubMed] [Google Scholar]
- 10.Cherepanov P., Maertens G., Proost P., Devreese B., Van Beeumen J., Engelborghs Y., De Clercq E., Debyser Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- 11.Maertens G., Cherepanov P., Pluymers W., Busschots K., De Clercq E., Debyser Z., Engelborghs Y. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J. Biol. Chem. 2003;278:33528–33539. doi: 10.1074/jbc.M303594200. [DOI] [PubMed] [Google Scholar]
- 12.Ganapathy V., Casiano C.A. Autoimmunity to the nuclear autoantigen DFS70 (LEDGF): what exactly are the autoantibodies trying to tell us? Arthritis Rheum. 2004;50:684–688. doi: 10.1002/art.20095. [DOI] [PubMed] [Google Scholar]
- 13.Izumoto Y., Kuroda T., Harada H., Kishimoto T., Nakamura H. Hepatoma-derived growth factor belongs to a gene family in mice showing significant homology in the amino terminus. Biochem. Biophys. Res. Commun. 1997;238:26–32. doi: 10.1006/bbrc.1997.7233. [DOI] [PubMed] [Google Scholar]
- 14.Stec I., Nagl S.B., van Ommen G.-J.B., den Dunnen J.T. The PWWP domain: a potential protein–protein interaction domain in nuclear proteins influencing differentiation? FEBS Letters. 2000;473:1–5. doi: 10.1016/s0014-5793(00)01449-6. [DOI] [PubMed] [Google Scholar]
- 15.Cherepanov P., Devroe E., Silver P.A., Engelman A. Identification of an evolutionarily-conserved domain in LEDGF/p75 that binds HIV-1 integrase. J. Biol. Chem. 2004;279:48883–48892. doi: 10.1074/jbc.M406307200. [DOI] [PubMed] [Google Scholar]
- 16.Vanegas M., Llano M., Delgado S., Thompson D., Peretz M., Poeschla E. Identification of the LEDGF/p75 HIV-1 integrase-interaction domain and NLS reveals NLS-independent chromatin tethering. J. Cell Sci. 2005;118:1733–1743. doi: 10.1242/jcs.02299. [DOI] [PubMed] [Google Scholar]
- 17.Singh D.P., Fatma N., Kimura A., Chylack J., Leo T., Shinohara T. LEDGF binds to heat shock and stress-related element to activate the expression of stress-related genes. Biochem. Biophys. Res. Comm. 2001;283:943–955. doi: 10.1006/bbrc.2001.4887. [DOI] [PubMed] [Google Scholar]
- 18.Busschots K., Vercammen J., Emiliani S., Benarous R., Engelborghs Y., Christ F., Debyser Z. The interaction of LEDGF/p75 with integrase is lentivirus-specific and promotes DNA binding. J. Biol. Chem. 2005;280:17841–17847. doi: 10.1074/jbc.M411681200. [DOI] [PubMed] [Google Scholar]
- 19.Maertens G., Cherepanov P., Debyser Z., Engelborghs Y., Engelman A. Identification and characterization of a functional nuclear localization signal in the HIV-1 integrase interactor LEDGF/p75. J. Biol. Chem. 2004;279:33421–33429. doi: 10.1074/jbc.M404700200. [DOI] [PubMed] [Google Scholar]
- 20.Aravind L., Landsman D. AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 1998;26:4413–4421. doi: 10.1093/nar/26.19.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Llano M., Vanegas M., Fregoso O., Saenz D., Chung S., Peretz M., Poeschla E.M. LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral preintegration complexes. J. Virol. 2004;78:9524–9537. doi: 10.1128/JVI.78.17.9524-9537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turlure F., Devroe E., Silver P.A., Engelman A. Human cell proteins and human immunodeficiency virus DNA integration. Front. Biosci. 2004;9:3187–3208. doi: 10.2741/1472. [DOI] [PubMed] [Google Scholar]
- 23.Bradley C.M., Craigie R. Seeing is believing: structure of the catalytic domain of HIV-1 integrase in complex with human LEDGF/p75. Proc. Natl Acad. Sci. USA. 2005;102:17543–17544. doi: 10.1073/pnas.0509078102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emiliani S., Mousnier A., Busschots K., Maroun M., Van Maele B., Tempe D., Vandekerckhove L., Moisant F., Ben-Slama L., Witvrouw M., et al. Integrase mutants defective for interaction with LEDGF/p75 are impaired in chromosome tethering and HIV-1 replication. J. Biol. Chem. 2005;280:25517–25523. doi: 10.1074/jbc.M501378200. [DOI] [PubMed] [Google Scholar]
- 25.Cherepanov P., Sun Z.-Y.J., Rahman S., Maertens G., Wagner G., Engelman A. Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75. Nature Struct. Mol. Biol. 2005;12:526–532. doi: 10.1038/nsmb937. [DOI] [PubMed] [Google Scholar]
- 26.Ciuffi A., Llano M., Poeschla E., Hoffmann C., Leipzig J., Shinn P., Ecker J.R., Bushman F. A role for LEDGF/p75 in targeting HIV DNA integration. Nature Med. 2005;11:1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- 27.Vandegraaff N., Devroe E., Turlure F., Silver P.A., Engelman A. Biochemical and genetic analyses of integrase-interacting proteins lens epithelium-derived growth factor (LEDGF)/p75 and hepatoma-derived growth factor related protein 2 (HRP2) in preintegration complex function and HIV-1 replication. Virology. 2006;346:415–426. doi: 10.1016/j.virol.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 28.Harris D., Engelman A. Both the structure and DNA binding function of the barrier-to-autointegration factor contribute to reconstitution of HIV type 1 integration in vitro. J. Biol. Chem. 2000;275:39671–39677. doi: 10.1074/jbc.M002626200. [DOI] [PubMed] [Google Scholar]
- 29.Goldhaber-Gordon I., Early M.H., Baker T.A. The terminal nucleotide of the Mu genome controls catalysis of DNA strand transfer. Proc. Natl Acad. Sci. USA. 2003;100:7509–7514. doi: 10.1073/pnas.0832468100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cherepanov P., Surratt D., Toelen J., Pluymers W., Griffith J., De Clercq E., Debyser Z. Activity of recombinant HIV-1 integrase on mini-HIV DNA. Nucleic Acids Res. 1999;27:2202–2210. doi: 10.1093/nar/27.10.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dorfman T., Luban J., Goff S.P., Haseltine W.A., Gottlinger H.G. Mapping of functionally important residues of a cysteine-histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 1993;67:6159–6169. doi: 10.1128/jvi.67.10.6159-6169.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fatma N., Singh D.P., Shinohara T., Chylack L.T., Jr Transcriptional regulation of the antioxidant protein 2 gene, a thiol-specific antioxidant, by lens epithelium-derived growth factor to protect cells from oxidative stress. J. Biol. Chem. 2001;276:48899–48907. doi: 10.1074/jbc.M100733200. [DOI] [PubMed] [Google Scholar]
- 33.Malmqvist M. BIACORE: an affinity biosensor system for characterization of biomolecular interactions. Biochem. Soc. Trans. 1999;27:335–340. doi: 10.1042/bst0270335. [DOI] [PubMed] [Google Scholar]
- 34.Rich R.L., Myszka D.G. BIACORE J: a new platform for routine biomolecular interaction analysis. J. Mol. Recognit. 2001;14:223–228. doi: 10.1002/jmr.535. [DOI] [PubMed] [Google Scholar]
- 35.Cherepanov P., Ambrosio A.L.B., Rahman S., Ellenberger T., Engelman A. From the Cover: structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc. Natl Acad. Sci. USA. 2005;102:17308–17313. doi: 10.1073/pnas.0506924102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh D.P., Kubo E., Takamura Y., Shinohara T., Kumar A., Chylack L.T.J., Fatma N. DNA binding domains and nuclear localization signal of LEDGF: contribution of two helix-turn-helix (HTH)-like domains and a stretch of 58 amino acids of the N-terminal to the trans-activation potential of LEDGF. J. Mol. Biol. 2006;355:379–394. doi: 10.1016/j.jmb.2005.10.054. [DOI] [PubMed] [Google Scholar]
- 37.Reeves R. Molecular biology of HMGA proteins: hubs of nuclear function. Gene. 2001;277:63–81. doi: 10.1016/s0378-1119(01)00689-8. [DOI] [PubMed] [Google Scholar]
- 38.Chen T., Tsujimoto N., Li E. The PWWP domain of Dnmt3a and Dnmt3b is required for directing DNA methylation to the major satellite repeats at pericentric heterochromatin. Mol. Cell. Biol. 2004;24:9048–9058. doi: 10.1128/MCB.24.20.9048-9058.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ge Y.-Z., Pu M.-T., Gowher H., Wu H.-P., Ding J.-P., Jeltsch A., Xu G.-L. Chromatin targeting of de novo DNA methyltransferases by the PWWP domain. J. Biol. Chem. 2004;279:25447–25454. doi: 10.1074/jbc.M312296200. [DOI] [PubMed] [Google Scholar]
- 40.Yoshida M., Horinouchi S. Trichostatin and leptomycin: inhibition of histone deacetylation and signal-dependent nuclear export. Ann. N.Y. Acad. Sci. 1999;886:23–35. doi: 10.1111/j.1749-6632.1999.tb09397.x. [DOI] [PubMed] [Google Scholar]
- 41.Liu J., Tan H., Rost B. Loopy proteins appear conserved in evolution. J. Mol. Biol. 2002;322:53–64. doi: 10.1016/s0022-2836(02)00736-2. [DOI] [PubMed] [Google Scholar]
- 42.LaCasse E.C., Lefebvre Y.A. Nuclear localization signals overlap DNA- or RNA-binding domains in nucleic acid-binding proteins. Nucleic Acids Res. 1995;23:1647–1656. doi: 10.1093/nar/23.10.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cokol M., Nair R., Rost B. Finding nuclear localization signals. EMBO Rep. 2000;1:411–415. doi: 10.1093/embo-reports/kvd092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huth J.R., Bewley C.A., Nissen M.S., Evans J.N., Reeves R., Gronenborn A.M., Clore G.M. The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif. Nature Struct. Biol. 1997;4:657–665. doi: 10.1038/nsb0897-657. [DOI] [PubMed] [Google Scholar]
- 45.Andrade M.A., Petosa C., O'Donoghue S.I., Muller C.W., Bork P. Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 2001;309:1–18. doi: 10.1006/jmbi.2001.4624. [DOI] [PubMed] [Google Scholar]
- 46.Rost B. PHD: predicting one-dimensional protein structure by profile-based neural networks. Methods Enzymol. 1996;266:283–296. doi: 10.1016/s0076-6879(96)66033-9. [DOI] [PubMed] [Google Scholar]
- 47.Letunic I., Copley R.R., Schmidt S., Ciccarelli F.D., Doerks T., Schultz J., Ponting C.P., Bork P. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 2004;32:D142–D144. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qiu C., Sawada K., Zhang X., Cheng X. The PWWP domain of mammalian DNA methyltransferase Dnmt3b defines a new family of DNA-binding folds. Nature Struct. Biol. 2002;9:217–224. doi: 10.1038/nsb759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kalpana G.V., Marmon S., Wang W., Crabtree G.R., Goff S.P. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science. 1994;266:2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]
- 50.Lee M.S., Craigie R. A previously unidentified host protein protects retroviral DNA from autointegration. Proc. Natl Acad. Sci. USA. 1998;95:1528–1533. doi: 10.1073/pnas.95.4.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Farnet C.M., Bushman F.D. HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell. 1997;88:483–492. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]
- 52.Hindmarsh P., Ridky T., Reeves R., Andrake M., Skalka A.M., Leis J. HMG protein family members stimulate human immunodeficiency virus type 1 and avian sarcoma virus concerted DNA integration in vitro. J. Virol. 1999;73:2994–3003. doi: 10.1128/jvi.73.4.2994-3003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mansharamani M., Graham D.R.M., Monie D., Lee K.K., Hildreth J.E.K., Siliciano R.F., Wilson K.L. Barrier-to-autointegration factor BAF binds p55 Gag and matrix and is a host component of human immunodeficiency virus type 1 virions. J. Virol. 2003;77:13084–13092. doi: 10.1128/JVI.77.24.13084-13092.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li L., Yoder K., Hansen M.S.T., Olvera J., Miller M.D., Bushman F.D. Retroviral cDNA integration: stimulation by HMG I family proteins. J. Virol. 2000;74:10965–10974. doi: 10.1128/jvi.74.23.10965-10974.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Engelman A. The roles of cellular factors in retroviral integration. Curr. Top. Microbiol. Immunol. 2003;281:209–238. doi: 10.1007/978-3-642-19012-4_6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.