Abstract
3′, 5′-Bisphosphate nucleotidase is a ubiquitous enzyme that converts 3′-phosphoadenosine-5′-phosphate to adenosine-5′-phosphate and inorganic phosphate. These enzymes are highly sensitive to sodium and lithium and, thus, perform a crucial rate-limiting metabolic step during salt stress in yeast. Recently, we have identified a bisphosphate nucleotidase gene (DHAL2) from the halotolerant yeast Debaryomyces hansenii. One of the unique features of Dhal2p is that it contains an N-terminal 54-amino-acid-residue hydrophobic extension. In this study, we have shown that Dhal2p exists as a cytosolic as well as a membrane-bound form and that salt stress markedly influences the accumulation of the latter form in the cell. We have demonstrated that the N-terminal hydrophobic region was necessary for the synthesis of the membrane-bound isoform. It appeared that an alternative translation initiation was the major mechanism for the synthesis of these two forms. Moreover, the two forms exhibit significant differences in their substrate specificity. Unlike the cytosolic form, the membrane-bound form showed very high activity against inositol-1,4-bisphosphate. Thus, the present study for the first time reports the existence of multiple forms of a bisphosphate nucleotidase in any organism.
3′, 5′-Bisphosphate nucleotidase (BPntase) is an important enzyme that catalyzes the removal of 3′ phosphate from 3′-phosphoadenosine-5′-phosphate (PAP) to form adenosine-5′-phosphate and inorganic phosphate (Pi). Hence, they are often referred as PAP phosphatase. These enzymes are highly sensitive to monovalent cations, particularly lithium. Lithium is one of the most effective drugs for the treatment of bipolar disorder, a severe, chronic mental illness that affects 1 to 3% of the population (35). Recent studies suggest that BPntases are important therapeutic targets of lithium. These enzymes are evolutionarily conserved and present in bacteria, yeast, plants, and animals (18, 21, 23, 26). In Saccharomyces cerevisiae, the gene HAL2/MET22 encodes the homologous enzyme, and it is an important determinant of halotolerance in this yeast (19). HAL2 was identified by functional assay as supporting the growth of yeast cells under high salinity stress conditions. Overexpression of this gene is known to improve salt tolerance in plants (3).
This gene is essential for sulfur assimilation in yeast. The inorganic sulfate is activated in two steps, consuming two ATP molecules (36), and is accumulated in the form of 3′-phosphoadenosine-5′-phosphosulfate (PAPS). Subsequently, PAPS is used up either by reduction to sulfite or by transferring its activated sulfate group to other molecules resulting in the formation of PAP. PAP is subsequently hydrolyzed to adenosine-5′-phosphate and inorganic phosphate by Hal2p (24). Hal2p is very sensitive to inhibition by lithium (50% inhibitory concentration, 0.1 mM) and sodium (50% inhibitory concentration, 20 mM) (23). During salt stress, the enzyme is inhibited in vivo, and PAP accumulates inside the cell. Accumulation of PAP is very toxic for the cell as this inhibits a number of metabolic enzymes involved in sulfur assimilation, RNA processing (12), and sulfotransferase reactions (2). It is proposed that the toxicity of lithium, at least in yeast, resulted from its deleterious effect on RNA processing. This is mediated by inhibition of RNase MRP by lithium ion and by the concurrent inhibition of another RNA processing enzyme, Xrn1p, due to the accumulation of PAP (12). Recently, both PAP and PAPS, as a ribonucleotide monomer mimic, have been shown to inhibit mammalian nucleoside diphosphate kinase (33).
Homologues of Hal2p from plants (18, 30, 31) and mammalian sources (21, 35) have been characterized. Some of these exhibit very high inositol polyphosphate-1-phosphatase (IPPase) activity and, therefore, are considered as enzymes having dual activity (21, 38, 39). These enzymes have been shown to function both in PAP metabolism as well as in phosphoinositide signaling. Despite their sequence similarities, homologous enzymes from the unicellular organism Escherichia coli (cysQ) and S. cerevisiae (Hal2p) do not have the IPPase activity (23, 26), contrasting with those from the multicellular organisms (31). Hal2p performs a metabolic reaction that is crucial for the survival of the yeast under high salt stress conditions (19, 34). Recently, we have identified an HAL2 homologue from Debaryomyces hansenii (1). This is one of the most halotolerant species of yeast and is regarded as a model for understanding salt tolerance (4). We have observed that the intrinsic salt tolerance of Dhal2p was much higher than that of the other homologues (1). One of the distinct features of the Dhal2p is that it contains a 54-amino-acid-residue hydrophobic stretch at its N terminus. However, the functional significance of this region remained unclear.
In this paper, we have demonstrated the existence of two distinct isoforms of Dhal2p in D. hansenii. One of them was cytosolic, whereas the other was present in the membrane. Interestingly, the membrane-bound form was accumulated in the cell only under salt stress. Our results clearly established that the N-terminal hydrophobic region was indispensable for the synthesis of this form. It appeared that an alternative translation initiation was the major mechanism for the synthesis of these two forms. Moreover, we have shown that the two forms also differed in their substrate specificity. Thus, our report is the first documentation of a membrane-bound bisphosphate nucleotidase in any organism.
MATERIALS AND METHODS
Materials.
Restriction and modifying enzymes used for DNA manipulations were obtained from New England Biolabs. A plasmid purification kit (QIAGEN), ECL Western blotting detection kit (Amersham Pharmacia Biotech), protease inhibitor cocktail (Roche), and X-ray film (Eastman Kodak) were obtained commercially. All other chemicals including PAP and PAP-agarose, were procured from Sigma Chemical Co.
Yeast strains, plasmids, and media.
Strains and plasmids used in this study are listed in Table 1. The yeast media used were YPD (1% yeast extract, 2% peptone, and 2% dextrose) and SD minimal medium (2% glucose and 0.67% yeast nitrogen base without amino acids; Difco). Only supplements that were required by the strains were added.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| RS16 | MATaura3-251, 328, 372 leu2-3,112 | 17 |
| RS1051 | MATaura3-251, 328, 372 leu2-3,112 hal2::URA3 | 19 |
| MTCC234 | D. hansenii | MTCC, India |
| Plasmids | ||
| pRS425 | LEU2 2μ | 8 |
| p414TEF | TRP1 CEN ARS1 | 22 |
| pRS-GFP | URA3 CEN ARS1 | 16 |
| pMA7 | LEU2 2μ DHAL2 | 1 |
| pMA14 | TRP1 CEN ARS1 DHAL2 (full-length) | 1 |
| pMA15 | TRP1 CEN ARS1 DHAL2 (truncated) | 1 |
| pMA23 | TRP1 CEN ARS1 DHAL2 (M3 ATG mutant) | This study |
| pMA24 | TRP1 CEN ARS1 DHAL2 (M1/M2 ATG mutant) | |
| pMA25 | URA3 CEN ARS1 DHAL2-GFP (M3 ATG mutant) | |
| pMA26 | URA3 CEN ARS1 DHAL2-GFP (M1/M2 ATG mutant) | |
| pMA27 | TRP1 CEN ARS1 DHAL2 (full-length; under HAL2 promoter) | |
| pMA28 | TRP1 CEN ARS1 DHAL2 (truncated; under HAL2 promoter) | |
| pMA29 | LEU2 2μ HAL2 | |
| pMA30 | TRP1 CEN ARS1 HAL2 (under HAL2 promoter) |
PCR amplification and construction of recombinant plasmids. (i) Constructs for Dhal2p expression in S. cerevisiae.
To express the full-length or the truncated Dhal2p, plasmids pMA14 and pMA15 were used (1). The plasmid pMA27 was constructed for expressing the full-length Dhal2p in a low-copy-number vector under the regulation of the S. cerevisiae HAL2 promoter. Based on the S. cerevisiae genome sequences, the forward (PBH36, 5′-TGGTGCAGATGGAGAGAGCTCGG-3′) and reverse (PBH37, 5′-AGTAAAGTGCTGATGTCTTCTGAG-3′) primers were designed to amplify the HAL2 promoter region. S. cerevisiae genomic DNA was used as a template for PCR. It was carried out with Vent polymerase (New England Biolabs) for 25 cycles (denaturation at 93°C for 30 s per cycle, annealing at 60°C for 40 s per cycle, and elongation at 72°C for 30 s per cycle). The amplified product was cloned into the SacI-SmaI site of pMA14 to replace the TEF promoter. Similarly, by cloning the amplified HAL2 promoter fragment into the SacI-SmaI site of pMA15, the plasmid pMA28 was constructed.
Cloning of S. cerevisiae HAL2 gene.
The S. cerevisiae HAL2 gene along with its promoter was amplified from genomic DNA using forward (PBH36, 5′-TGGTGCAGATGGAGAGAGCTCGG-3′) and reverse (PBH28, 5′-GTTCTCGAGTTAGGCGTTTCTTGACTGAAT-3′) primers. The primer PBH28 was modified to incorporate an XhoI restriction site (underlined). This fragment was cloned at the SacI-XhoI site of plasmids pRS425 (8) and p414TEF (22) to obtain plasmids pMA29 and pMA30, respectively.
Constructs for expression of ATG mutants of Dhal2p in S. cerevisiae
To introduce a mutation at either methionine M1/M2 or M3 ATG in DHAL2 (see Fig. 2A), a PCR-based overlap extension method was followed (20). In the mutant forms, the ATG codon was changed to AAG. To obtain the M1/M2 ATG mutant, a 1.2-kb fragment was amplified from the plasmid pMA14 using a mutagenic forward (PBH38, 5′-GGGAAGTTAAGACTAAAGAAATACTTTACG-3′) primer and the reverse PBH18 primer. It was subsequently cloned into p414TEF vector at the SmaI and PstI site to obtain plasmid pMA24. To introduce a mutation at the M3 ATG, two separate PCRs utilizing the primer pair consisting of the forward PBH17 and reverse mutagenic (PBH40, 5′-GCTGGTATTGTAGACTTTGTTGCTTTG-3′) primers and the pair consisting of the forward mutagenic PBH39 (5′-CAAAGCAACAAAGTCTACAATACCAGC-3′) and reverse PBH18 primers were performed to obtain overlapping mutated products from plasmid pMA14 as the template. These were combined in a third PCR using the primers PBH17 and PBH18. The amplified fragment (1.2 kb) was cloned into p414TEF at the SmaI and PstI sites to construct plasmid pMA23. Mutated codons are shown in bold in the mutagenic primers. All the mutations were confirmed by DNA sequencing.
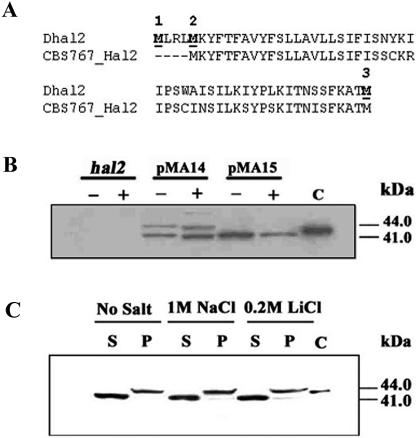
FIG. 2.
Western analysis of Dhal2p expression in S. cerevisiae. (A) Comparison of the N-terminal region of Dhal2p of our clone (Dhal2) with the homologous sequence from D. hansenii strain CBS767 (CBS767_Hal2). Potential initiator methionine residues are numbered and shown in bold. (B) Immunoblot showing expression of the full-length and the truncated forms of Dhal2p in S. cerevisiae. S. cerevisiae strain RS1051 (hal2) or RS1051 carrying plasmid pMA14 (full-length DHAL2) or pMA15 (truncated DHAL2) were grown without (−) or with (+) 1 M NaCl, and total protein extracts from these cultures were made as described in Materials and Methods. Total protein extracts (20 μg) were subjected to SDS-PAGE, followed by immunoblotting with anti-Dhal2p antibody. In lane C, the purified recombinant truncated Dhal2p was loaded as control. (C) Immunoblot showing localization of Dhal2p in S. cerevisiae. S. cerevisiae strain RS1051 carrying plasmid pMA14 was grown with or without salt as indicated. Cytosolic (S) and membrane (P) fractions from these cultures were made as described in Materials and Methods. A total of 20 μg of protein from each fraction was subjected to immunoblot analysis using anti-Dhal2p antibody. Purified recombinant truncated Dhal2p was loaded as control in lane C. The immunoblot shown is representative of three different experiments.
Constructs for Dhal2p localization.
For the expression of the truncated Dhal2-green fluorescent protein (GFP) fusion protein, a 1.7-kb fragment (TEF promoter plus DHAL2 open reading frame [ORF] having a mutation in the M1/M2 ATG) was PCR amplified from plasmid pMA24 using the forward (PBH44, 5′-CCGCTCGAGCGGATAACAATTTCACAC-3′) and reverse (PBH45, 5′-CCGCTCGAGTTGCCTTTAATACATCACTA-3′) (XhoI site incorporated [underlined]) primers. This fragment was digested with XhoI and SacI and cloned at the SacI-XhoI site in plasmid pRS-GFP (16). The resulting plasmid was named pMA26. Similarly, for the expression of the full-length Dhal2-GFP fusion protein, a 1.7-kb fragment (TEF promoter plus DHAL2 ORF having a mutation in the M3 ATG) was PCR amplified from plasmid pMA23 using primers PBH44 and PBH45. It was cloned at the SacI-XhoI site in plasmid pRS-GFP to obtain plasmid pMA25. Positive clones were confirmed by nucleotide sequencing.
Preparation of cell extract and immunoblot analysis.
Cell extract was made as described previously (40). Briefly, overnight cultures of Debaryomyces hansenii strain MTCC234 were reinoculated in 50 ml of YPD medium and grown at 30°C to early log phase (A600 of ∼0.6 to 0.8). Cells were harvested by centrifugation and resuspended in 50 ml of YPD medium with salt or without salt as a control and incubated for another 2.5 h. Cells were harvested, washed with water, and resuspended in 1 ml of sorbitol buffer (300 mM sorbitol, 100 mM NaCl, 5 mM MgCl2, 10 mM Tris-Cl, pH 7.5, 10 mM EDTA, 0.05% phenylmethylsulfonyl fluoride [PMSF]) containing protease inhibitor cocktail (Roche). Glass beads (425 to 600 μm) were added to the mixture, and the cells were broken by vortexing. Unbroken cells and debris were removed by centrifugation (500 × g for 5 min). The postnuclear supernatant was further fractionated into a crude membrane pellet (P) and cytosolic (S) fractions by centrifugation at 55,000 rpm in a TLA 100.2 rotor (Beckman) for 1 h at 4°C. The pellet was resuspended in one-fifth of the total volume of sorbitol buffer to normalize membrane and cytosolic fractions to approximately equal cell equivalents. Membrane and cytosolic fractions from S. cerevisiae transformants were prepared similarly except that SD minimal medium was used for growing the cultures.
Total cell extract of S. cerevisiae culture was prepared as described previously (11). Briefly, overnight cultures of strain RS1051 expressing different forms of Dhal2p were reinoculated and allowed to grow to logarithmic phase (A600 of ∼0.6 to 0.8). Cells were harvested by centrifugation and resuspended in SD minimal medium with salt or without salt as a control and incubated for an additional 2.5 h. Cells were harvested by centrifugation and resuspended in buffer containing protease inhibitors (50 mM Tris-Cl, pH 7.5, 1% sodium deoxycholate, 1% triton X-100, 0.1% sodium dodecyl sulfate [SDS], 0.05% PMSF, 0.05 μg/ml leupeptin). Glass beads (425 to 600 μm) were added to the mixture, and the cells were broken by vortexing. The resulting homogenate was centrifuged for 5 min at 13,000 rpm and the supernatant fraction was collected.
Immunoblotting was performed to check the expression as well as the membrane localization of the full-length Dhal2p. Protein concentration was estimated using Bradford reagent with bovine serum albumin as a standard. Approximately 20 μg of protein was resolved by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred at 120 V for 1 h to a Hybond ECL nitrocellulose membrane (Amersham Pharmacia Biotech) in a mini transblot apparatus (Bio-Rad) using Tris-glycine buffer (48 mM Tris, 39 mM glycine, and 20% methanol, pH 8.3). Blots were probed with anti-Dhal2p antibody (1) at a dilution of 1:2,000 in TBS buffer (20 mM Tris-Cl, pH 7.6, 137 mM NaCl) containing 5% skim milk. This was followed by secondary (horseradish peroxidase-conjugated) antibody and processed with an ECL Plus Western detection system per the manufacturer's protocol. In all cases, after the stripping step, blots were reprobed with preimmune serum to verify the results.
Affinity purification of Dhal2p by PAP-agarose.
D. hansenii culture at logarithmic phase was induced with 0.2 M LiCl for 2.5 h. Following centrifugation, the cells were resuspended in 1.0 ml of sucrose buffer (1.2% sucrose, 10 mM Tris-Cl, pH 7.6, 20 mM KCl, 1 mM EDTA, pH 8.0, 0.05% PMSF, and protease inhibitor cocktail). The crude membrane pellet and the cytosolic fractions were prepared as described previously. The membrane fraction was solubilized with sucrose buffer containing 1% Triton X-100. Dhal2p was purified from the cytosolic as well as the solubilized membrane fractions following the method described by Albert et al. (2). Briefly, each fraction was incubated with PAP-agarose beads (Sigma) for 30 min at 4°C. Beads were washed twice, and Dhal2p was eluted with 100 μl of buffer (10 mM EDTA, pH 8.0, 20 mM Tris-Cl, pH 7.6, 5 mM β-mercaptoethanol, and 20% sucrose). To purify the M1/M2 ATG (pMA23) or M3 ATG (pMA24) mutant of Dhal2p from S. cerevisiae, cultures were grown to logarithmic phase. The crude extract was prepared by resuspending the cell pellet in 1.0 ml of sucrose buffer containing 1% Triton X-100. Total cell lysate was incubated with PAP-agarose beads (Sigma) for 30 min at 4°C, and Dhal2p isoforms were purified by following a method similar to that described above. To confirm affinity purification, immunoblotting was performed as described previously.
Enzyme assay.
The nucleotidase activity of the enzyme was determined by quantifying the Pi released from the substrate using a colorimetric method (5). Briefly, a standard assay was conducted in a 100-μl reaction mixture including 50 mM Tris-morpholinethanesulfonic acid (pH 7.5), 1.0 mM substrate (PAP or inositol-1,4-bisphosphate), 5.0 mM magnesium acetate, and 0.2 μg of the purified protein. The mixture was incubated at 30°C for 30 min. The enzymatic reaction was stopped by adding an acidic solution (25 μl) of malachite green, ammonium molybdate, and Tween 20. The activity was measured as the amount of Pi liberated that produced a phosphomolybdate malachite complex detected at 650 nm in a microplate reader. The values obtained were corrected by subtracting the blank readings obtained by hydrolysis of the substrate in the absence of the enzyme. Nanomoles of Pi released were calculated from a standard curve with sodium dihydrogen phosphate made with each set of experiments. Enzyme activity is expressed as nanomoles of Pi liberated/min/mg of protein, and the data are presented in the form of the means ± standard deviations.
Fluorescence microscopy.
Fluorescence microscopy was performed on an inverted LSM510 META laser scanning confocal microscope (Carl Zeiss) fitted with a Plan-Apochromat ×100 (numerical aperture, 1.4) oil immersion objective. All measurements were performed with living nonfixed cells. For detection of the Dhal2-GFP fusion protein, the 488-nm line of an argon ion laser was directed over an HFT UV/488 beam splitter, and fluorescence was detected using an NFT 490 beam splitter in combination with a BP 505 to 530 band pass filter. S. cerevisiae strain RS16 expressing either the full-length Dhal2-GFP (plasmid pMA25) or the truncated Dhal2-GFP (pMA26) was grown to logarithmic phase (A600 of ∼0.6 to 0.8) and observed under the microscope. Cells were also treated with 0.2 M LiCl for 150 min or brefeldin A (10 μg/ml) for 30 min prior to visualization. Images obtained were processed using Adobe Photoshop version 5.5.
RESULTS
Expression of the DHAL2 gene from a single-copy vector confers a high level of salt tolerance.
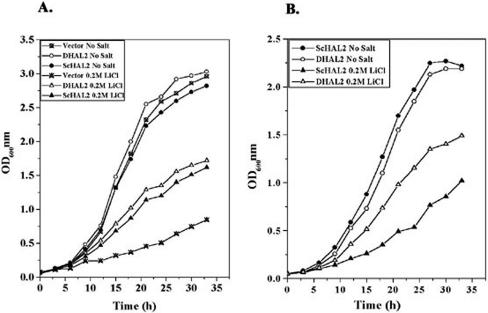
The enzyme bisphosphate nucleotidase is an important determinant of halotolerance in yeast. Recently, we cloned a bisphosphate nucleotidase gene (DHAL2) from D. hansenii strain MTCC234 (1). An increase in copy number of the homologous gene (HAL2) in S. cerevisiae was found to improve its halotolerance (19). We therefore compared the potential of DHAL2 with HAL2 to improve the halotolerance of S. cerevisiae. For this, the growth pattern of strain RS16 (17) harboring either DHAL2 (pMA7) or HAL2 (pMA29) in a multicopy vector was checked. Our results clearly indicated that strain RS16 expressing DHAL2 grew remarkably well in the presence of salt (Fig. 1A). However, the improvement in growth exhibited by DHAL2 under salt stress was marginally better than that shown by HAL2 (Fig. 1A). We next compared the level of halotolerance between S. cerevisiae expressing DHAL2 or HAL2 from a low-copy-number vector under the regulation of the HAL2 promoter. For this, plasmids pMA27 and pMA30 were transformed into the S. cerevisiae hal2 mutant strain RS1051. Our results showed that the salt tolerance conferred by DHAL2 was significantly higher than that by HAL2 under these conditions (Fig. 1B), and it was quite similar to that observed with DHAL2 present in the multicopy vector (Fig. 1A). Thus, it appeared that, unlike the S. cerevisiae homologue, the DHAL2 gene could improve salt tolerance of S. cerevisiae when present in a single-copy vector. This could be due to the higher intrinsic salt tolerance exhibited by Dhal2p.
FIG. 1.
Comparison of salt tolerance exhibited by the DHAL2 gene and S. cerevisiae HAL2 (ScHAL2). (A) Growth curve of strain RS16 carrying plasmid pRS425, pMA29 (ScHAL2), or pMA7 (DHAL2). Cultures were grown to early log phase (A600 of ∼0.5) and reinoculated at A600 of 0.05 in leucine minimal medium with or without 0.2 M LiCl. Growth of the cultures was followed by their absorbance at 600 nm. (B) Growth curve of strain RS1051 carrying plasmid pMA27 (DHAL2) or pMA30 (ScHAL2) in a low-copy-number vector. Cultures were grown to early log phase (A600 of ∼0.5) and reinoculated at A600 of 0.05 in methionine minimal medium with or without 0.2 M LiCl. Growth of the cultures was followed by their absorbance at 600 nm. The experiment was repeated three times with similar results.
N-terminal hydrophobic region is essential for the membrane localization of Dhal2p.
The nucleotide sequence of the cloned DHAL2 revealed that the predicted Dhal2p contained a unique N-terminal stretch that was highly hydrophobic (1). Recently, the genome sequence of D. hansenii strain CBS767 has been published (14). Analysis of the genomic sequences suggested that CBS767_Hal2 also contained a similar N-terminal region (Fig. 2A). However, the functional importance of this region has not been elucidated. To determine whether this region is translated in vivo, the two plasmids pMA14 and pMA15 were constructed by cloning the DHAL2 gene in the yeast expression vector p414TEF. In pMA14, the full-length ORF starting from +1 ATG was placed under the TEF promoter. It should therefore be capable of giving rise to the full-length form of the protein. By contrast, the plasmid pMA15 was designed to allow the production of the truncated form of the protein initiating from the M3 methionine (without the first 54 amino acids). These plasmids were individually transformed in S. cerevisiae strain RS1051, which is an hal2 mutant and therefore does not express the native Hal2p. The presence of Dhal2p in these transformants was checked by immunoblotting (see Materials and Methods). As expected, in the cells harboring pMA15, only a single band of 41 kDa was observed. In the case of plasmid pMA14, two bands, 44 kDa and 41 kDa, were observed in the Western blot (Fig. 2B). Computer analysis (27) of the Dhal2p sequence suggested that a possible signal peptide cleavage site could be present between residues 22 to 23 (numbering based on the full-length form of Dhal2p). In fact, the 44-kDa band was comigrating with the recombinant His-tagged truncated Dhal2p included as a control (Fig. 2B). We therefore reasoned that this band could possibly correspond to the processed form of the full-length Dhal2p. Our result suggested that the expression of DHAL2 in S. cerevisiae produced two distinct isoforms.
The existence of the N-terminal hydrophobic stretch in Dhal2p prompted us to examine the intracellular localization of the two isoforms. For this purpose, strain RS1051 transformed with plasmid pMA14 was grown in SD minimal medium with or without salt. The postnuclear supernatant was fractionated by centrifugation at 100,000 × g for 1 h at 4°C to prepare membrane and soluble fractions. These fractions were analyzed by immunoblotting (see Materials and Methods). From the blot it was evident that the 44-kDa form was present only in the membrane fractions, whereas the 41-kDa form could be seen only in the cytosolic fractions (Fig. 2C). Thus, our result clearly demonstrated the existence of a membrane-bound form of Dhal2p, and the N-terminal hydrophobic region was essential for its membrane localization.
Membrane-bound form of Dhal2p appeared due to alternative translation initiation.
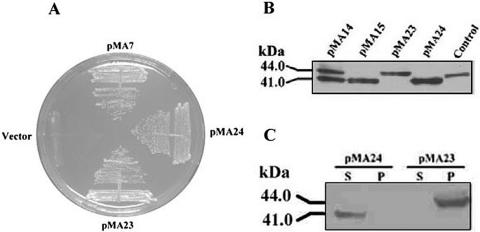
In the previous experiment, we showed that the cloning of the full-length DHAL2 ORF under TEF promoter produces two isoforms. Since the full-length DHAL2 ORF contains three in-frame ATG codons, the alternative use of translation initiation could be responsible for the expression of both forms of Dhal2p. To investigate this possibility, mutations at the potential initiation codons were introduced. In the construct pMA24, both M1 and M2 ATG codons were mutated to AAG, and by replacing the M3 ATG codon with AAG, we obtained pMA23. These plasmids were separately transformed into S. cerevisiae strain RS1051. Both mutants could complement the methionine auxotrophy of strain RS1051 (Fig. 3A). We next examined the expression of Dhal2p in these transformants by Western blot analysis. In the cells carrying the M1/M2 ATG mutant, only the truncated form of Dhal2p (41 kDa) was observed, whereas with the M3 ATG mutant, only the full-length form (44 kDa) was detected (Fig. 3B). As expected, these two forms also differed in their subcellular localization, as evidenced by the immunoblot (Fig. 3C). These results convincingly demonstrated that for the truncated cytosolic form of Dhal2p, M3 ATG served as the start codon. For the full-length membrane-bound form, either the M1 or M2 ATG was the initiation codon. Thus, alternative translation initiation could be the major mechanism for the synthesis of different isoforms of Dhal2p.
FIG. 3.
Expression profiles of ATG mutants of Dhal2p in S. cerevisiae. (A) Complementation of methionine auxotrophy exhibited by RS1051 harboring plasmid pMA7 (Dhal2p), pMA23 (Dhal2p M3 ATG mutant), or pMA24 (Dhal2p M1/M2 ATG mutant). (B) Immunoblot showing expression of ATG mutants of Dhal2p in S. cerevisiae. Total protein extract (20 μg) from strain RS1051 harboring plasmid pMA23 or pMA24 was subjected to immunoblot analysis with anti-Dhal2p antibody. For comparison, protein extracts from cells with plasmid pMA14 (expressing both full-length and truncated Dhal2p) and plasmid pMA15 (expressing only truncated Dhal2p) were loaded. Purified recombinant truncated Dhal2p is shown as Control. The blot shown is a representative of three different experiments. (C) Immunoblot showing localization of ATG mutants of Dhal2p in S. cerevisiae. Cytosolic (S) and membrane (P) fractions from strain RS1051 harboring plasmid pMA23 or pMA24 were subjected to immunoblot analysis using anti-Dhal2p antibody. The experiment was repeated three times with similar results.
Full-length Dhal2p exhibits a high level of IPPase activity.
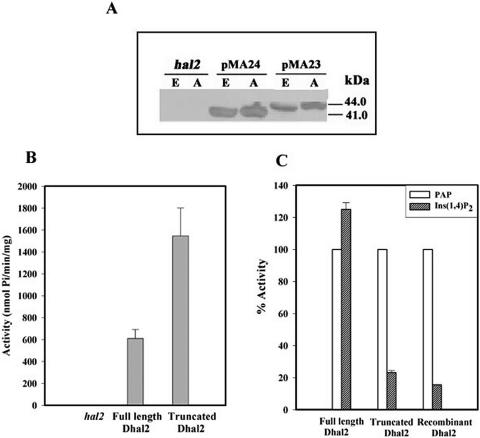
PAP is a substrate for Hal2p or its homologues, and therefore these enzymes display specific binding to a PAP-agarose matrix (2). Previously, we have shown that strain RS1051 harboring plasmid pMA24 expressed the truncated form of Dhal2p, whereas with plasmid pMA23 only the full-length form was observed. To determine the PAP-hydrolyzing ability of these isoforms, affinity purification utilizing PAP-agarose was carried out. The presence of Dhal2p isoforms in the purified fractions was confirmed by immunoblotting (Fig. 4A). Enzyme assays performed with these fractions showed that both full-length and truncated Dhal2p could efficiently hydrolyze PAP though the activity of the full-length form was lower compared to that of the truncated form (Fig. 4B). As a control, the total cell extract from the parent strain (RS1051) was also subjected to affinity purification, and this fraction did not show any activity. Some of the Hal2p homologues showed very high levels of IPPase activity. Earlier we have reported that the truncated, recombinant Dhal2p exhibited a low level of activity against inositol-1,4-bisphosphate (1). We therefore examined the activity of the full-length form against this substrate. Surprisingly, the full-length Dhal2p showed higher activity against inositol-1,4-bisphosphate than PAP (Fig. 4C). In contrast, the activity of the truncated Dhal2p was quite similar to that of the recombinant truncated Dhal2p observed earlier. Most importantly, the affinity-purified fraction from control strain RS1051 did not show any activity against either of these substrates. Thus, the full-length Dhal2p exhibited dual activity, like SAL1/FRY1 and RnPIP (21, 38).
FIG. 4.
Affinity purification and enzymatic activity of ATG mutants of Dhal2p. Total protein extracts from strain RS1051 (hal2) or strain RS1051 with pMA23 (M3 ATG mutant) or pMA24 (M1/M2 ATG mutant) were subjected to affinity purification using PAP-agarose (see Materials and Methods). (A) Immunoblot of total protein extracts (E) and affinity-purified fractions (A) with anti-Dhal2p antibody The immunoblot shown is representative of three different experiments. (B) An equal amount of protein (0.2 μg) from each fraction was incubated with PAP, and the amount of Pi released was measured as described in Materials and Methods. The PAP nucleotidase activity of each fraction is expressed as nmol of Pi/min/mg. (C) Comparison of activity of each fraction against PAP and inositol-1,4-bisphosphate. The activity of each form against inositol-1,4-bisphosphate is expressed as a percentage of the activity of the particular form against PAP. As a control, the activity of the recombinant truncated Dhal2p was measured. Results of three independent experiments are presented and error bars represent means ± standard deviations.
Subcellular localization of Dhal2p.
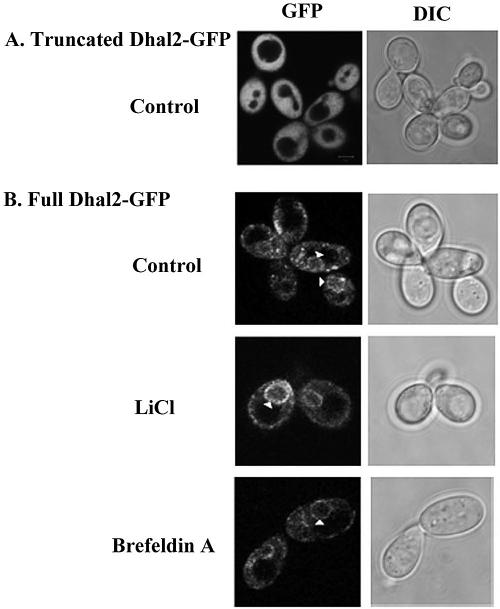
Mutation of the in-frame ATG codons, described above, resulted in expression of either the truncated or the full-length Dhal2p separately. To determine the subcellular localization of the different isoforms of Dhal2p, these mutant constructs were fused to GFP (see Materials and Methods). Plasmid pMA26 was made to express only the truncated Dhal2-GFP fusion protein, whereas plasmid pMA25 was made to produce only the full-length Dhal2-GFP fusion protein. Both the constructs complemented the methionine auxotrophy of the hal2 mutant, indicating that the fusion proteins have preserved the biological function of Dhal2p (data not shown). S. cerevisiae strain RS16 transformed with either plasmid pMA25 or pMA26 was grown to logarithmic phase (A600 of ∼0.5) and examined by fluorescence microscopy. In cells expressing the truncated Dhal2-GFP fusion protein, strong GFP fluorescence was visible throughout the cytoplasm (Fig. 5A). A similar fluorescence pattern was also observed in the cells treated with salt (data not shown). However, in the cells expressing the full-length Dhal2-GFP fusion protein, fluorescence was observed in the perinuclear region as a ring-like structure and in the intracellular aspect of cell surface that is typical for endoplasmic reticulum (ER)-located protein (32). The fluorescence pattern remained unchanged with brefeldin A treatment, also indicating that this could be located in the ER. Moreover, the addition of salt did not have any effect on the localization of the fusion protein (Fig. 5B).
FIG. 5.
Subcellular localization of Dhal2p by fluorescence microscopy. (A) Localization of truncated Dhal2-GFP fusion protein. Strain RS16 expressing the fusion protein was grown to logarithmic phase in SD minimal medium, and GFP fluorescence was viewed under a microscope. (B) Localization of full-length Dhal2-GFP fusion protein. Strain RS16 expressing the fusion protein were grown to logarithmic phase. Cells were either untreated (control) or treated with LiCl or brefeldin A as indicated prior to visualization (see Materials and Methods). The arrow indicates the perinuclear localization of full-length Dhal2-GFP fusion protein as a ring-like structure. Bar, 5 μm.
Dhal2 full-length protein is accumulated in the membrane upon salt stress.
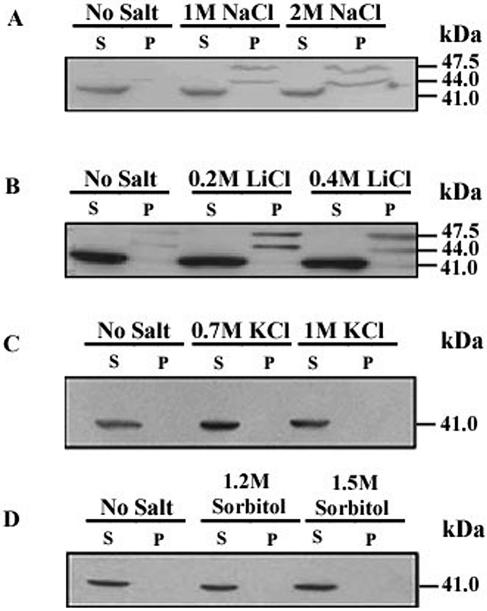
The expression of DHAL2 in S. cerevisiae clearly indicated the existence of a membrane-bound isoform of Dhal2p. This prompted us to examine whether the membrane-bound form is also expressed in D. hansenii. For this, cultures of D. hansenii strain MTCC234 were grown in the presence or absence of salt. The membrane and the cytosolic fractions from these cells were analyzed by immunoblotting as described previously. In the Western blot, a prominent band of 41 kDa was observed in the cytosolic fractions (Fig. 6A). From the molecular size, it appeared that this band could be a truncated form of Dhal2p initiating from the M3 methionine, thereby suggesting the cytosolic localization of Dhal2p. In S. cerevisiae, Hal2 nucleotidase is also known to be a cytosolic protein (19). Surprisingly, two additional bands of 44 kDa and 47.5 kDa were observed in the membrane fractions. These bands were detected only in the cells grown in the presence of 1.0 M or 2.0 M sodium chloride (Fig. 6A). When cells were grown in the presence of 0.2 M or 0.4 M lithium chloride, similar results were obtained (Fig. 6B). Interestingly, the membrane-bound forms could not be detected upon the addition of either KCl or sorbitol (Fig. 6C and D). Therefore, the accumulation of Dhal2p in the membrane fractions was possibly due to the salt toxicity response and not to the high osmolarity of the medium.
FIG. 6.
Analysis of DHAL2 expression in D. hansenii. Immunoblots showing localization of Dhal2p. Membrane (P) and cytosolic (S) fractions from D. hansenii strain MTCC234 grown with or without NaCl (A), LiCl (B), KCl (C) and sorbitol (D) at indicated concentrations were made as described in Materials and Methods. A total of 20 μg of protein from each fraction was subjected to SDS-PAGE followed by immunoblotting with anti-Dhal2p antibody. The experiment was repeated five times with similar results.
To ascertain that the membrane proteins highlighted in the immunoblot were indeed the isoforms of Dhal2p, we have conducted affinity purification using a PAP-agarose matrix. For this, membrane and cytosolic fractions were made from D. hansenii cells grown in the presence of salt. Dhal2p isoforms present in these fractions were pulled down by PAP-agarose. We next performed immunoblot analysis of affinity-purified fractions (see Materials and Methods). As expected, one band corresponding to the truncated Dhal2p was observed in the fraction purified from cytosol. However, in the fraction purified from the membrane, a single band of 44 kDa was detected (Fig. 7, inset). Absence of the 47.5-kDa band indicated that it was not a Dhal2p isoform. The detection of this band in the earlier blots (Fig. 6A and B) could be due to the cross-reactivity of our antibody. Alternatively, the 47.5-kDa protein could be a precursor that did not bind to PAP-agarose. An enzyme assay, using the affinity-purified fractions from the membrane as well as from the cytosol, further confirmed that both forms of Dhal2p could efficiently utilize PAP as a substrate (Fig. 7). Interestingly, the expression of the two isoforms of Dhal2p was also observed in D. hansenii strain CBS767. Moreover, in this strain also the membrane-bound form of Dhal2p appeared only in response to salt stress (data not shown). Thus, our results unequivocally established that the two forms of Dhal2p were present in D. hansenii and that the membrane-associated 44-kDa form was accumulated in the cell in response to the salt stress.
FIG. 7.
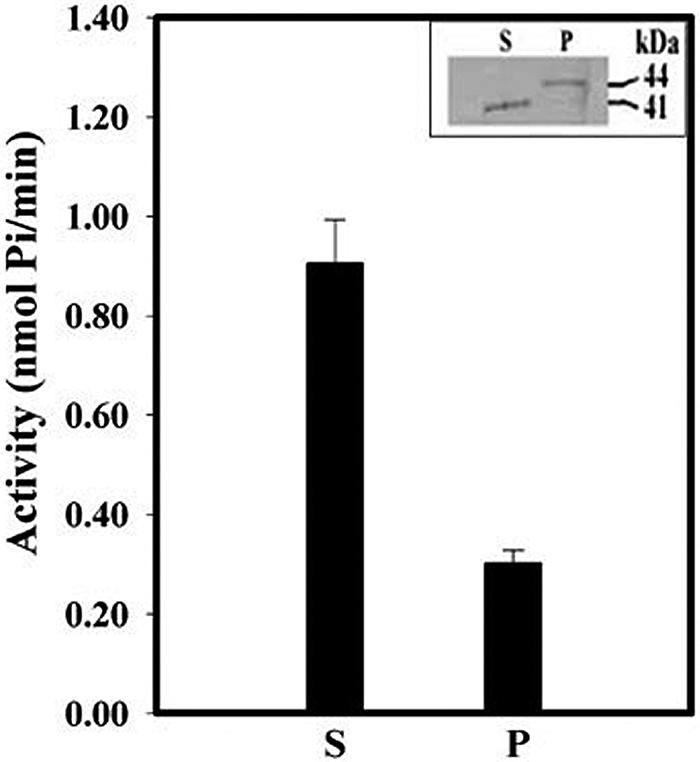
Enzyme activity of full-length and truncated forms of Dhal2p purified from D. hansenii. Cytosolic (S) and membrane (P) fractions from D. hansenii strain MTCC234 grown with 0.2 M LiCl were subjected to affinity purification by PAP-agarose (see Materials and Methods). The presence of Dhal2p in the purified fractions was checked by immunoblotting with anti-Dhal2p antibody (inset). PAP nucleotidase activity of the affinity-purified fractions was determined using 1.0 mM PAP as a substrate (see Materials and Methods). The assay was performed three times.
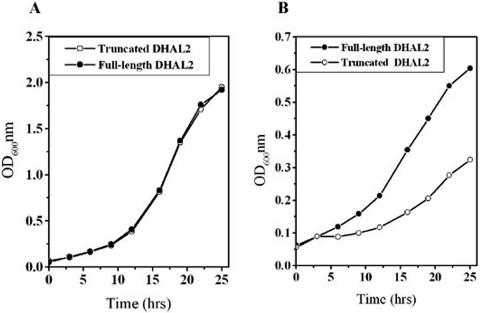
We further investigated whether the membrane-bound form has any role in the growth of yeast cells under high salt stress conditions. For this, two plasmids were constructed by cloning either the truncated (pMA28) or the full-length (pMA27) DHAL2 ORF under the regulation of S. cerevisiae HAL2 promoter in a low-copy-number vector. These plasmids were transformed into S. cerevisiae strain RS1051. Cells harboring the plasmid pMA28 expressed only the truncated form of Dhal2p. In contrast, both the truncated and full-length forms of Dhal2p were observed in the cells carrying plasmid pMA27 (data not shown). In the absence of salt, the growth pattern of the cells expressing both the isoforms was indistinguishable from those expressing only the truncated form of Dhal2p (Fig. 8A). In contrast, strain RS1051 harboring plasmid pMA27 grew much better in the presence of 0.4 M LiCl (Fig. 8B). Our result thus confirmed that the full-length form of Dhal2p plays an important physiological role in yeast under high salt stress conditions.
FIG. 8.
Growth pattern of S. cerevisiae expressing Dhal2p under high salt stress conditions. Cultures of strain RS1051 expressing the full-length DHAL2 ORF (pMA27) or the truncated DHAL2 ORF (pMA28) under the regulation of HAL2 promoter was grown to early log phase (A600 of ∼0.5) and reinoculated at A600 of 0.05. Growth of the cultures was followed by measuring their absorbance at 600 nm. (A) Growth curve in methionine minimal medium. (B) Growth curve in methionine minimal medium containing 0.4 M LiCl. The experiment was repeated three times with similar results.
DISCUSSION
D. hansenii represents one of the most halotolerant species of yeast. However, the molecular basis of this remarkable trait is poorly understood. Recent studies suggested that HAL2 is one of the most important determinants of salt tolerance in S. cerevisiae. It encodes a BPntase that is very sensitive to sodium and lithium, and therefore it is considered as a crucial rate-limiting metabolic step under salt stress (34). Surprisingly very little information is available about the homologous enzymes from salt-tolerant organisms. Recently we cloned the DHAL2 gene from D. hansenii by phenotypic complementation of an hal2 mutation in S. cerevisiae (1). The nucleotide sequence of the clone revealed that it encoded a protein of 420 amino acid residues. This protein showed substantial homology with Hal2p. One of the striking features of Dhal2p was that the N-terminal 54 amino acid residues (preceding M3) did not have any homology with other yeast homologues. In fact, comparison of protein sequences with other known homologues revealed that this region is unique to D. hansenii. Further analysis indicated that this region is highly hydrophobic. Expression of Dhal2p in E. coli is toxic to the cell. However, a high level of expression was achieved when this region was deleted. This observation led us to believe that the full-length Dhal2p could be a membrane protein. The present study was undertaken to test this hypothesis.
When Dhal2p was expressed in S. cerevisiae under control of the TEF promoter, two forms, one 41 kDa and the other 44 kDa, were observed. The 41-kDa form was found in the cytosol, whereas the 44-kDa form was present exclusively in the membrane fraction (Fig. 2B). These results clearly indicated that the two forms were the products of the cloned DHAL2 gene (Fig. 2B). Moreover, the N-terminal hydrophobic region is necessary for the synthesis of the membrane-bound isoform as the deletion of this region resulted in the synthesis of the cytoplasmic form alone. This region contained three methionine residues that could serve as potential initiators (1). Utilizing site-directed mutagenesis, we have clearly demonstrated that an alternative translation initiation was the major mechanism that regulates the synthesis of these two forms. This form of regulation has been previously documented in yeast. A number of genes in S. cerevisiae such as LEU4, FUM1, VAS1, TRM1, and HTS1 (6, 7, 9, 15, 37) encode two protein species. Like DHAL2, these genes also have ORFs with more than one in-frame ATG codon near the 5′ ends. The smaller protein is produced from 3′ ATG, whereas the larger form utilizes the 5′ ATG as the initiation codon. Interestingly, in these examples the two forms of the protein are also associated with different intracellular locations, cytosol and mitochondria.
It was evident from the Western blot analysis and biochemical assay that the two isoforms of Dhal2p also existed in D. hansenii. The 41-kDa form was present only in the cytoplasm, whereas the 44-kDa form was associated with the membrane fraction, presumably in the ER (Fig. 5 and 6). Although a number of homologues from several species have been characterized, this is the first report of a membrane-bound form of a BPntase. In D. hansenii the membrane-bound form was observed only during salt stress (Fig. 6). Moreover, the S. cerevisiae cells expressing both the isoforms exhibited a higher level of salt tolerance than those expressing only the cytosolic form of Dhal2p (Fig. 8). These results clearly suggested that the membrane-bound form played an important role under high salt stress conditions.
Finally, the most interesting and novel finding of this study was that the two forms differed not only in their subcellular localization but also in their substrate specificity. The cytosolic form was a typical BPntase, which was similar to the recombinant truncated Dhal2p (1). In contrast, the membrane-bound form was like a dually active BPntase prevalent in multicellular organisms that was also active against inositol-1,4-bisphosphate (21). This type of enzyme has been suggested to play an important role in inositol signaling. In Arabidopsis thaliana, SAL1/FRY1, a negative regulator of abscisic acid and stress signaling, exhibits both BPntase and IPPase activity (38). Mutations in this gene rendered the plants more sensitive to abscisic acid and to damage by low temperature, drought, or salt stress. The importance of IPPase activity in these functions has been demonstrated. Interestingly, the expression of SAL1 has been shown to modulate the Na+ efflux system (ENA) in S. cerevisiae, and the IPPase activity of SAL1 is involved in this process (31).
Our results indicate that D. hansenii accumulates the full-length Dhal2p in the ER in response to high salt stress conditions. What physiological advantage does this organism achieve by adopting such a mechanism? Inositol is a key metabolic sensor, and inositol levels regulate important ER functions such as membrane biosynthesis (28). In S. cerevisiae, the role of inositol polyphosphates in maintaining cell wall integrity, salt stress tolerance, endocytosis, and mRNA export has also been elaborated (10, 13, 29). Recent studies indicated that the salt stress markedly influenced the level of inositol mono- and polyphosphate in S. cerevisiae (25). A number of important regulators of phosphoinositide signaling in yeast are located in the ER. Therefore, it is possible that the full-length Dhal2p plays an important role in maintaining the inositol homeostasis under salt stress. Thus, our study illustrates that the modulation of subcellular localization and activity of Dhal2p could be an important adaptive mechanism for growth of D. hansenii under high salt stress conditions.
Acknowledgments
We thanks R. Serrano for yeast strains RS16 and RS1051 and P. A. Silver for plasmid pRS-GFP used in this study. The authors thank Chaaya Iyengar for critical reading of the manuscript.
This work was supported in part by a research grant from the Department of Biotechnology, New Delhi, India. M.A. is the recipient of a senior research fellowship from the Council of Scientific and Industrial Research, New Delhi, India.
REFERENCES
- 1.Aggarwal, M., P. K. Bansal, and A. K. Mondal. 2005. Molecular cloning and biochemical characterization of a 3′(2′),5′-bisphosphate nucleotidase from Debaryomyces hansenii. Yeast 22:457-470. [DOI] [PubMed] [Google Scholar]
- 2.Albert, A., L. Yenush, M. R. Gill-Mascarell, P. L. Rodriguez, S. Patel, M. Martinez-Ripell, T. L. Blundell, and R. Serrano. 2000. X-ray structure of yeast Hal2p, a major target of lithium and sodium toxicity, and identification of framework interactions determining cation sensitivity. J. Mol. Biol. 295:927-938. [DOI] [PubMed] [Google Scholar]
- 3.Arrillaga, I., R. Gil-Mascarell, C. Gisbert, E. Sales, C. Montesinos, R. Serrano, and V. Moreno. 1998. Expression of the yeast HAL2 gene in tomato increases the in vitro salt tolerance of transgenic progenies. Plant Sci. 136:219-2268. [Google Scholar]
- 4.Bansal, P. K., and A. K. Mondal. 2000. Isolation and sequence of the HOG1 homologue from Debaryomyces hansenii by complementation of the hog1 delta strain of Saccharomyces cerevisiae. Yeast 16:81-88. [DOI] [PubMed] [Google Scholar]
- 5.Baykov, A. A., O. A. Evtushenko, and S. M. Avaeva. 1988. Inorganic pyrophosphatase as a label in heterogeneous enzyme immunoassay. Anal. Biochem. 171:266-270. [DOI] [PubMed] [Google Scholar]
- 6.Beltzer, J. P., L. F. L. Chang, A. E. Hinkkanen, and G. B. Kohlhaw. 1986. Structure of yeast LEU4. The 5′ flanking region contains features that predict two modes of control and two productive translation starts. J. Biol. Chem. 261:5160-5167. [PubMed] [Google Scholar]
- 7.Chatton, B., P. Walter, J. P. Ebel, F. Lacroute, and F. Fasiolo. 1988. The yeast VAS1 gene encodes both mitochondrial and cytoplasmic valyl-tRNA synthetases. J. Biol. Chem. 263:52-57. [PubMed] [Google Scholar]
- 8.Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero, and P. Heiter. 1992. Multifunctional yeast-high-copy-number shuttle vectors. Gene 110:119-122. [DOI] [PubMed] [Google Scholar]
- 9.Chui, M. I., T. L. Mason, and G. R. Fink. 1992. HTS1 encodes both the cytoplasmic and mitochondrial histidyl-tRNA synthetase of Saccharomyces cerevisiae: mutations alter the specificity of compartmentation. Genetics 132:987-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooke, F. T., S. K. Dove, R. K. McEwen, G. Painter, A. B. Holmes, M. N. Hall, R. H. Michell, and P. J. Parker. 1998. The stress-activated phosphatidylinositol 3-phosphate 5-kinase Fab1p is essential for vacuole function in S. cerevisiae. Curr. Biol. 8:1219-1222. [DOI] [PubMed] [Google Scholar]
- 11.Davenport, K. R., M. Sohaskey, Y. Kamada, and D. E. Levin. 1995. A second osmosensing signal transduction pathway in yeast. J. Biol. Chem. 270:30157-30161. [DOI] [PubMed] [Google Scholar]
- 12.Dichtl, B., A. Stevens, and D. Tollervey. 1997. Lithium toxicity in yeast is due to the inhibition of RNA processing enzymes. EMBO J. 16:7184-7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubois, E., B. Scherens, F. Vierendeels, M. M. Ho, F. Messenguy, and S. B. Shears. 2002. In Saccharomyces cerevisiae, the inositol polyphosphate kinase activity of Kcs1p is required for resistance to salt stress, cell wall integrity, and vacuolar morphogenesis. J. Biol. Chem. 277:23755-23763. [DOI] [PubMed] [Google Scholar]
- 14.Dujon, B., D. Sherman, G. Fischer, P. Durrens, S. Casaregola, I. Lafontaine, et al. 2004. Genome evolution in yeasts. Nature 430:35-44. [DOI] [PubMed] [Google Scholar]
- 15.Ellis, S. R., A. K. Hopper, and N. C. Martin. 1989. Amino-terminal extension generated from an upstream AUG codon increases the efficiency of mitochondrial import of yeast N2,N2-dimethylguanosine-specific tRNA methyltransferases. Mol. Cell. Biol. 9:1611-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferrigno, P., F. Posas, D. Koepp, H. Saito, and P. A. Silver. 1998. Regulated nucleo/cytoplasmic exchange of HOG1 MAPK requires the importin beta homologs NMD5 and XPO1. EMBO J. 17:5606-5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaxiola, R., I. F. de Larrinoa, J. M. Villalba, and R. Serrano. 1992. A novel and conserved salt induced protein is an important determinant of salt tolerance in yeast. EMBO J. 11:3157-3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gil-Mascarell, R., J. M. Lopez-Coronado, J. M. Belles, R. Serrano, and P. L. Rodriguez. 1999. The Arabidopsis thaliana HAL2-like gene family includes a novel sodium-sensitive phosphatase. Plant J. 17:373-383. [DOI] [PubMed] [Google Scholar]
- 19.Glaser, H. V., D. Thomas, H. Gaxiola, F. Montrichard, Y. Surdin-Kerjan, and R. Serrano. 1993. Salt tolerance and methionine biosynthesis in Saccharomyces cerevisiae involves a putative phosphatase gene. EMBO J. 12:3105-3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 21.Lopez-Coronado, J. M., J. M. Belles, F. Lesage, R. Serrano, and P. L. Rodriguez. 1999. A novel mammalian lithium-sensitive enzyme with a dual enzymatic activity, 3′-phosphoadenosine 5′-phosphate phosphatase and inositol-polyphosphate 1-phosphatase. J. Biol. Chem. 274:16034-16039. [DOI] [PubMed] [Google Scholar]
- 22.Mumberg, D., R. Muller, and M. Funk. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156:119-122. [DOI] [PubMed] [Google Scholar]
- 23.Murguia, J. R., J. M. Belles, and R. Serrano. 1995. A salt sensitive 3′(2′),5′-bisphosphate nucleotidase involved in sulfate activation. Science 267:232-234. [DOI] [PubMed] [Google Scholar]
- 24.Murguia, J. R., J. M. Belles, and R. Serrano. 1996. The yeast HAL2 nucleotidase is an in vivo target of salt toxicity. J. Biol. Chem. 271:29029-29033. [DOI] [PubMed] [Google Scholar]
- 25.Navarro-Avino, J. P., J. M. Belles, and R. Serrano. 2003. Yeast inositol mono- and trisphosphate levels are modulated by inositol monophosphatase activity and nutrients. Biochem. Biophys. Res. Comm. 302:41-45. [DOI] [PubMed] [Google Scholar]
- 26.Neuwald, A. F., B. R. Krishnan, I. Brikun, S. Kulakauskas, K. Suziedelis, T. Tomcsanyi, T. E. Leyh, and D. E. Berg. 1992. cysQ, a gene needed for cysteine synthesis in Escherichia coli K-12 only during aerobic growth. J. Bacteriol. 174:415-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 28.Nunnari, J., and P. Walter. 1996. Regulation of organelle biogenesis. Cell 84:389-394. [DOI] [PubMed] [Google Scholar]
- 29.Odom, A. R., A. Stahlberg, S. R. Wente, and J. D. York. 2000. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science 287:2026-2029. [DOI] [PubMed] [Google Scholar]
- 30.Peng, Z., and D. P. S. Verma. 1995. A rice HAL2-like gene ecodes a Ca+-sensitive 3′(2′)5′-diphosphonucleoside 3′(2′)-phosphohydrolase and complements yeast met22 and Escherichia coli cysQ mutations. J. Biol. Chem. 270:29105-29110. [DOI] [PubMed] [Google Scholar]
- 31.Quintero, F. J., B. Garciadeblas, and A. Rodriguez-Navarro. 1996. The SAL1 gene of Arabidopsis, encoding an enzyme with 3′(2′),5′-bisphosphate nucleotidase and inositol polyphosphate 1-phosphatase activities, increases salt tolerance in yeast. Plant Cell 8:529-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rose, M. D., L. M. Misra, and J. P. Vogel. 1989. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell 57:1121-1221. [DOI] [PubMed] [Google Scholar]
- 33.Schneider, B., Y. W. Xu, J. Janin, M. Veron, and D. Deville-Bonne. 1998. 3′-Phosphorylated nucleotides are tight binding inhibitors of nucleoside diphosphate kinase activity. J. Biol. Chem. 273:28773-28778. [DOI] [PubMed] [Google Scholar]
- 34.Serrano, R. 1996. Salt tolerance in plants and microorganisms: toxicity targets and defense responses. Int. Rev. Cytol. 165:1-52. [DOI] [PubMed] [Google Scholar]
- 35.Spiegelberg, B. D., J. P. Xiong, J. J. Smith, R. F. Gu, and J. D. York. 1999. Cloning and characterization of a mammalian lithium sensitive bisphosphate 3′-nucleotidase inhibited by Inosiotol-1,4-bisphosphate. J. Biol. Chem. 274:13619-13628. [DOI] [PubMed] [Google Scholar]
- 36.Thomas, D., and Y. Surdin-Kerjan. 1997. Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 61:503-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu, M., and A. Tzagoloff. 1987. Mitochondrial and cytoplasmic fumarases in Saccharomyces cerevisiae are encoded by a single nuclear gene FUM1. J. Biol. Chem. 262:12275-12282. [PubMed] [Google Scholar]
- 38.Xiong, L., B. Lee, M. Ishitani, H. Lee, C. Zhang, and J. K. Zhu. 2001. FIERY1 encoding an inositol polyphosphate 1-phosphatase is a negative regulator of abscisic acid and stress signaling in Arabidopsis. Genes Dev. 15:1971-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yenush, L., J. M. Belles, J. M. Lopez-Coronado, R. Gil-Mascarell, R. Serrano, and P. L. Rodriguez. 2000. A novel target of lithium therapy. FEBS Lett. 467:321-325. [DOI] [PubMed] [Google Scholar]
- 40.Zhao, L., S. Lobo, X. Dong, A. D. Ault, and R. Deschenes. 2002. Erf4p and Erf2p form an endoplasmic reticulum-associated complex involved in the plasma membrane localization of yeast Ras proteins. J. Biol. Chem. 277:49352-49359. [DOI] [PubMed] [Google Scholar]