Abstract
Epstein-Barr virus (EBV) is a human herpesvirus capable of establishing a latent state in B lymphocytes. The product of the immediate-early BZLF1 gene, Zta, is a transcriptional transactivator essential for viral DNA amplification and virion production. Previously, we identified a negative cis-acting element within the BZLF1 promoter termed ZV. ZV contains the sequence 5′-CAGGTA-3′ located at nucleotides −17 to −12 relative to the transcription initiation site. It sequence specifically binds a cellular factor, ZVR. Based on sequence binding specificity, we postulated that ZVR may be zinc finger E-box binding factor (ZEB) or a related zinc finger/homeodomain family member. We show here by immunoshift assays that ZVR and human ZEB specifically cross-react with an antibody to δEF1, the chicken homolog of ZEB. Competition electrophoretic mobility shift assays confirmed that ZEB binds to the ZV element with the same binding specificity as ZVR. Overexpression of ZEB in either B-lymphocytic DG75 cells or mammary epithelial MCF-7 cells repressed Zta-induced activation of the BZLF1 promoter four- to fivefold via the ZV site. Thus, we conclude that the previously identified cellular repressor ZVR is, in fact, ZEB. We also present evidence that other cellular factors likely affect the transcriptional activity of ZEB. Lastly, we identify a ZEB-binding site within the promoter of the lytic BRLF1 gene of EBV. We postulate that ZEB likely plays an important role in regulating the life cycle of EBV.
Epstein-Barr virus (EBV) is a human gammaherpesvirus estimated to infect 90% of the world's population (24, 25). Primary infection and replication of the virus are thought to occur in the oropharyngeal epithelium (24, 25, 30, 45). EBV can also infect B lymphocytes, where it establishes a latent infection (1, 26, 40). Latent infection by EBV is associated with a number of malignancies. These include Burkitt's lymphoma (24, 25), some cases of Hodgkin's disease (1, 24), nasopharyngeal carcinoma (9) and gastric carcinoma (reference 46 and references cited therein). However, lytic replication of EBV may also be associated with human diseases in addition to infectious mononucleosis, with the presence of antibodies to EBV lytic antigens correlating with increased risk for nasopharyngeal carcinoma (7) and hairy leukoplakia (3, 48, 49, 53).
Once EBV has established a latent infection, only a small subset of EBV's approximately 100 genes are expressed: EBV-encoded early antigens (EBNAs) 1 to 6, latent membrane proteins (LMP) 1 and 2, two EBV-encoded small nuclear RNAs, and the BamHIA transcripts (24, 25). The expression of a subset of these genes is sufficient to immortalize B cells and maintain the viral genome as an episome (27). However, reactivation of the viral lytic phase can occur, resulting in replication of the viral genome to a higher copy number and production of infectious virions. Treatment of certain viral-genome-harboring B cells and epithelial cells with chemical reagents (4, 12, 23, 33, 56), serum factors (2), transforming growth factor β1 (15), and Ca2+ ionophores (10) or cross-linking of surface anti-immunoglobulin (6, 19, 44, 47) can lead to this latent-lytic switch in the expression of EBV's genes. These factors likely initiate EBV's lytic replication cycle by signaling transcriptional activation of the promoters of two of EBV's intermediate-early genes, BZLF1 and BRLF1 (reference 11 and references cited therein). The BZLF1 gene product, Zta, functions as a transcriptional transactivator of viral genes essential for lytic replication (13, 31, 39, 54) and binds to the viral origin of lytic replication, ori-Lyt (16, 21, 31, 41, 42). In addition, mutational analysis of the EBV genome confirmed that the BRLF1 gene product, Rta, is required for replication of the EBV genome (11). Thus, regulation of the BZFL1 and BRLF1 promoters is crucial to the life cycle of the virus.
The BZLF1 gene is not expressed during latency, probably due to active repression of its promoter. Nucleotides (nt) −221 to +12 relative to the transcription initiation site of the BZLF1 promoter are sufficient for both its responses to exogenous inducers (6, 12, 13) and silencing in their absence via cis-acting elements (28, 32, 34, 43) (Fig. 1A). However, the identities of the cellular sequence-specific binding factors that recognize these silencing elements remain unknown. Recently, we identified a silencing element, ZV, which maps to nt −17 to −12 of the BZLF1 promoter, and showed that it sequence specifically binds a cellular factor termed the ZV regulator (ZVR) (28). We now report the identity of ZVR as the zinc finger E-box binding factor (ZEB) and show that ZEB can, indeed, function as a transcriptional repressor of the BZLF1 promoter.
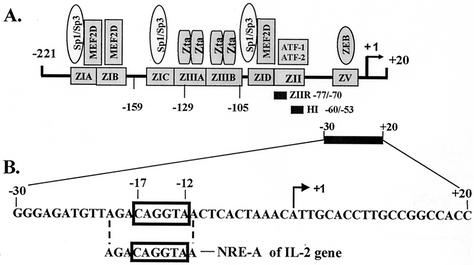
FIG. 1.
Schematic representation of the cis-acting elements and their binding factors present within the −221 to +20 region of the BZLF1 promoter. (A) Three distinct regions exist: ZIA and ZID are A-T-rich regions that bind transcription factors Sp1/Sp3 and/or myocyte enhancer factor 2D (MEF2D); ZII shares homology with the consensus CRE/AP-1 binding site; and ZIIIA and ZIIIB bind the BZLF1-encoded protein, Zta. The boxed region from nt −17 to −12 denotes the previously identified (28) negative cis-acting element referred to as ZV, shown here to bind the zinc finger E-box binding protein, ZEB. Nucleotide numbers are presented relative to the transcription initiation site indicated by a rightward arrow. (B) A blowup indicates the sequence of the nt −30 to + 20 region of the BZLF1 promoter. The negative regulatory element NRE-A, present in the IL-2 gene, is shown below ZV's extended sequence, 5′-AGACAGGTAA-3′.
MATERIALS AND METHODS
Cells.
All cell lines were maintained at 37°C in a humidified 5% CO2 atmosphere. The EBV-negative Burkitt's lymphoma cell line DG75 and the human mammary tumor epithelial cell line MCF-7 were grown as described previously (28). The human embryonic kidney cell line 293 was grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 100 U of penicillin and streptomycin per ml.
Plasmids.
The luciferase reporter plasmid WTZpluc contains the −221 to +12 region of the BZLF1 promoter, relative to the transcription initiation site, cloned into the KpnI and HindIII sites of the pGL3 basic luciferase vector (Promega Corp., Madison, Wisc.) (28). The luciferase reporter plasmid −12CZpluc is identical to WTZpluc except for an A→C mutation at nt −12, a sequence previously shown to be defective in binding ZVR (28). The expression plasmid pCiZEB, generously provided by Douglas Dean, encodes full-length human ZEB under the control of the cytomegalovirus (CMV) promoter. It also contains a FLAG sequence immediately upstream of the ATG translation initiation codon of the ZEB open reading frame (36). The expression plasmid pCMV-BZLF1, generously provided by Bill Sugden, contains the BZLF1 open reading frame encoding Zta under the control of the CMV promoter in a retroviral background (21).
Western blots.
Cells from the 293 cell line were transfected with pCiZEB (6 μg/100-mm diameter dish) by the Mirus TransIT-LT1 technique (Mirus Corp., Madison, Wisc.). After incubation for 48 h, whole-cell extract was prepared as described previously (29). Nuclear extracts (NE) were prepared from DG75 and MCF-7 cells as described previously (28). To examine the extracts for the presence of ZEB, 100 μg of protein was resolved by sodium dodecyl sulfate-8% polyacrylamide gel electrophoresis. The proteins in the gel were electroblotted onto a nitrocellulose membrane. The membranes were probed with either a monoclonal antibody directed against the FLAG epitope (Sigma Chemical Co., St. Louis, Mo.) or a rabbit polyclonal antiserum raised against a synthetic peptide corresponding to amino acids 8 through 21 of the chicken homolog of ZEB (δEF1), a gift from Michel Sanders (5). This peptide is identical in sequence to that of amino acids 42 through 55 of ZEB. The retained antibodies were probed with anti-rabbit immunoglobulin G-horseradish peroxidase and detected by enhanced chemiluminescence (Amersham Pharmacia Biotech, Piscataway, N.J.).
EMSAs.
Electrophoretic mobility shift assays (EMSAs) were performed as previously described (28), with radiolabeled double-stranded DNA corresponding to the −30 to +20 region of the BZLF1 promoter (Fig. 1) serving as the probe and with either whole-cell extract made from pCiZEB-transfected 293 cells or NE made from DG75 cells serving as the protein source. Immunoshift EMSAs were performed by incubation of the reaction mixture with the ZEB-specific antiserum prior to the addition of the radiolabeled probe. Competition EMSAs were performed as previously described (28), with the indicated double-stranded oligonucleotides serving as competitors.
Transient transfections and luciferase assays.
MCF-7 and DG75 cells grown in 12-well or 100-mm dishes, respectively, were transfected by using Mirus TransIT LT1 reagent (Mirus Corp.) with the indicated amounts of appropriate plasmid DNAs (see Fig. 5 and 6). Seventy-two hours later, the transfected cells were harvested, washed twice with phosphate-buffered saline, and resuspended in 150 μl of luciferase assay cell lysis buffer (Promega Corp.). Luciferase activities were determined according to the manufacturer's protocol and normalized to protein concentrations determined by a modified Bradford assay (Bio-Rad Laboratories, Hercules, Calif.).
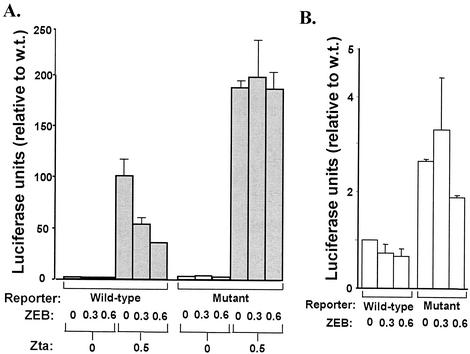
FIG. 5.
ZEB represses Zta-induced transcription of the BZLF1 promoter in DG75 cells. DG75 cells were cotransfected with the reporter plasmids WTZpluc (wild-type) or −12CZpluc (mutant) and the indicated amounts (in micrograms per 100-mm dish) of the ZEB expression plasmid pCiZEB and the Zta expression plasmid pCMV-BZFL1. After incubation for 72 h, the cells were harvested and luciferase activities were determined, with normalization to the concentration of protein present in each extract. Data are presented relative to the activity observed with the wild-type reporter in the absence of either Zta or exogenous ZEB. The bars show the means ± standard errors of the means of assays performed in triplicate. These data are representative of the results obtained from a typical experiment.
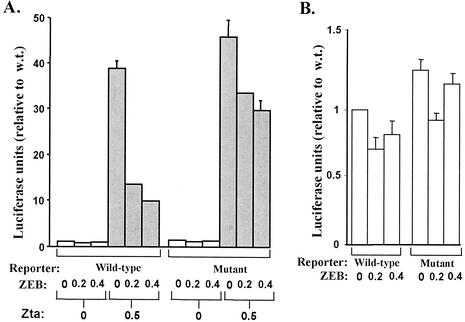
FIG. 6.
ZEB represses Zta-induced transcription of the BZLF1 promoter in MCF-7 cells. MCF-7 cells were cotransfected with the reporter plasmids WTZpluc (wild-type) or −12CZpluc (mutant) and the indicated amounts (in micrograms per well of a 12-well dish) of the ZEB expression plasmid pCiZEB and the Zta expression plasmid pCMV-BZFL1. After incubation for 72 h, the cells were harvested and luciferase activities were determined, with normalization to the concentration of protein present in each extract. Data are presented relative to the activity observed with the wild-type reporter in the absence of both Zta and exogenous ZEB. The bars show the means ± standard errors of the means of assays performed in triplicate. These data are representative of the results obtained from a typical experiment.
RESULTS
ZVR is ZEB.
The silencing element within the BZLF1 promoter named ZV contains the sequence 5′-CAGGTA-3′ (28). This sequence is identical to the sequence of a silencing element called NRE-A found in the promoter region of the interleukin-2 (IL-2) gene (50) (Fig. 1B). A zinc finger homeodomain protein termed ZEB was identified as the trans-acting factor that mediates repression of the IL-2 gene through this site (51). Therefore, we speculated that the cellular sequence-specific factor ZVR might well be ZEB.
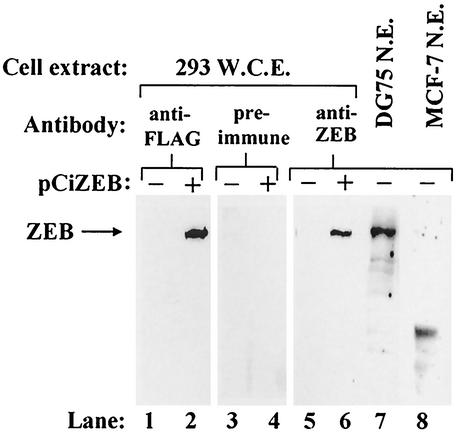
To test this hypothesis, we synthesized ZEB by transfection of pCiZEB, a ZEB expression plasmid, into cells of the 293 cell line. Analysis of whole-cell extract made from these transfected cells by immunoblotting with an antibody directed against the FLAG epitope present at the amino-terminal end of the protein confirmed that full-length ZEB had accumulated efficiently in these cells (Fig. 2, lane 2). This recombinant protein also immunologically cross-reacted with a ZEB-specific antiserum (Fig. 2, lane 6), confirming its identity as ZEB. Importantly, DG75 cells contain a protein that both comigrates with ZEB and cross-reacts with the ZEB-specific antiserum (Fig. 2, lane 7). Therefore, we conclude that ZEB is naturally present in this human B-lymphocytic cell line in which EBV can replicate.
FIG. 2.
DG75, a human B-lymphocytic cell line, contains ZEB, but MCF-7, a human mammary carcinoma epithelial cell line, does not. Cells of the 293 line were transiently transfected with pCiZEB, a plasmid expressing FLAG-tagged ZEB. Forty-eight h later, the cells were harvested and whole-cell extract (W.C.E.) was prepared. Untransfected cells of the 293 line were processed similarly in parallel. Approximately 100 μg of extract per lane was loaded, and the proteins were resolved by electrophoresis in an sodium dodecyl sulfate-8% polyacrylamide gel (lanes 1 to 6). Approximately 100 μg of DG75 and MCF-7 NE was loaded in lanes 7 and 8, respectively. The proteins were transferred to a filter and probed with anti-FLAG antibody (lanes 1 and 2), preimmune serum (lanes 3 and 4), or a ZEB-specific antiserum, anti-δEF1, made against the chicken homolog of ZEB (lanes 5 to 8).
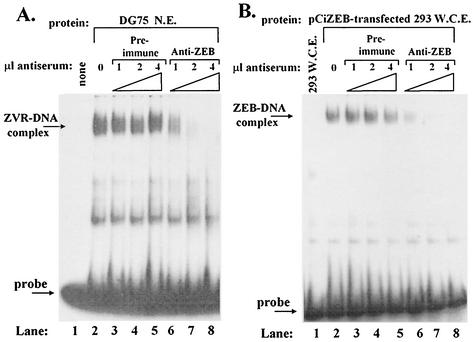
To determine whether ZVR is, indeed, ZEB or a closely related family member, we performed EMSAs which utilized NE obtained from DG75 cells that had been incubated with preimmune serum or the ZEB-specific antiserum prior to the addition of the radiolabeled oligonucleotide corresponding to the −30 to +20 region of the BZLF1 promoter (Fig. 3A). The presence of the preimmune serum had no effect on the formation or mobility of the ZVR-DNA complex (Fig. 3A, lanes 3 to 5 versus lane 2). However, the presence of the ZEB-specific antiserum inhibited the formation of the ZVR-DNA complex in a dose-dependent manner (Fig. 3A, lanes 6 to 8 versus lane 2). Thus, we conclude that ZVR is either ZEB or a protein closely related to ZEB.
FIG. 3.
ZVR is ZEB or a closely related protein. (A) Immunoshift EMSAs were performed by incubation of approximately 10 μg per lane of protein from a DG75 NE with radiolabeled probe corresponding to the −30 to +20 region of the BZLF1 promoter in the presence of the indicated amounts of the indicated antisera added prior to the probe, followed by electrophoresis in a 4% native polyacrylamide gel. (B) Immunoshift assays were performed as described in the legend for panel A, except the protein source was whole-cell extract obtained from untransfected cells of the 293 line (lane 1) or cells of the 293 line that were transfected with pCiZEB (lanes 2 to 8).
To test directly whether ZEB can bind the ZV-containing region of the BZLF1 promoter, we performed EMSAs using the above-analyzed whole-cell extract obtained from pCiZEB-transfected cells of the 293 line as the protein source. As predicted, incubation of the probe composed of the −30 to +20 region with this ZEB-containing extract resulted in the generation of a protein-DNA complex that migrated with the same mobility as the ZVR complex (compare Fig. 3B, lane 2, with Fig. 3A, lane 2). Preincubation of the extract with the ZEB-specific antiserum inhibited formation of the protein-DNA complex (Fig. 3B, lanes 6 to 8), while preincubation with the preimmune serum had no effect on either the mobility of the complex or the amount shifted (Fig. 3B, lanes 3 to 5). These findings are similar to those reported by Yasui et al. for a study involving ZEB-specific antiserum, an NRE-A probe, ZEB, and the NRE-A-binding activity present in T cells (51). Thus, we conclude that ZEB can bind the −30 to +20 region of the BZFL1 promoter.
ZEB sequence specifically binds the ZV site.
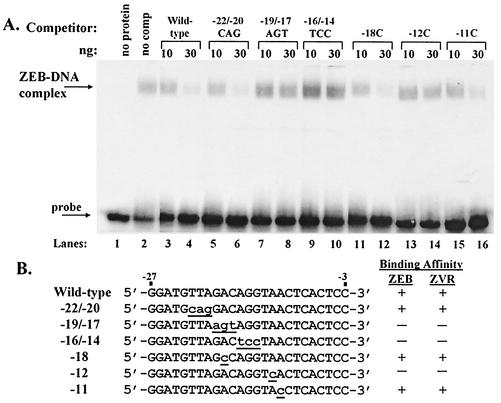
To determine whether ZEB recognizes the same sequence element within the BZFL1 promoter as does ZVR, i.e., the ZV site at nt −12 to −17, we performed competition EMSAs using as competitors many of the same sets of unlabeled double-stranded oligonucleotides that were previously employed to identify ZVR's DNA-binding specificity (28) (Fig. 4). As previously found for ZVR (28), all of the oligonucleotides containing sequence alterations within the region from nt −12 to −17 failed to compete for the binding of ZEB, while all of the oligonucleotides containing sequence alterations lying outside of this region interfered with the binding of ZEB to the radiolabeled wild-type DNA from the probe composed of the −30 to +20 region as efficiently as did the wild-type oligonucleotide (Fig. 4A; summarized in Fig. 4B). Thus, ZEB recognizes, with similar affinity, the same bases within the BZFL1 promoter as does ZVR, further confirming the identity of ZVR as ZEB.
FIG. 4.
ZEB has the same DNA-binding specificity as ZVR for the ZV site. (A) Competition EMSAs performed by incubation of 30 μg of whole-cell extract prepared from pCiZEB-transfected cells of the 293 line, with the radiolabeled DNA corresponding to the −30 to +20 region of the BZLF1 promoter serving as the probe and 10 or 30 ng of the indicated unlabeled 25-bp oligonucleotide serving as the competitor. Lane 2, no competitor added. (B) Nucleotide sequences of the oligonucleotides used as competitors in the competition EMSAs shown in panel A and summary of the data obtained with them using ZEB-containing extract (panel A) versus DG75 NE (28).
Binding of ZEB to the ZV site represses transcription from the BZLF1 promoter.
We had previously reported that the presence of the ZV site plays crucial roles in repression of the BZFL1 promoter and its activation by inducers (28). One hypothesis is that the ZV site provides these functions by binding ZEB. As a first test of the validity of this hypothesis, we cotransfected DG75 cells with WTZpluc, a luciferase reporter plasmid whose expression is driven by the presence of the −221 to +12 region of the BZLF1 promoter, and various amounts of the ZEB expression plasmid pCiZEB. To test whether observed effects of overexpression of ZEB were occurring via binding to the ZV site, we cotransfected DG75 cells in parallel with pCiZEB and −12CZpluc, a variant of WTZpluc containing a single base substitution mutation at nt −12 that inactivates binding by ZEB (Fig. 4B). As previously observed (28), the basal activity of the wild-type BZLF1 promoter in DG75 cells was extremely low (Fig. 5A), likely due in part to ZEB being endogenously present in these cells at fairly high levels (Fig. 2, lane 7). Thus, the addition of exogenous ZEB had little, if any, effect on expression (Fig. 5B). As expected, the presence of the −12C mutation led to a two- to threefold increase in expression of the reporter (Fig. 5B). This activity was not significantly affected by exogenous ZEB (Fig. 5B) because neither endogenous nor exogenous ZEB can bind to the mutant promoter.
Zta, the product of the BZLF1 gene, is a very strong transcriptional transactivator of the BZLF1 promoter (13). Thus, as expected, cotransfection of DG75 cells with pCMV-BZFL1, a plasmid expressing Zta, led to an approximately 100-fold activation of transcription from the wild-type BZLF1 promoter (Fig. 5A). Interestingly, this high-level expression of the BZLF1 promoter was repressed two- to threefold by exogenous ZEB (Fig. 5A, wild-type). By contrast, Zta-activated transcription from the mutant promoter was unaffected by exogenous ZEB (Fig. 5A, mutant). Thus, we conclude that ZEB can function as a repressor of the BZFL1 promoter in DG75 cells by binding the ZV site.
To examine the effects of exogenous ZEB in the absence of the complication of endogenous ZEB, we repeated these cotransfection experiments using MCF-7 cells. MCF-7 cells are a mammary epithelial cell line that lacks ZVR DNA-binding activity (28) and ZEB, based on both immunoblot assays (Fig. 2, lane 8) and immunoshift assays performed with the ZEB-specific antiserum and the probe composed of the −30 to +20 region (data not shown). Again, we found that ZEB had little, if any, effect on expression of either WTZpluc or −12CZpluc in the absence of inducers (Fig. 6B). However, the 40-fold activation of transcription of the wild-type promoter by Zta was repressed three- to fourfold by exogenous ZEB (Fig. 6A, wild type). On the other hand, the 45-fold activation of transcription of the mutant promoter by Zta was reduced at most 40% by exogenous ZEB (Fig. 6A, mutant). This latter modest reduction could have been due to indirect effects of ZEB overexpression in these cells or the −12 mutation allowing some residual binding when ZEB is present at nonphysiological concentrations. Regardless, these data confirm the conclusion that ZEB can function as a sequence-specific repressor of the BZLF1 promoter.
The promoter of the BRLF1 gene of EBV also contains a binding site for ZEB.
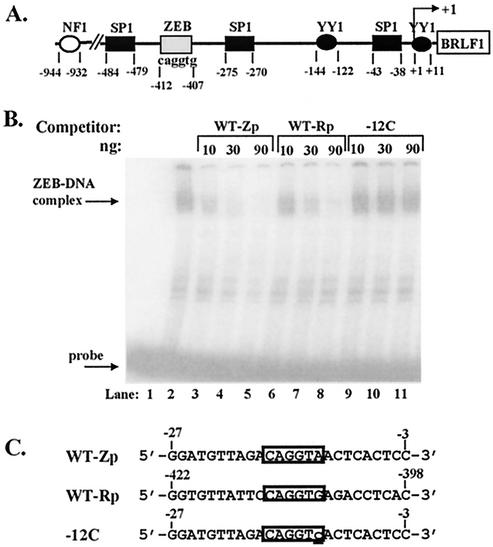
Like the BZLF1 gene, the BRLF1 gene also encodes a strong transcriptional transactivator (8, 22, 39) that is required for replication of EBV (11). If ZEB's activity is crucial for controlling the lytic cycle, the BRLF1 promoter may be regulated, in part, by ZEB as well. To begin to test the validity of this hypothesis, we looked for a putative ZEB DNA-binding sequence, 5′-CAGGTG/A-3′, within the BRLF1 promoter. One putative site exists approximately 400 bp upstream of BRLF1's transcription initiation site (Fig. 7A). Competition EMSAs performed with the oligonucleotides shown in Fig. 7C confirmed that this sequence within the BRLF1 promoter does, indeed, bind ZEB and that it does so with an affinity similar to that of the ZV site within the BZLF1 promoter (Fig. 7B, lanes 6 to 8 versus lanes 3 to 5). Thus, ZEB may well be a global regulator of EBV's lytic genes.
FIG. 7.
ZEB DNA-binding site is also present in the BRLF1 promoter. (A) Sequence of the promoter region of the EBV lytic gene BRLF1. Boxes and ovals indicate previously identified sites for known trans-acting factors. Numbers are nucleotides relative to the site of transcription initiation indicated by the rightward arrow. (B) Competition EMSAs performed with whole-cell extract made from pCiZEB-transfected cells of the 293 line serving as the protein source, radiolabeled DNA corresponding to the −30 to +20 region of the BZLF1 promoter serving as the probe, and the indicated amount of the indicated unlabeled oligonucleotide serving as the competitor. Arrows indicate the ZEB-DNA complex and free probe. (C) Sequences of the oligonucleotides used as competitors in panel B. The WT-Zp oligonucleotide corresponds to nt −27 to −3 of the BZLF1 promoter; the boxed sequence indicates the ZEB DNA-binding site identified in this paper. The WT-Rp oligonucleotide corresponds to nt −422 to −398 of the BRLF1 promoter relative to the site of transcription initiation; the boxed region indicates the likely location of the ZEB DNA-binding site identified in panel B. The −12C oligonucleotide corresponds to the −12C mutant variant of WT-Zp that fails to bind ZEB (Fig. 4); the substitution mutation is indicated by the lowercase underlined c.
DISCUSSION
We previously identified a cellular factor, ZVR, which regulates expression of the BZLF1 promoter by sequence-specifically binding to an element, ZV, located at nt −17 to −12 relative to the transcription initiation site (28). We now report the identity of ZVR as ZEB or a closely related factor. We show here that a ZEB-specific antiserum interferes with the formation of both ZEB-ZV and ZVR-ZV protein-DNA complexes (Fig. 3) and that the DNA-binding specificities of ZEB and ZVR are identical (Fig. 4). We further show that the binding of ZEB can specifically repress Zta-activated transcription from the BZLF1 promoter via binding to the ZV site (Fig. 5 and 6). Lastly, we identify a high-affinity ZEB DNA-binding site located within the BRLF1 promoter of EBV (Fig. 7). Thus, we conclude that ZEB functions as a sequence-specific repressor of the BZLF1 and, possibly, BRLF1 genes, which play crucial roles in regulation of the lytic cycle of EBV.
ZVR is ZEB.
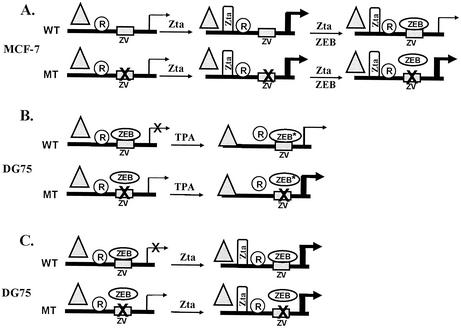
Identification of ZEB as a repressor of the BZLF1 promoter parallels the identification of ZEB as a silencer of the IL-2 promoter. First, the sequence of the ZV site is identical to that of the negative regulatory element NRE-A present within the promoter of the IL-2 gene (Fig. 1B). Yasui et al. subsequently showed the NRE-A DNA-binding activity to be ZEB (51), as we have now shown for ZVR (Fig. 3 and 4). Lastly, we demonstrated in transient cotransfection experiments that ZEB represses Zta-activated transcription from the BZLF1 promoter by binding to the ZV site (Fig. 5 and 6), as depicted by the model in Fig. 8A.
FIG. 8.
Models depicting repression activities of ZEB. (A) Repression of Zta-induced activation of the BZLF1 promoter in ZEB-negative MCF-7 cells by exogenous ZEB binding to the ZV site in wild-type (WT) but not the mutant (MT) promoter (Fig. 6). (B) Treatment of ZEB-positive DG75 cells with TPA plus ionomycin leads to alterations in the activities or amounts of ZEB, activators, and other repressors of the BZLF1 promoter, resulting in modest activation of the wild-type promoter and superinduction of the mutant promoter (see reference 28 for details). (C) Zta induces similarly the wild-type and ZV site-mutant BZLF1 promoters in DG75 cells (Fig. 5). Relative levels of transcription are depicted by the thickness of the arrows; an X through the arrow denotes minimal transcription. Rectangles depict ZV sites; an X through them indicates a mutation inactivating binding by ZEB. Triangles, cellular sequence-specific positive transcription factors activated by signaling through TPA; R, unknown cellular repressors inactivated by signaling through TPA. ZEB*, enhanced ZEB repressor activity.
ZEB is a member of a family of proteins that contain both a zinc finger(s) and a homeodomain (20). ZEB was originally cloned as a factor that bound to an E-box sequence, 5′-CAGGTG-3′, located within the immunoglobulin H enhancer, repressing its transcription (17). ZEB has also been shown to function as a transcriptional repressor of genes involved in regulation of muscle differentiation (reference 37 and references cited therein) and the tumor suppressor gene p73 (14). The Drosophila homolog of ZEB, zfh-1, represses somatic and cardiac muscle differentiation in Drosophila embryos (38). Thus, the identification of ZEB as a repressor of the BZLF1 promoter is consistent with its previously reported activities.
Transcriptional activity of ZEB is a target of signaling pathways.
We previously reported that BZLF1 promoter mutants that are defective in binding ZVR exhibit superinduction of transcription following treatment of DG75 cells with inducers (28); i.e., TPA (tetradecanoyl phorbol acetate) induces transcription from the mutant promoter relative to its basal activity significantly more than it does from the wild-type promoter. However, the −12C mutant promoter did not exhibit superinduction by Zta in the experiments shown here; i.e., Zta activated transcription from the mutant promoter approximately 75-fold relative to its basal activity, similar to the 100-fold activation observed with the wild-type promoter (Fig. 5). One hypothesis to explain this difference is that the presence of TPA plus ionomycin leads to elevation of the efficiency of repression by ZEB in addition to signaling activators of the BZLF1 promoter (Fig. 8B). Indeed, treatment of DG75 cells with TPA plus ionomycin increases the binding activity of ZVR (28), shown here to be ZEB. Whether this increase is the result of TPA-plus-ionomycin-induced posttranslational modifications or enhanced expression of ZEB remains to be determined. Since Zta likely acts only directly via binding the BZFL1 promoter, not indirectly via signaling pathways that might alter the activities of ZEB and other cellular transcription factors, one might not expect to observe superinduction by Zta (Fig. 8C).
Regulation of EBV lytic-cycle genes.
Despite considerable effort to identify repressors of EBV replication, few have been found to date. Expression of the BZLF1 and BRLF1 promoters can be repressed by the cellular factor YY1 (34, 55). However, binding of YY1 to its cognate cis-acting site present within the BZLF1 promoter depends on the presence of the EBV genome (34), suggesting that inhibition of viral replication requires modification of YY1 by some viral activity. Thus, control of lytic replication and the establishment of a latent state are, in part, dictated by the virus. Niller et al. (35) have identified a binding site for Sp1-NF1 as another possible transcriptional repressor element within the BZLF1 promoter. We have shown here that ZEB also represses transcription of the BZFL1 promoter via binding to a specific site on this promoter. In addition, we identified a ZEB DNA-binding site within the BRLF1 promoter (Fig. 7). Thus, regulation of EBV's lytic-cycle genes by ZEB may also be fairly global. Whether ZEB actually represses transcription from the BRLF1 promoter via binding to this site remains to be determined.
What role does repression of lytic-cycle gene expression play in regulation of the life cycle of EBV? Two conditions are necessary for the EBV genome to exist in a latent state in a cell. The first is maintenance of the viral genome as an episome that replicates synchronously with the cellular genome. This condition is met by the presence of an oriP replicon in cis and EBNA1, an early viral gene product, in trans (52). The second condition is that lytic-cycle replication remains repressed. Early evidence suggested that cellular repressors dictate the life cycle of EBV. For example, Glaser and Nonoyama (18) found that the fusion of HR-1, a lymphoblastoma cell line that spontaneously produces EBV, to D98, a variant of HeLa cells, led to inhibition of expression of EBV lytic-cycle genes even though the HR-1/D98 hybrid still contained EBV genomes. More recently, it has been shown that viral replication is dependent on the cell type, with epithelial cells being more permissive for expression of lytic-cycle genes and viral replication than lymphoid cells (22, 39, 54). For example, Rta, the product of the BRLF1 gene, activates expression of itself and the BZLF1 gene in epithelial cells but not in lymphoid cells (22, 54). Activation is the consequence of an indirect mechanism mediated via a signal transduction pathway (8). Yet, despite the cell type differences in expression of lytic-cycle genes, the signaling pathways and the essential trans-acting factors are intact in both cell types (54). Cell type-specific differences in repressors and the complex array of factors that modulate their activities likely explain the differences observed in lytic-cycle gene expression.
Finally, what role do repressors, either virally directed or constitutive, play in regulating the life cycle of EBV and, perhaps, the pathologies associated with EBV? Future work dictates elucidation of the negative, cis-acting elements within the lytic-cycle genes, their cognate trans-acting repressors, and the pathways mediating their activities. In addition, generation of EBV genomes harboring mutations that relieve repression of the lytic-cycle genes should provide insights regarding the roles repressors play in regulation of the life cycle of EBV.
Acknowledgments
We thank Bill Sugden for DG75 cells and plasmid pCMV-BZLF1, Douglas Dean for plasmid pCiZEB, and Michel Sanders for the δEF1 antiserum. We are grateful to Greg Kennedy and members of the Mertz laboratory for helpful discussions and comments on the manuscript.
This work was supported by Public Health Service research grants CA22443 and CA07175 from the National Cancer Institute.
REFERENCES
- 1.Babcock, G. J., L. L. Decker, M. Volk, and D. A. Thorley-Lawson. 1998. EBV persistence in memory B cells in vivo. Immunity 9:395-404. [DOI] [PubMed] [Google Scholar]
- 2.Bauer, G., P. Hofler, and H. Zur Hausen. 1982. Epstein-Barr virus induction by serum factor. I. Induction and cooperation with additional inducers. Virology 121:184-194. [DOI] [PubMed] [Google Scholar]
- 3.Becker, J., U. Leser, M. Marschall, A. Langford, W. Jilg, H. Gelderblom, P. Reichart, and H. Wolf. 1991. Expression of proteins encoded by Epstein-Barr virus trans-activator genes depends on the differentiation of epithelial cells in oral hairy leukoplakia. Proc. Natl. Acad. Sci. USA 88:8332-8336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borras, A. M., J. L. Strominger, and S. H. Speck. 1996. Characterization of the ZI domains in the Epstein-Barr virus BZLF1 promoter: role in phorbol ester induction. J. Virol. 70:3894-3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chamberlain, E. M., and M. M. Sanders. 1999. Identification of the novel player δEF1 in estrogen transcriptional cascades. Mol. Cell. Biol. 19:3600-3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daibata, M., S. H. Speck, C. Mulder, and T. Sairenji. 1994. Regulation of the BZLF1 promoter of Epstein-Barr virus by second messengers in anti-immunoglobulin-treated B cells. Virology 198:446-454. [DOI] [PubMed] [Google Scholar]
- 7.Dardari, R., M. Khyatti, A. Benider, H. Jouhadi, A. Kahlain, C. Cochet, A. Mansouri, B. El Gueddari, A. Benslimane, and I. Joab. 2000. Antibodies to the Epstein-Barr virus transactivator protein (ZEBRA) as a valuable biomarker in young patients with nasopharyngeal carcinoma. Int. J. Cancer 86:71-75. [DOI] [PubMed] [Google Scholar]
- 8.Darr, C. D., A. Mauser, and S. Kenney. 2001. Epstein-Barr virus immediate-early protein BRLF1 induces the lytic form of viral replication through a mechanism involving phosphatidylinositol-3 kinase activation. J. Virol. 75:6135-6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de-The, G. 1997. Nasopharyngeal carcinoma, p. 935-968. In A. S. Evans and R. A. Kaslow (ed.), Viral infections of humans, 4th ed. Plenum Medical Book Co., New York, N.Y.
- 10.Faggioni, A., C. Zompetta, S. Grimaldi, G. Barile, L. Frati, and J. Lazdins. 1986. Calcium modulation activates Epstein-Barr virus genome in latently infected cells. Science 232:1554-1556. [DOI] [PubMed] [Google Scholar]
- 11.Feederle, R., M. Kost, M. Baumann, A. Janz, E. Drouet, W. Hammerschmidt, and H. J. Delecluse. 2000. The Epstein-Barr virus lytic program is controlled by the co-operative functions of two transactivators. EMBO J. 19:3080-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flemington, E., and S. H. Speck. 1990. Identification of phorbal ester responsive elements in the promoter of Epstein-Barr virus putative lytic switch gene BZLF1. J. Virol. 64:1217-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flemington, E., and S. H. Speck. 1990. Autoregulation of Epstein-Barr virus putative lytic switch gene BZLF1. J. Virol. 64:1227-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fontemaggi, G., A. Gurtner, S. Strano, Y. Higashi, A. Sacchi, G. Piaggio, and G. Blandino. 2001. The transcriptional repressor ZEB regulates p73 expression at the crossroad between proliferation and differentiation. Mol. Cell. Biol. 21:8461-8470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukuda, M., K. Ikuta, K. Yanagihara, M. Tajima, H. Kuratsune, T. Kurata, and T. Sairenja. 2001. Effect of transforming growth factor-β1 on the cell growth and Epstein-Barr virus reactivation in EBV-infected epithelial cell lines. Virology 288:109-118. [DOI] [PubMed] [Google Scholar]
- 16.Gao, Z., A. Krithivas, J. E. Finan, O. J. Semmes, S. Zhou, Y. Wang, and S. D. Hayward. 1998. The Epstein-Barr virus lytic transactivator Zta interacts with the helicase-primase replication proteins. J. Virol. 72:8559-8567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Genetta, T., D. Ruezinsky, and T. Kadesch. 1994. Displacement of an E-box-binding repressor by basic helix-loop-helix proteins: implications for B-cell specificity of the immunoglobulin heavy-chain enhancer. Mol. Cell. Biol. 14:6153-6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glaser, R., and M. Nonoyama. 1974. Host cell regulation of induction of Epstein-Barr virus. J. Virol. 14:174-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldfeld, A. E., P. Liu, S. Liu, E. K. Flemington, J. L. Strominger, and S. H. Speck. 1995. Cyclosporin A and FK506 block induction of the Epstein-Barr virus lytic cycle by anti-immunoglobin. Virology 209:225-229. [DOI] [PubMed] [Google Scholar]
- 20.Gray, S., and M. Levine. 1996. Transcriptional repression in development. Curr. Opin. Cell Biol. 8:358-364. [DOI] [PubMed] [Google Scholar]
- 21.Hammerschmidt, W., and B. Sugden. 1988. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell 55:427-433. [DOI] [PubMed] [Google Scholar]
- 22.Holley-Guthrie, E. A., E. B. Quinlivan, E. Mar, and S. Kenney. 1990. The Epstein-Barr virus (EBV) BMRF1 promoter for early antigen (EA-D) is regulated by the EBV transactivators, BRLF1 and BZLF1, in a cell-specific manner. J. Virol. 64:3753-3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kallin, B., J. Luka, and G. Klein. 1979. Immunochemical characterization of Epstein-Barr virus-associated early and late antigens in n-butyrate-treated P3HR-1 cells. J. Virol. 32:710-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kieff, E., and D. Liebowitz. 1990. Epstein-Barr virus and its replication, p. 1889-1920. In B. N. Fields, D. M. Knipe, R. M. Chanock, M. S. Hirsh, J. L. Melnick, T. P. Monath, B. Roizman, and S. E. Straus (ed.), Fields virology, 2nd ed. Raven Press, New York, N.Y.
- 25.Kieff, E., and D. Liebowitz. 1991. Epstein-Barr virus and its replication, p. 897-928. In B. Fields and D. M. Knipe (ed.), Fundamentals in virology, 2nd ed. Raven Press, New York, N.Y.
- 26.Kieff, E. 1996. Epstein-Barr virus and its replication, p. 2343-2396. In B. N. Fields, D. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa.
- 27.Knutson, J. C., and B. Sugden. 1989. Immortalization of B lymphocytes by Epstein-Barr virus: what does the virus contribute to the cell? Adv. Viral Oncol. 8:151-172. [Google Scholar]
- 28.Kraus, R. J., S. J. Mirocha, H. M. Stephany, J. R. Puchalski, and J. E. Mertz. 2001. Identification of a novel element involved in regulation of the lytic switch BZLF1 gene promoter of Epstein-Barr virus. J. Virol. 75:867-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kraus, R. J., L. Shadley, and J. E. Mertz. 2001. Nuclear factor 1 family members mediate repression of the BK virus late promoter. Virology 287:89-104. [DOI] [PubMed] [Google Scholar]
- 30.Li, Q. X., L. S. Young, G. Niedobitek, C. W. Dawson, M. Birkenbach, F. Wang, and A. B. Rickinson. 1992. Epstein-Barr virus infection and replication in a human epithelial cell system. Nature 356:347-350. [DOI] [PubMed] [Google Scholar]
- 31.Lieberman, P. M., J. M. Hardwick, J. Sample, G. S. Hayward, and S. D. Hayward. 1990. The Zta transactivator involved in induction of lytic cycle gene expression in Epstein-Barr virus-infected lymphocytes binds to both AP-1 and ZRE sites in target promoter and enhancer regions. J. Virol. 64:1143-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, P., S. Liu, and S. H. Speck. 1998. Identification of a negative cis element within the ZII domain of the Epstein-Barr virus lytic switch BZLF1 gene promoter. J. Virol. 72:8230-8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luka, J., B. Kallin, and G. Klein. 1979. Induction of the Epstein-Barr virus (EBV) cycle in latently infected cells by n-butyrate. Virology 94:228-231. [DOI] [PubMed] [Google Scholar]
- 34.Montalvo, E. A., M. Cottam, S. Hill, and Y.- J. Wang. 1995. YY1 binds to and regulates cis-acting negative elements in the Epstein-Barr virus BZLF1 promoter. J. Virol. 69:4158-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niller, H. H., D. Salamon, J. Uhlig, S. Ranf, M. Granz, F. Schwarzmann, H. Wolf, and J. Minarovits. 2002. Nucleoprotein structure of immediate-early promoters Zp and Rp and of oriLyt of latent Epstein-Barr virus genomes. J. Virol. 76:4113-4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Postigo, A. A., and D. C. Dean. 1997. ZEB, a vertebrate homolog of Drosophila zfh-1, is a negative regulator of muscle differentiation. EMBO J. 16:3935-3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Postigo, A. A., and D. C. Dean. 1999. Independent repressor domains in ZEB regulate muscle and T-cell differentiation. Mol. Cell. Biol. 19:7961-7971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Postigo, A. A., E. Ward, J. B. Skeath, and D. C. Dean. 1999. zfh-1, the Drosophila homologue of ZEB, is a transcriptional repressor that regulates somatic myogenesis. Mol. Cell. Biol. 19:7255-7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ragoczy, T., L. Heston, and G. Miller. 1998. The Epstein-Barr virus Rta protein activates lytic cycle genes and can disrupt latency in B lymphocytes. J. Virol. 72:7978-7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rickinson, A. B., and E. Kieff. 1996. Epstein-Barr virus, p. 2397-2446. In B. N. Fields, D. Knipe, and P. M. Howley (ed.), Fields virology, 3rd. ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa.
- 41.Sarisky, R. T., Z. Gao, P. M. Liebermann, E. D. Fixman, G. S. Hayward, and S. D. Hayward. 1996. A replication function associated with the activation domain of the Epstein-Barr virus Zta transactivator. J. Virol. 70:8340-8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schepers, A., D. Pich, and W. Hammerschmidt. 1993. A transcription factor with homology to AP-1 family links RNA transcription and DNA replication in the lytic cycle of Epstein-Barr virus. EMBO J. 12:3921-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwarzmann, F., N. Prang, B. Reichelt, B. Rinkes, S. Haist, M. Marschall, and H. Wolf. 1994. Negative cis-acting elements in the distal part of Epstein-Barr virus trans-activator gene BZLF1. J. Gen. Virol. 75:1999-2006. [DOI] [PubMed] [Google Scholar]
- 44.Shimizu, N., and K. Takada. 1993. Analysis of the BZLF1 promoter of Epstein-Barr virus: identification of an anti-immunoglobin response sequence. J. Virol. 67:3240-3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sixby, J. W., J. G. Nedrud, N. Raab-Traub, R. A. Hanes, and J. S. Pagano. 1984. Epstein-Barr virus replication in oropharyngeal epithelial cells. N. Engl. J. Med. 310:1225-1230. [DOI] [PubMed] [Google Scholar]
- 46.Tajima, M., M. Komuro, and K. Okinaga. 1998. Establishment of Epstein-Barr virus-positive human epithelial cell lines. Jpn. J. Cancer Res. 89:262-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tovey, M. G., G. Lenoir, and J. Begon-Lours. 1978. Activation of latent Epstein-Barr virus by antibody to human IgM. Nature 276:270-272. [DOI] [PubMed] [Google Scholar]
- 48.Walling, D. M., A. M. Flaitz, C. M. Nichols, S. D. Hudnall, and K. Adler-Storthz. 2001. Persistent productive Epstein-Barr virus replication in normal epithelial cell in vivo. J. Infect. Dis. 184:1499-1507. [DOI] [PubMed] [Google Scholar]
- 49.Webster-Cyriaque, J., J. Middeldorp, and N. Raab-Traub. 2000. Hairy leukoplakia: an unusual combination of transforming and permissive Epstein-Barr virus infections. J. Virol. 74:7610-7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williams, T. M., D. Moolten, J. Burlein, J. Romano, R. Bhaerman, A. Godillot, M. Mellon, F. J. Rauscher III, and J. A. Kant. 1991. Identification of a zinc finger protein that inhibits IL-2 gene expression. Science 254:1791-1794. [DOI] [PubMed] [Google Scholar]
- 51.Yasui, D. H., T. Genetta, T. Kadesch, T. M. Williams, S. L. Swain, L. V. Tsui, and B. T. Huber. 1998. Transcriptional repression if the IL-2 gene in Th cells by ZEB. J. Immunol. 160:4433-4440. [PubMed] [Google Scholar]
- 52.Yates, J. L., N. Warren, and B. Sugden. 1985. Stable replication of plasmids derived from Epstein-Barr virus in a variety of mammalian cells. Nature 313:812-815. [DOI] [PubMed] [Google Scholar]
- 53.Young, L. S., R. Lau, M. Rowe, G. Niedobitek, G. Packham, F. Shanahan, D. T. Rowe, D. Greenspan, J. S. Greenspan, A. B. Rickinson, and P. J. Farrell. 1991. Differentiation-associated expression of the Epstein-Barr virus BZLF1 transactivator protein in oral hairy leukoplakia. J. Virol. 65:2868-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zalani, S., E. Holley-Guthrie, and S. Kenney. 1996. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc. Natl. Acad. Sci. USA 93:9194-9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zalani, S., A. Coppage, E. Holley-Guthrie, and S. Kenney. 1997. The cellular YY1 transcription factor binds a cis-acting negatively regulated element in the Epstein-Barr virus BRLF1 promoter. J. Virol. 71:3268-3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.zur Hausen, H., F. J. O'Neill, U. K. Freese, and E. Hecker. 1978. Persisting oncogenic herpesvirus induced by the tumor promoter TPA. Nature 272:373-375. [DOI] [PubMed] [Google Scholar]