Abstract
Vacuoles in filamentous fungi are highly pleomorphic and some of them, e.g., tubular vacuoles, are implicated in intra- and intercellular transport. In this report, we isolated Aovam3, the homologue of the Saccharomyces cerevisiae VAM3 gene that encodes the vacuolar syntaxin, from Aspergillus oryzae. In yeast complementation analyses, the expression of Aovam3 restored the phenotypes of both Δvam3 and Δpep12 mutants, suggesting that AoVam3p is likely the vacuolar and/or endosomal syntaxin in A. oryzae. FM4-64 [N-(3-triethylammoniumpropyl)-4-(p-diethylaminophenyl-hexatrienyl)pyridinium dibromide] and CMAC (7-amino-4-chloromethylcoumarin) staining confirmed that the fusion protein of enhanced green fluorescent protein (EGFP) with AoVam3p (EGFP-AoVam3p) localized on the membrane of the pleomorphic vacuolar networks, including large spherical vacuoles, tubular vacuoles, and putative late endosomes/prevacuolar compartments. EGFP-AoVam3p-expressing strains allowed us to observe the dynamics of vacuoles with high resolutions, and moreover, led to the discovery of several new aspects of fungal vacuoles, which have not been discovered so far with conventional staining methods, during different developmental stages. In old hyphae, EGFP fluorescence was present in the entire lumen of large vacuoles, which occupied most of the cell, indicating that degradation of cytosolic materials had occurred in such hyphae via an autophagic process. In hyphae that were not in contact with nutrients, such as aerial hyphae and hyphae that grew on a glass surface, vacuoles were composed of small punctate structures and tubular elements that often formed reticulum-like networks. These observations imply the presence of so-far-unrecognized roles of vacuoles in the development of filamentous fungi.
Vacuoles are acidic compartments that are important for metabolite storage and cytosolic ion and pH homeostasis (for a review, see reference 18). In the unicellular yeast Saccharomyces cerevisiae, roles of and transport pathways to vacuoles have been extensively studied. Especially, many studies have been focused on the syntaxin family t-SNAREs (target organelle soluble N-ethyl-maleimide-sensitive factor [NSF] attachment protein receptors), which are central molecules of intracellular vesicular transport. t-SNAREs that reside on the target organelle membrane mediate the fusion of transport vesicles with the organelles via the specific interaction with vesicle SNAREs on the transport vesicles (29). S. cerevisiae Pep12p is the syntaxin family t-SNARE on the late endosome/prevacuolar compartment and receives Golgi-derived vesicles (3). Vam3p is the vacuolar membrane syntaxin that regulates the vesicular traffic to and the homotypic fusion of vacuoles (44). Since S. cerevisiae Vam3p localizes exclusively on the vacuolar membrane, Vam3p is frequently used for visualization of vacuoles (45, 46) and as a marker of vacuoles in subcellular fractionation experiments. AtVam3p is a Vam3p-homologue protein in Arabidopsis thaliana (30), and its fusion protein with green fluorescent protein (GFP) has been successfully used for the observation of vacuolar dynamics in A. thaliana (41) and Nicotiana tabacum (22).
On the other hand, characteristic aspects of the vacuoles in multicellular fungi were eventually identified using microscopic approaches in the past decade. First, vacuoles in filamentous fungi display approximately predictable pleomorphism in the different regions of the mycelia; vacuoles are typically rare or absent in the apical region, whereas ovoid-spherical vacuoles are present in the subapical region, tubular vacuoles in the more distal region, and large spherical vacuoles in the basal zone (14). Second is the presence of highly motile tubular vacuoles that can be visualized by vacuolar fluorescent probes such as 6-carboxyfluorescein diacetate (CFDA) derivatives (2, 6, 7, 34, 42) or N-(3-triethylammoniumpropyl)-4-(p-diethylaminophenyl-hexatrienyl)pyridinium dibromide (FM4-64) (10) and by the fusion protein of carboxypeptidase Y (CPY) with enhanced GFP (EGFP) (23, 24). These characteristic vacuolar morphologies indicate that the vacuoles may have particular roles in filamentous fungi. In fact, the tubular vacuoles that were originally found in a mycorrhizal fungus Pisolithus tinctorius (34) were implicated in intra- and intercellular transport of nutrients (reviewed in reference 2). In spite of these intriguing observations, however, molecular genetics studies on vacuoles in filamentous fungi have been limited (25, 26, 39). Therefore, identification and investigation of the proteins involved in vacuolar morphogenesis are now crucial for further understanding of the molecular mechanisms regulating pleomorphic vacuoles in filamentous fungi.
In this report, we developed Aspergillus oryzae strains that express the fusion protein of EGFP with AoVam3p, the Vam3p homologue in A. oryzae. As EGFP-AoVam3p localized on the vacuolar membrane, these strains enabled us to observe vacuolar membrane dynamics in A. oryzae with high resolution.
MATERIALS AND METHODS
Plasmids and strains.
Primer pairs vam3 Bsr-N (5′-CTGTACATGTATTTCGACCGTCTTAGT-3′) and vam3 Bsr-C (5′-TGTACATTATCCAATAGTAGCCGCCAG-3′) were designed (BsrGI sites are underlined) based on the expressed sequence tag sequence of a putative VAM3 homologue gene in A. oryzae (Aovam3). The Aovam3 cDNA (0.8 kb) was amplified with these primer pairs, using the A. oryzae RIB40 cDNA library as a template. The egfp gene (0.7 kb) was fused to the 5′ end of the resultant Aovam3 cDNA in frame, resulting in an egfp-Aovam3 cDNA fusion gene. Two plasmids for the expression of the fusion gene were subsequently constructed. The plasmid pUEGFP-VAM carries the 0.6-kb amyB promoter, followed by the 1.5-kb egfp-Aovam3 fusion gene, the 0.3-kb nos3′ terminator, and the 5.5-kb niaD gene encoding a nitrate reductase as a transformation marker. This plasmid was introduced to A. oryzae niaD300 (niaD−) strain with the A. oryzae transformation procedure (17), yielding UEV strains. The plasmid pBNVPEV carries the 1.3-kb Aovam3 promoter followed by the 1.5-kb egfp-Aovam3 fusion gene, the 0.3-kb nos3′ terminator, and the 5.5-kb niaD marker. For the generation of Aovam3 conditional expression strains, TPVIIs, a DNA fragment that contained Aovam3 5′ flanking region followed by Aspergillus nidulans sC marker encoding an ATP sulfurylase, A. oryzae thiA promoter driving Aovam3 cDNA, and Aovam3 3′ flanking region, was introduced into A. oryzae NS4 strain (niaD−, sC−). The gene replacement of Aovam3 by thiA promoter-driven Aovam3 cDNA was confirmed by PCR and Southern analysis (data not shown). The plasmid pBNVPEV was introduced to one of the conditional expression strains, the TPVII118 strain, yielding TPVEV strains. Southern analysis of the genomic DNA of TPVEV1, 2, 3, and 4 revealed that single copies of the plasmid had been inserted homologously at the niaD locus in the TPVEV1, 2, and 3 strains, while a few extra copies of the plasmid (probably one or two additional copies, judged by the signal intensity) had been inserted in the TPVEV4 strain (data not shown).
For yeast complementation analyses, deletion strains of EUROSCARF (http://www.rz.uni-frankfurt.de) constructed from S. cerevisiae BY4741 (MATa his3Δ1 leu2Δ0 met5Δ0 ura3Δ0) (5) were used. Aovam3 cDNA was amplified using the Vam3 N-term (5′-CATGTATTTCGACCGTCTTAG-3′) and Vam3 C-term (5′-TTATCCAATAGTAGCCGCCAG-3′) primers and subsequently inserted downstream of the GAL1 promoter of the pYES2 plasmid. The obtained plasmid, pYESVAM, was introduced into the yeast Δvam3 strain Y02362 (BY4741 vam3Δ::KanMX4) and Δpep12 strain Y01812 (BY4741 pep12Δ::KanMX4). pYES2 alone was also introduced into the Δvam3 and Δpep12 strains to obtain the control strains.
Culture conditions.
Czapek-Dox medium (CD; 0.3% NaNO3, 0.2% KCl, 0.1% KH2PO4, 0.05% MgSO4 · 7H2O, 0.002% FeSO4 · 7H2O, 2% glucose, pH 5.5) was used for microscopic observations. M medium [0.2% NH4Cl, 0.1% (NH4)2SO4, 0.05% KCl, 0.05% NaCl, 0.1% KH2PO4, 0.05% MgSO4 · 7H2O, 0.002% FeSO4 · 7H2O, 2% glucose, pH 5.5] was used for comparison of phenotypes between TPVII118 and TPVEV strains. Thiamine hydrochloride (Sigma Chemical Co., St. Louis, MO) was added for observation of TPVEV strains at a concentration of 10 μM. DPY (2% dextrin, 1% polypeptone, 0.5% yeast extract, 0.5% KH2PO4, 0.05% MgSO4 · 7H2O) was used for observation of vacuoles in germinating conidia.
A YPGal plate (1% yeast extract, 2% Bacto-peptone, and 2% galactose) was used to compare the growth of yeast strains. For the CFDA staining of yeast vacuoles, SGal medium (0.67% yeast nitrogen base without amino acids, 2% galactose, with required nutrients) was used.
Microscopic equipment.
For routine microscopy we used an Olympus System microscope model BX52 (Olympus, Tokyo, Japan) equipped with a UPlanApo 100× objective lens (1.35 numerical aperture) (Olympus). A GFP filter (495/20 nm excitation, 510 nm dichroic, 530/35 nm emission) (Chroma Technologies, Brattleboro, VT) or a U-MWIB filter cube (460 to 490 nm excitation, 505 nm dichroic, >515 nm emission) (Olympus) was used for observation of EGFP fluorescence. A DsRed filter (570/20 nm excitation, 590 nm dichroic, 630/60 nm emission) (Chroma Technologies) and a BH-DMU (330 to 385 nm excitation, 400 nm dichroic, >420 nm emission) UV excitation cube (Olympus) were used to observe the fluorescence of N-(3-triethylammoniumpropyl)-4-(p-diethylaminophenyl-hexatrienyl)pyridinium dibromide (FM4-64) (Molecular Probes Inc., Eugene, OR) and 7-amino-4-chloromethylcoumarin (CMAC) (Molecular Probes Inc.), respectively. Images were analyzed with MetaMorph software (Molecular Devices Co., Sunnyvale, CA).
Confocal microscopy was performed with an IX70 inverted microscope (Olympus) equipped with 100× and 40× Neofluor objective lenses (1.30 numerical aperture), a Sapphire 488-20, 20-mW diode laser (Coherent, Santa Clara, CA), a GFP filter, a CSU21 confocal scanning system (Yokogawa Electronics, Tokyo, Japan), an AP imager camera (Hamamatsu Photonics, Hamamatsu, Japan), and an image intensifier unit (Hamamatsu Photonics). Images were analyzed with IPlab software (Scanalytics, Fairfax, VA).
Culture conditions for microscopy.
For observations of cover glass cultures, approximately 103 conidia were inoculated in 100 μl of CD on coverslips that were placed in sealed petri dishes and incubated for 20 h at 30°C.
For observations with the inverted confocal microscope, approximately 103 conidia were inoculated in 100 μl of CD in a 35-mm glass base dish (Asahi Techno Glass, Chiba, Japan), which is a small petri dish whose bottom was made with cover glass, and incubated for 18 to 72 h at 30°C.
Observations of aerial and glass surface hyphae (see Results for definition) were performed essentially as described by Ishi et al. (15) with slight adaptations. In brief, approximately 1-mm-thick CD containing 2% agar was excised as a square of 1 cm by 1 cm and placed on a glass slide. Conidia were inoculated on the side surface of the CD agar medium, and hyphae were allowed to grow in lateral directions by preventing upward-directed growth with a cover glass. The culture was incubated at 30°C for up to 4 days in a petri dish containing a wet paper towel to prevent drying.
For observation of vacuoles in germinating conidia, fresh conidia collected from 5- to 6-day cultures in a CD plate were suspended in DPY at a concentration of 107/ml. The cultures were incubated at 30°C with shaking for ∼6 h.
Staining of vacuoles.
FM4-64 staining was performed as follows. Cultivation media of coverslip cultures were replaced with CD containing 8 μM FM4-64, and the cultures were incubated for 10 min at 30°C. The media containing the dye were replaced twice with fresh media without dye, and the cultures were incubated at 30°C for another 10 min, followed by microscopy observations. CMAC staining of vacuoles was performed as described previously (23). Dual staining with FM4-64 and CMAC was carried out as follows. Cover glass cultures were first incubated in CD containing 10 μM CMAC for 15 min, followed by incubation for another 15 min in CD containing both 10 μM CMAC and 8 μM FM4-64. After being washed with fresh dye-free medium, the cover glass cultures were observed.
CFDA staining of yeast vacuoles was performed as described by Roberts et al. (28).
Measurement of apical extension rate.
Conidia of RIB40, the A. oryzae wild-type strain, were inoculated in 100 μl of CD in glass base dishes and incubated for 20 h at 30°C. The culture media were then replaced with either CD (control) or CD containing 8 μM FM4-64 and incubated for 30 min. The culture dishes were then placed on a Thermo Plate (Tokai Hit Co., Shizuoka, Japan) set at 30°C and observed with a 40× objective lens. Hyphae at the mycelial periphery were selected for measurement, since other hyphae lying in the growth direction might have influenced hyphal growth. Two pictures that were taken at intervals of 10 min were overlaid, and the distance between the respective hyphal tips was defined as the hyphal elongation length per 10 min.
RESULTS
Cloning and characterization of Aovam3, the VAM3 homologue gene in A. oryzae.
The cDNA of the S. cerevisiae VAM3 homologue gene in A. oryzae was cloned using a PCR-based approach. To obtain the genomic DNA of the gene, plaque hybridization for A. oryzae genomic DNA library (13) was performed essentially as described previously (21). Sequence analysis revealed that the gene contains a single intron and encodes a putative protein product of 271 amino acids (DDBJ nucleotide sequence database accession no. AB232045). This gene was designated Aovam3, since this is the only gene in the A. oryzae genome database (20) that encodes a putative protein displaying high sequence identity to S. cerevisiae Vam3p (25.2%). In addition, AoVam3p has characteristic features of the syntaxin family t-SNAREs, such as a SNARE motif and a predicted transmembrane domain at the carboxy terminus. Notably, the AoVam3p sequence also showed an equal level of identity to S. cerevisiae Pep12p (27.1%), a t-SNARE in the late endosome/prevacuolar compartment (3). Thus, as was predicted in other filamentous fungi (12), A. oryzae possesses only one gene corresponding to either VAM3 or PEP12.
As Aovam3 showed sequence similarity to both VAM3 and PEP12, we were interested in determining whether Aovam3 was also functionally homologous to these genes. To test this, we performed complementation analyses for S. cerevisiae deletion mutants. Deletion of VAM3 causes a growth defect in the presence of a high concentration of calcium (36). As shown in Fig. 1a, however, expression of the Aovam3 cDNA restored the growth in the presence of 250 mM CaCl2 (Fig. 1a). Vacuolar fragmentation of the Δvam3 strain (44) was also complemented by the expression of the Aovam3 cDNA (Fig. 1b). On the other hand, deletion of PEP12 causes a temperature sensitivity (3) and defects in maturation of vacuolar hydrolytic enzymes (8). We therefore tested the complementation of the CPY activity (data not shown) by APE assay (16) and the growth at 37°C (Fig. 1c). In both cases, the expression of the Aovam3 cDNA restored the defects in the Δpep12 mutant. These results indicate that AoVam3p is likely the vacuolar and/or endosomal t-SNARE in A. oryzae.
FIG. 1.
Complementation of S. cerevisiae Δvam3 and Δpep12 phenotypes by the expression of Aovam3 cDNA. (a) The indicated yeast strains were streaked on the YPGal plate containing either 100 mM or 250 mM CaCl2 and grown for 3 days. (b) Vacuoles of the indicated strains were stained with 10 μM CFDA. Large developed vacuoles were observed in the wild-type (WT) strain and the Δvam3 strain that expressed Aovam3 cDNA, while Δvam3 strain that only possessed the empty vector showed vacuolar fragmentation. Bars, 10 μm. (c) The indicated yeast strains were streaked on the YPGal plate and incubated at either 25°C or 37°C for 3 days.
Generation of EGFP-AoVam3p-expressing strains.
For the expression of AoVam3p fused at its amino terminus with EGFP (EGFP-AoVam3p) in A. oryzae, three strains, UEV1, TPVEV1, and TPVEV4, were generated (Table 1). In all three strains, EGFP was fused at the amino terminus of AoVam3p and therefore should locate on the cytosolic side. UEV1 carries the plasmid pUEGFP-VAM and overexpresses EGFP-AoVam3p under the strong, inducible promoter of amyB gene that encodes an α-amylase (37). UEV1 displayed no distinguishable differences in morphology compared with the control strain in glucose medium (data not shown) in which the amyB promoter was induced at the intermediate level (37), suggesting that overexpression of EGFP-AoVam3p does not impair the integrity of the mycelium. In some cases, however, overexpression and fusion of a t-SNARE with EGFP result in mislocalization or leakage of the fusion protein to other organelles (4). We therefore tested whether the expression of EGFP-AoVam3p can complement the phenotypes of the Aovam3 conditional expression strain (J.-Y. Shoji, M. Arioka, and K. Kitamoto, unpublished results) to determine whether the fusion protein is functional. The TPVII118 strain, an Aovam3 conditional expression strain, expresses Aovam3 under the thiamine-regulatable thiA promoter (35) and is defective in aerial hyphal formation, so that its mycelium shows a rough periphery in the Aovam3-repressed condition. These phenotypes were complemented in TPVEV strains that expressed EGFP-AoVam3p under the control of the Aovam3 promoter on a TPVII118 background (Fig. 2). This observation confirmed that the fusion protein is functional. Southern analysis of TPVEV1, 2, 3, and 4 revealed that single copies of the plasmid were inserted homologously at the niaD locus in the TPVEV1, 2, and 3 strains, while a few extra copies of the plasmid (probably one or two additional copies, judged by the signal intensity) had been inserted in the TPVEV4 strain (data not shown). Therefore, the expression level of EGFP-AoVam3p is similar to that of authentic AoVam3p in TPVEV1, while the fusion protein is slightly overexpressed in TPVEV4 and is evidently overexpressed in UEV1.
TABLE 1.
Genotypes of strains used in this study
| Name | Parental strain | Genotype | Expression level of EGFP-AoVam3p |
|---|---|---|---|
| RIB40 | Wild type | ||
| niaD300 | RIB40 | niaD− | |
| NS4 | niaD300 | niaD−sC− | |
| TPVII118a | NS4 | niaD−sC− ΔAovam3::(PthiA-Aovam3 sC) | |
| UEV1 | niaD300 | niaD−::(PamyB-egfp-Aovam3 niaD) | Overexpressed |
| TPVEV1 | TPVII118 | niaD−::(PAovam3-egfp-Aovam3 niaD) sC− ΔAovam3::(PthiA-Aovam3 sC) | Similar to that of the authentic Aovam3 |
| TPVEV4 | TPVII118 | niaD−::(PAovam3-egfp-Aovam3 niaD) sC− ΔAovam3::(PthiA-Aovam3 sC) | Slightly overexpressed |
TPVII118 is an Aovam3 conditional expression mutant in which the entire Aovam3 coding sequence is replaced by A. nidulans sC and A. oryzae thiA promoter followed by Aovam3 cDNA.
FIG. 2.

Complementation of phenotypes in an Aovam3 conditional expression mutant by the expression of EGFP-AoVam3p. Approximately 102 conidia of either TPVII118, an Aovam3 conditional expression mutant that expresses Aovam3 under the control of thiamine-regulatable thiA promoter, and TPVEV strains that express EGFP-AoVam3p under the Aovam3 promoter on a TPVII118 background were inoculated and grown for 4 days in M medium (thiA promoter induced, left) or M medium containing 10 μM thiamine (thiA promoter repressed, right).
EGFP-AoVam3p localized on the vacuolar membrane.
In all three strains, EGFP-AoVam3p localized on small punctate structures in the apical region (Fig. 3b and e) and on tubular vacuoles in more distal compartments (Fig. 4). In the basal region of hyphae (Fig. 3a, c, d, f, and g), the membrane of large developed vacuoles and a few small punctate structures were observed with EGFP-AoVam3p (Fig. 3f and g). As the localization pattern of EGFP-AoVam3p was fundamentally similar in these strains (Fig. 3a to d and data not shown), we present the representative results of one of these transformants in the following studies, although essentially similar results were obtained for all three strains tested.
FIG. 3.
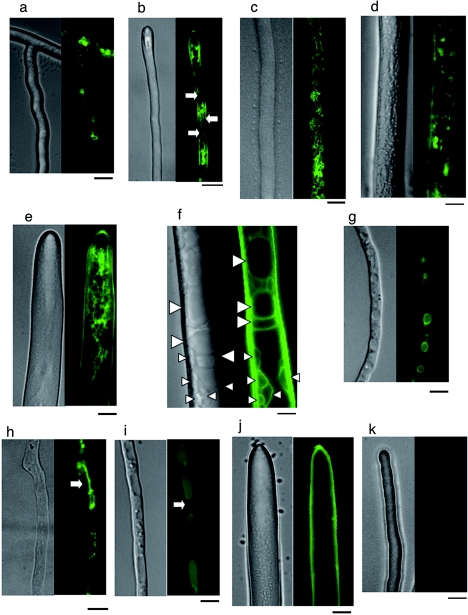
FM4-64 and CMAC staining of EGFP-AoVam3p-expressing strains. Cover glass cultures of UEV1, TPVEV1, and TPVEV4 grown for 20 h at 30°C were stained with either 8 μM FM4-64 (a to d) or 10 μM CMAC (e, f). (a) Basal region in UEV1. (b, c) Apical region and basal region, respectively, in TPVEV1. (d) Basal region in TPVEV4. (e, f) Apical and basal regions, respectively, in TPVEV1. (g) Dual staining of TPVEV1 with FM4-64 and CMAC. Images of DIC, FM4-64, EGFP, and CMAC fluorescence are shown. Arrowheads (f, g) indicate putative late endosomes/prevacuolar compartments visualized by EGFP-AoVam3p that are not stained with CMAC. Bars, 5 μm.
FIG. 4.
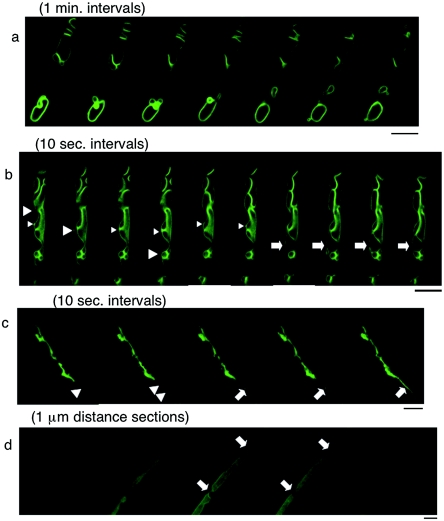
Confocal microscopy of EGFP-AoVam3p-expressing strains. UEV1 was grown in glass base dishes for 20 h (a to c) or 48 h (d) and observed with inverted confocal laser scanning microscopy. (a) Time-lapse images of large vacuoles taken at intervals of 1 min. Vacuoles changed in size, position, and number over time. (b) Time-lapse images of vacuoles taken at intervals of 10 s. Moving punctate structures (large arrowheads), putative late endosomes/prevacuolar compartments (small arrowheads), and tubular vacuoles (arrows) that replaced the punctate structures are shown. (c) Time-lapse images of a tubular-vesicular cluster taken at intervals of 10 s. Moving punctate structures (arrowheads) and a tubular vacuole (arrows) that replaced the punctate structures were also observed here. (d) Distance sections (1 μm) of vacuoles in the basal region. The entire lumen was occupied with EGFP fluorescence. Tubular vacuoles (arrows) interconnected these vacuoles. Bars, 5 μm.
We next performed FM4-64 and CMAC staining of UEV1, TPVEV1, and TPVEV4 to compare the staining patterns of the dyes with EGFP fluorescence. In S. cerevisiae, FM4-64 stains endosomal compartments such as endosomes and vacuoles (43). In filamentous fungi, some arguments remain on its means of internalization and possible cytotoxic effects (6, 40). FM4-64 is believed to be internalized by endocytosis and accumulates in endosomal organelles (10). CMAC permeates into the cytoplasm due to its hydrophobic character and is converted to a cell-impermeant conjugate with glutathione by glutathione S-transferase. In fungi, CMAC accumulates in vacuoles, presumably via the action of glutathione pumps on the vacuolar membrane.
As some reports pointed out possible toxic effects of FM4-64 and subsequent artifactual results (6, 40), we first investigated the effects of the dye on A. oryzae cultures. The growth rates of hyphal tips in CD medium in the presence of 8 μM FM4-64 (1.01 ± 0.16 μm min−1, n = 8) were similar to those in the absence of FM4-64 (0.98 ± 0.19 μm min−1, n = 8). Vacuolar morphology judged by EGFP-AoVam3p was not affected by the presence of FM4-64 (Fig. 3). We also carried out dual staining of cover glass cultures with FM4-64 and CMAC. In the mycorrhizal fungus P. tinctorius, FM4-64 showed good labeling of vacuolar membranes in only dead or damaged hyphae and was scarcely observed on the membrane of intact vacuoles that accumulated CMAC (6). However, this was not the case in A. oryzae, and the membranes of most CMAC-positive vacuoles were stained by FM4-64 (Fig. 3g). From these results, we concluded that FM4-64 did not affect mycelial physiology in A. oryzae under our experimental conditions.
FM4-64 and EGFP-AoVam3p colocalized on the membrane of large developed vacuoles as well as on small punctate structures throughout hyphae (Fig. 3a to d). In extreme apices, where FM4-64 showed a diffuse and uneven staining pattern, however, EGFP fluorescence was normally absent (Fig. 3b). The diffuse and uneven fluorescence of FM4-64 seems to correspond to putative endosomes and a cloud of stained endocytic vesicles that are most rapidly stained by FM4-64 (10). In conclusion, EGFP-AoVam3p localizes in endocytic compartments, vacuoles, and possibly endosomes, but is likely absent in the early endocytic compartments.
The staining pattern of CMAC also showed good coincidence with EGFP-AoVam3p (Fig. 3e and f). The membrane of vacuoles that were stained by CMAC was always visualized by EGFP-AoVam3p (Fig. 3e and f). However, there were some small punctate structures visualized by EGFP-AoVam3p, often positioned adjacent to large vacuoles, that showed no or only faint staining by CMAC (Fig. 3f and g). As FM4-64 also stained them (Fig. 3g), these punctate structures are endocytic compartments. These observations led us to speculate that these structures are late endosomes/prevacuolar compartments, since they have common properties with the class E compartments, enlarged late endosomes/prevacuolar compartments in yeast class E vps mutants (27), in that they can be visualized by a vacuolar membrane protein and endocytic tracers and that they reside next to vacuoles. Overall, EGFP-AoVam3p localizes on the membrane of large and small vacuoles stained by both FM4-64 and CMAC. In addition, the fusion protein also localized on the putative late endosomes/prevacuolar compartments that were stained by FM4-64 but not by CMAC.
Confocal microscopy of EGFP-AoVam3p-expressing strains.
To further study the vacuolar morphology in A. oryzae, we performed confocal microscopy on EGFP-AoVam3p-expressing strains grown in glass base dishes. The use of glass base dishes allowed a longer incubation period than the use of cover glass cultures, and cultures grown for up to 72 h were observed. Time-lapse imaging of EGFP-AoVam3p revealed the dynamics of pleomorphic vacuolar networks in A. oryzae (Fig. 4a to c; see image series in the supplemental material). Even the large developed vacuoles that seemed to be rather stable over a short time period changed in number, size, and position in minutes, most probably reflecting fusion and fission of vacuoles (Fig. 4a; see series A in the supplemental material). In basal hyphae, where many vacuoles formed clusters, small vesicle-like structures occasionally moved between large vacuoles (Fig. 4b). A tubular vacuole was then formed from a small punctate structure (Fig. 4b) and replaced the vesicle-like structures to interconnect the spherical vacuoles directly (Fig. 4b; see series B in the supplemental material). In the subapical region, where clusters of small punctate structures and tubular vacuoles formed a continuum, reversible transformations between the punctate structures and tubular vacuoles were often observed. (Fig. 4c; see series C in the supplemental material).
Observation with confocal microscopy highlighted the localization of EGFP-AoVam3p on the membrane (Fig. 4a and b). In old hyphae that were often observed in 2-day cultures in glass base dishes, however, membrane localization of the fusion protein was not clear (Fig. 4d). In all sections of 1-μm distance taken with a confocal microscope, EGFP fluorescence was observed throughout the vacuolar lumen and was not restricted to the membrane. Such topological changes in EGFP fluorescence localization should require budding of the vacuolar membrane into the lumen. Since vacuoles often occupied the greater part of cells in these hyphae (Fig. 4d), internalization of EGFP-AoVam3p into the vacuolar lumen is likely achieved via an autophagic process (19). Interestingly, these vacuoles were often interconnected by tubular vacuoles (Fig. 4d).
Vacuoles in various developmental stages.
We next observed vacuoles in several developmental stages, conidial germination, and aerial hyphae. In conidia just after inoculation (Fig. 5, 0 h), small punctate vacuoles (<1 μm) and clouds of fluorescence were observed with EGFP-AoVam3p. At 2 h after inoculation, larger vacuoles (>1 μm) eventually emerged (Fig. 5), concomitant with conidial swelling. Vacuoles enlarged further, and at 3 to 4 h after inoculation, large spherical vacuoles (2 to 5 μm) appeared in swollen or germinating conidia (Fig. 5), accompanied by a few small punctate structures. In germ tubes, large vacuoles were normally absent and small punctate structures were often observed.
FIG. 5.
Vacuolar morphology in germinating conidia. DIC images (left) and EGFP fluorescence (right) of conidia from TPVEV1 at the time indicated after inoculation into DPY medium. Large vacuoles (>1 μm) that eventually developed in swelling conidia approximately 2 h after inoculation are indicated by arrows. Fluorescence at the cell periphery was also detected in a control strain that did not express EGFP and was therefore judged as autofluorescence. Bars, 5 μm.
We next observed hyphae that were not in contact with nutrients. Conidia were inoculated on the side surface of approximately 1-mm-thick CD agar, and hyphae were allowed to grow in lateral directions by preventing upward-directed growth with a cover glass. Under these conditions, three kinds of hyphae that grew laterally were observed, hyphae that grew in/on the agar medium and two kinds of hyphae that were not in contact with nutrients, hyphae that grew in the air (aerial hyphae) and hyphae that grew on the cover glass surface (glass surface hyphae). These hyphae almost always had yellowish fluorescence at the cell wall or plasma membrane and at hyphal tips, even in the wild-type strain that does not express EGFP (Fig. 6j and k, respectively). Based on this observation, fluorescence at the cell periphery in Fig. 6 was considered to be autofluorescence. In the wild-type strain, green organelle-like fluorescence inside cells, such as that shown in Fig. 6a to i, was never observed (Fig. 6j and k). In glass surface hyphae that were judged by their thickness of <4 μm, large vacuoles (>2 μm) were rather rare and were restricted to the region near the agar medium, where liquid enclosed the hyphae. Instead, small vacuoles (500 nm to 1 μm in diameter) were predominant (Fig. 6a to c). In addition, tubular vacuoles interconnecting the small vacuoles were more often observed in the glass surface hyphae (Fig. 6b) than in hyphae that were in contact with nutrients. Such tubular-vesicular clusters were sometimes so dense that they showed a rather reticulum-like appearance (Fig. 6c).
FIG. 6.
Vacuolar morphologies in hyphae that are not in contact with nutrients. Conidia of TPVEV4 were inoculated on the side of approximately 1-mm-thick CD containing 2% agar, and hyphae were allowed to grow in lateral directions for 48 h (a to e, g, h) or 72 h (f, i) by preventing the upward-directed growth with a cover glass. Pictures of the glass surface hyphae and the aerial hyphae were prepared under the respective identical conditions of exposure time and contrast enhancement. In all figures, fluorescence at the cell wall/membrane is judged as autofluorescence (see text). (a to c) Glass surface hyphae; (d to f) aerial hyphae; (g to i) hyphae in/on the agar medium. (a) Small vacuoles that are predominant in a glass surface hypha. (b) Clusters of small vacuoles interconnected with tubular vacuoles (arrows). (c) Tubular-vesicular vacuoles with a reticulum-like appearance. (d) Punctate vacuoles in an aerial hypha. (e) Tubular-reticular networks in an aerial hypha. (f) Large vacuoles in an aerial hypha that were observed in 3-day cultures. Some of these vacuoles moved toward the hyphal tips (large arrowheads), while others remained immobile (small arrowheads). (g) Large vacuoles in the agar medium. (h) A tubular vacuole in agar medium. (i) Vacuoles that are interconnected with tubular vacuoles in the basal region. The lumen of these vacuoles was entirely occupied with EGFP fluorescence. (j) Autofluorescence in a glass surface hypha of RIB40 strain that does not express EGFP-AoVam3p. (k) Autofluorescence in an aerial hypha of RIB40. As the pictures were taken with a monochromic camera and subsequently pseudocolored, yellowish autofluorescence appears green on this figure. Bars, 5 μm.
In aerial hyphae that were distinguished from glass surface hyphae by their thickness of >5 μm, vacuoles were present as small punctate structures (Fig. 6d). Tubular vacuoles that extended in the longitudinal direction and had a reticular appearance were also observed in aerial hyphae (Fig. 6e) but at a lower frequency than glass surface hyphae. There were also aerial hyphae that had entirely strong yellowish autofluorescence and no greenish EGFP fluorescence, suggesting that some aerial hyphae had begun to die in the 2-day cultures. Interestingly, large vacuoles were present in aerial hyphae in 3- to 4-day cultures (Fig. 6f). Some of these vacuoles flew (approximately 5 μm s−1) toward the hyphal tips (Fig. 6f), but others positioned near the plasma membrane remained immobile (Fig. 6f, compare with the differential interference contrast [DIC] image that was taken several seconds later). As these hyphae were often enclosed in liquid droplets, the physical condition of such hyphae may differ from that of authentic aerial hyphae. In hyphae in/on the agar medium, in contrast, the vacuolar morphology was identical to that observed in cover glass and glass base dish cultures. Large developed vacuoles (Fig. 6g) and tubular vacuoles (Fig. 6 h), vacuoles whose lumen was occupied with EGFP fluorescence and were interconnected with tubular vacuoles (Fig. 6i), were observed. In conclusion, vacuoles showed characteristic morphology in hyphae that were not in contact with nutrients, with vacuoles forming highly reticular networks composed of tubular vacuoles, and with small punctate vacuoles.
DISCUSSION
Thus far, studies on vacuoles in filamentous fungi have made great progress in highlighting vacuolar morphology using fluorescence and electron microscopy. These studies identified the presence of highly pleomorphic dynamic vacuoles, such as tubular vacuoles, in filamentous fungi (reviewed in reference 2). In this report, we established three strains expressing the fusion protein of EGFP with AoVam3p, a putative vacuolar t-SNARE in Aspergillus oryzae. In contrast to conventional methods, in which the vacuolar lumen is stained with either CFDA derivatives (2, 6, 7, 34, 42) or CPY-EGFP (23, 24), EGFP-AoVam3p visualizes vacuolar membranes and therefore allowed us to observe vacuolar membrane dynamics at high resolution easily and reproducibly. Furthermore, several aspects of fungal vacuoles that had not been paid much attention previously, such as vacuoles in aerial and glass surface hyphae, as well as the internalization of EGFP fluorescence in the vacuolar lumen in old hyphae, were demonstrated for the first time using our observation system.
Does AoVam3p localize and act as a t-SNARE on both vacuoles and late endosomes/prevacuolar compartments?
As Aovam3 is the only obvious homologue of yeast VAM3 and PEP12 in A. oryzae, it is possible that AoVam3p acts as the t-SNARE on both the vacuoles and the late endosomes/prevacuolar compartments. In our yeast complementation analysis, Aovam3 was capable of compensating the functions of both VAM3 and PEP12, suggesting that AoVam3p is most likely the vacuolar and/or endosomal t-SNARE in A. oryzae. We cannot yet conclude, however, that AoVam3p plays the roles of both Vam3p and Pep12p in A. oryzae, since in S. cerevisiae, overexpression of Vam3p complements a pep12 null mutant and, at least in part, vice versa (11). Moreover, Vam3p can complement a pep12 null mutant even not overexpressed when its dileucine sorting signal is mutated (9). The dileucine sorting signal of Vam3p is required for its delivery from the Golgi apparatus to the vacuole via the alkaline phosphatase pathway that bypasses the late endosome/prevacuolar compartment. Mutations in the dileucine signal lead to missorting of Vam3p to the CPY pathway, the delivery route from the Golgi apparatus to the vacuole via the late endosome/prevacuolar compartment, and Vam3p that partially localizes on the late endosome/prevacuolar compartment compensates for the function of Pep12p (9). These results imply that if a Vam3p/Pep12p homologue can localize on both the vacuole and the late endosome/prevacuolar compartment, it can compensate for the roles of both Vam3p and Pep12p. Therefore, it was of importance to determine the localization of AoVam3p in A. oryzae. As revealed by FM4-64 and CMAC staining, EGFP-AoVam3p localized on CMAC-negative, nonvacuolar structures in addition to vacuoles. These CMAC-negative structures resembled class E vps compartments, enlarged late endosomes/prevacuolar compartments in yeast class E vps mutants (27), in that they can be visualized with vacuolar membrane proteins, they are adjacent to vacuoles, and they are endocytic compartments. Taken together, it is apparent that EGFP-AoVam3p localizes on late endosomes/prevacuolar compartments in addition to vacuoles. Exploration for possible candidates of late endosome/prevacuolar compartment-resident proteins will be necessary for the further identification of late endosomes/prevacuolar compartments in filamentous fungi.
Enlargement of vacuoles in germinating conidia.
Besides their roles in hyphal growth, fungal vacuoles may also be important in several developmental stages. Consistent with this, our observation demonstrated that swelling of conidia was accompanied by vacuolar enlargement. Moreover, in germinating conidia of Colletotrichum graminicola, lipid droplets are taken up and degraded by vacuoles (31), accompanied by vacuolar expansion (32). Further investigations of possible roles of vacuoles (e.g., storage degradation and cell expansion) may help us to understand the mechanism of conidial germination in filamentous fungi.
What is the role of tubular vacuoles in A. oryzae?
Motile tubular vacuoles were first discovered in the mycorrhizal fungus P. tinctorius by electron microscopy and CFDA staining (34), and it has been proposed that they are endosomal compartments involved in intra- and intercellular transport (for a detailed discussion, see reference 2). We observed that a tubular vacuole was formed from small punctate structures and replaced vesicle-like structures. This result is consistent with the previous report on CFDA staining of vacuoles (34) and supports the notion that tubular vacuoles transport materials such as nutrients. In mycorrhizal fungi, there is an urgent need for bidirectional transport of nutrients; phosphorus and nitrogen compounds must be transported from hyphal tips to the plant interface, while carbon compounds that are supplied by plants should move to hyphal tips, where metabolic activity is the highest (2). Consistent with this hypothesis, tubular vacuoles of P. tinctorius contain phosphorus compounds (6). Given that tubular vacuoles are transport compartments, then why do they also exist in other filamentous fungi that do not have such a dependency on nutrients from other organisms? We found that tubular vacuoles interconnected large vacuoles in old hyphae that were often evident in 2- to 3-day cultures. As these vacuoles occupied the greater part of compartments and included EGFP fluorescence in the entire lumen, autophagic degradation of cytosolic materials and subsequent internalization of the vacuolar membrane into the lumen had possibly occurred in such compartments. An attractive explanation for a role of these tubular vacuoles is that they transport the degraded materials in old hyphae to more fresh and active regions in the mycelium. We also discovered that tubular vacuoles and clusters of small punctate vacuoles were prominent in hyphae that were not in contact with nutrients, especially in glass surface hyphae. Since these hyphae should have a more urgent requirement for nutrient transport, it is reasonable to speculate that tubular vacuoles develop more extensively in these hyphae and mediate transport of nutrients from hyphae that are in contact with media.
To date, nothing is known about the molecular machinery involved in the formation of tubular vacuoles, except the possible involvement of certain dynamin-like GTPases for tubule formation (14). AoVam3p is the first protein to be discovered that locates on the membrane of tubular vacuoles. As AoVam3p is a putative vacuolar t-SNARE that may mediate the heterotypic and homotypic fusion of vacuoles, it is quite possible that AoVam3p has important roles in regulating fungal pleomorphic vacuolar networks: fusion of small moving structures and tubular vacuoles with spherical vacuoles may be mediated by AoVam3p. The functional analysis of AoVam3p and its possible interacting partners will lead to better understanding of the precise mechanisms and roles of vacuolar networks in filamentous fungi.
Aerial hyphae of filamentous fungi.
Although filamentous fungi can grow in liquid and on solid substrates, they are perhaps best known for their fuzzy aerial appearance. However, studies on these aerial structures have so far mainly focused on conidiation (see reference 1 for a review), and the specific phenomenon of aerial hyphae is only known for hydrophobins (38). To the best of our knowledge, our research is the first report on the observation of an organelle in the aerial hyphae of filamentous fungi. As our data demonstrated, the vacuolar morphology in aerial hyphae visualized by EGFP-AoVam3p was essentially different in appearance from that of hyphae in the medium. We assume that vacuolar morphology is characteristically regulated in aerial and glass surface hyphae to adapt to the unusual environment in terms of nutrients and osmotic pressure. As mentioned above, vacuoles showed a highly reticular-tubular appearance in aerial hyphae. As nutrients and/or materials for cell elongation should be delivered to aerial hyphae from basal compartments, vacuoles possibly mediate intra- and intercellular transport in these hyphae.
The characteristic vacuolar morphology is probably just an aspect of aerial hyphae, and other physical activities such as metabolism and morphogenesis are also differentially regulated in these hyphae. To date, essentially nothing is known about the molecular mechanisms regulating the metabolism and morphology of aerial hyphae, presumably because, at least in part, they are not well developed or clearly recognizable in A. nidulans, a model filamentous fungus. In A. oryzae, aerial hyphae grow approximately 5 mm in 2 days under our conditions and are therefore easily observed using microscopy. Further investigation of aerial hyphae in A. oryzae will compensate for the lack of this feature in A. nidulans and bring new insights to fungal physiology.
Supplementary Material
Acknowledgments
This study was supported by a grant-in-aid for scientific research (B) (no. 15380058) to K.K. from the Ministry of Education, Culture, Sports, Science, and Technology, Japan, and a program for the promotion of basic research activities for innovative biosciences of the Bio-Oriented Technology Research Advancement Institution.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Adams, T. H., J. K. Wieser, and J. H. Yu. 1998. Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 62:35-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashford, A. E. 1998. Dynamic pleiomorphic vacuole systems: are they endosomes and transport compartments in fungal hyphae? Adv. Bot. Res. 28:119-159. [Google Scholar]
- 3.Becherer, K. A., S. E. Rieder, S. D. Emr, and E. W. Jones. 1996. Novel syntaxin homologue, Pep12p, required for the sorting of lumenal hydrolases to the lysosome-like vacuole in yeast. Mol. Biol. Cell 7:579-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black, M. W., and H. R. Pelham. 2000. A selective transport route from Golgi to late endosomes that requires the yeast GGA proteins. J. Cell Biol. 151:587-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 6.Cole, L., D. A. Orlovich, and A. E. Ashford. 1998. Structure, function, and motility of vacuoles in filamentous fungi. Fungal Genet. Biol. 24:86-100. [DOI] [PubMed] [Google Scholar]
- 7.Cole, L., G. J. Hyde, and A. E. Ashford. 1997. Uptake and compartmentalisation of fluorescent probes by Pisolithus tinctorius hyphae: evidence for an anion transport mechanism at the tonoplast but not for fluid-phase endocytosis. Protoplasma 199:18-29. [Google Scholar]
- 8.Cowles, C. R., W. B. Snyder, C. G. Burd, and S. D. Emr. 1997. Novel Golgi to vacuole delivery pathway in yeast: identification of a sorting determinant and required transport component. EMBO J. 16:2769-2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darsow, T., C. G. Burd, and S. D. Emr. 1998. Acidic di-leucine motif essential for AP-3-dependent sorting and restriction of the functional specificity of the Vam3p vacuolar t-SNARE. J. Cell Biol. 142:913-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischer-Parton, S., R. M. Parton, P. C. Hickey, J. Dijksterhuis, H. A. Atkinson, and N. D. Read. 2000. Confocal microscopy of FM4-64 as a tool for analysing endocytosis and vesicle trafficking in living fungal hyphae. J. Microsc. 198:246-259. [DOI] [PubMed] [Google Scholar]
- 11.Götte, M., and D. Gallwitz. 1997. High expression of the yeast syntaxin-related Vam3 protein suppresses the protein transport defects of a pep12 null mutant. FEBS Lett. 411:48-52. [DOI] [PubMed] [Google Scholar]
- 12.Gupta, G. D., and I. B. Heath. 2002. Predicting the distribution, conservation, and functions of SNAREs and related proteins in fungi. Fungal Genet. Biol. 36:1-21. [DOI] [PubMed] [Google Scholar]
- 13.Hirozumi, K., H. Nakajima, M. Machida, M. Yamaguchi, K. Takeo, and K. Kitamoto. 1999. Cloning and characterization of a gene (arpA) from Aspergillus oryzae encoding an actin-related protein required for normal nuclear distribution and morphology of conidiophores. Mol. Gen. Genet. 262:758-767. [DOI] [PubMed] [Google Scholar]
- 14.Hyde, G. J., D. Davies, L. Cole, and A. E. Ashford. 2002. Regulators of GTP-binding proteins cause morphological changes in the vacuoles system of the filamentous fungus, Pisolithus tinctorius. Cell Motil. Cytoskeleton 51:133-146. [DOI] [PubMed] [Google Scholar]
- 15.Ishi, K., J. Maruyama, H. Nakajima, and K. Kitamoto. 2005. Multinucleate conidia are not formed in conidia but through migration of plural nuclei from phialide in Aspergillus oryzae. Biosci. Biotechnol. Biochem. 69:747-754. [DOI] [PubMed] [Google Scholar]
- 16.Jones, E. W. 1977. Proteinase mutants of Saccharomyces cerevisiae. Genetics 85:23-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kitamoto, K. 2002. Molecular biology of the Koji molds. Adv. Appl. Microbiol. 51:129-153. [DOI] [PubMed] [Google Scholar]
- 18.Klionsky, D. J., P. K. Herman, and S. D. Emr. 1990. The fungal vacuole: composition, function, and biogenesis. Microbiol. Rev. 54:266-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klionsky, D. J., and Y. Ohsumi. 1999. Vacuolar import of proteins and organelles from the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:1-32. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi, T., K. Abe, K. Asai, K. Gomi, P. R. Juvvadi, M. Kato, K. Kitamoto, M. Takeuchi, and M. Machida. Genomics of Aspergillus oryzae. Appl. Mycol. Biotechnol., in press. [DOI] [PubMed]
- 21.Kuroki, Y., P. R. Juvvadi, M. Arioka, H. Nakajima, and K. Kitamoto. 2002. Cloning and characterization of vmaA, the gene encoding a 69-kDa catalytic subunit of the vacuolar H+-ATPase during alkaline pH mediated growth of Aspergillus oryzae. FEMS Microbiol. Lett. 209:277-282. [DOI] [PubMed] [Google Scholar]
- 22.Kutsuna, N., F. Kumagai, M. H. Sato, and S. Hasezawa. 2003. Three-dimensional reconstruction of tubular structure of vacuolar membrane throughout mitosis in living tobacco cells. Plant Cell Physiol. 44:1045-1054. [DOI] [PubMed] [Google Scholar]
- 23.Ohneda, M., M. Arioka, H. Nakajima, and K. Kitamoto. 2002. Visualization of vacuoles in Aspergillus oryzae by expression of CPY-EGFP. Fungal Genet. Biol. 37:29-38. [DOI] [PubMed] [Google Scholar]
- 24.Ohneda, M., M. Arioka, and K. Kitamoto. 2005. Isolation and characterization of Aspergillus oryzae vacuolar protein sorting mutants. Appl. Environ. Microbiol. 71:4856-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohsumi, K., M. Arioka, H. Nakajima, and K. Kitamoto. 2002. Cloning and characterization of a gene (avaA) from Aspergillus nidulans encoding a small GTPase involved in vacuolar biogenesis. Gene 291:77-84. [DOI] [PubMed] [Google Scholar]
- 26.Oka, M., J. Maruyama, M. Arioka, H. Nakajima, and K. Kitamoto. 2004. Molecular cloning and functional characterization of avaB, a gene encoding Vam6p/Vps39p-like protein in Aspergillus nidulans. FEMS Microbiol. Lett. 232:113-121. [DOI] [PubMed] [Google Scholar]
- 27.Raymond, C. K., I. Howald-Stevenson, C. A. Vater, and T. H. Stevens. 1992. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell 3:1389-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts, C. J., C. K. Raymond, C. T. Yamashiro, and T. H. Stevens. 1991. Methods for studying the yeast vacuole. Methods Enzymol. 194:644-661. [DOI] [PubMed] [Google Scholar]
- 29.Rothman, J. E. 1994. Mechanisms of intracellular protein transport. Nature 372:55-63. [DOI] [PubMed] [Google Scholar]
- 30.Sato, M. H., N. Nakamura, Y. Ohsumi, H. Kouchi, M. Kondo, I. Hara-Nishimura, M. Nishimura, and Y. Wada. 1997. The AtVAM3 encodes a syntaxin-related molecule implicated in the vacuolar assembly in Arabidopsis thaliana. J. Biol. Chem. 272:24530-24535. [DOI] [PubMed] [Google Scholar]
- 31.Schadeck, R. J., B. Leite, and D. F. Buchi. 1998. Lipid mobilization and acid phosphatase activity in lytic compartments during conidium dormancy and appressorium formation of Colletotrichum graminicola. Cell Struct. Funct. 23:333-340. [DOI] [PubMed] [Google Scholar]
- 32.Schadeck, R. J., M. A. Randi, D. F. Buchi, and B. Leite. 2003. Vacuolar system of ungerminated Colletotrichum graminicola conidia: convergence of autophagic and endocytic pathways. FEMS Microbiol. Lett. 218:277-283. [DOI] [PubMed] [Google Scholar]
- 33.Reference deleted.
- 34.Shepherd, V. A., D. A. Orlovich, and A. E. Ashford. 1993. A dynamic continuum of pleiomorphic tubules and vacuoles in growing hyphae of a fungus. J. Cell Sci. 104:495-507. [DOI] [PubMed] [Google Scholar]
- 35.Shoji, J. Y., J. Maruyama, M. Arioka, and K. Kitamoto. 2005. Development of Aspergillus oryzae thiA promoter as a tool for molecular biological studies. FEMS Microbiol. Lett. 244:41-46. [DOI] [PubMed] [Google Scholar]
- 36.Srivastava, A., and E. W. Jones. 1998. Pth1/Vam3p is the syntaxin homolog at the vacuolar membrane of Saccharomyces cerevisiae required for the delivery of vacuolar hydrolases. Genetics 148:85-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tada, S., K. Gomi, K. Kitamoto, K. Takahashi, G. Tamura, and S. Hara. 1991. Construction of a fusion gene comprising the Taka-amylase A promoter and the Escherichia coli beta-glucuronidase gene and analysis of its expression in Aspergillus oryzae. Mol. Gen. Genet. 229:301-306. [DOI] [PubMed] [Google Scholar]
- 38.Talbot, N. J. 1997. Growing into the air. Curr. Biol. 7:78-81. [DOI] [PubMed] [Google Scholar]
- 39.Tarutani, Y., K. Ohsumi, M. Arioka, H. Nakajima, and K. Kitamoto. 2001. Cloning and characterization of Aspergillus nidulans vpsA gene which is involved in vacuolar biogenesis. Gene 268:23-30. [DOI] [PubMed] [Google Scholar]
- 40.Torralba, S., and I. B. Heath. 2002. Analysis of three separate probes suggests the absence of endocytosis in Neurospora crassa hyphae. Fungal Genet. Biol. 37:221-232. [DOI] [PubMed] [Google Scholar]
- 41.Uemura, T., S. H. Yoshimura, K. Takeyasu, and M. H. Sato. 2002. Vacuolar membrane dynamics revealed by GFP-AtVam3 fusion protein. Genes Cells 7:743-753. [DOI] [PubMed] [Google Scholar]
- 42.Uetake, Y., T. Kojima, T. Ezawa, and M. Saito. 2002. Extensive tubular vacuole system in an arbuscular mycorrhizal fungus, Gigaspora margarita. New Phytol. 154:761-768. [DOI] [PubMed] [Google Scholar]
- 43.Vida, T. A., and S. D. Emr. 1995. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128:779-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wada, Y., N. Nakamura, Y. Ohsumi, and A. Hirata. 1997. Vam3p, a new member of syntaxin related protein, is required for vacuolar assembly in the yeast Saccharomyces cerevisiae. J. Cell Sci. 110:1299-1306. [DOI] [PubMed] [Google Scholar]
- 45.Wang, L., A. J. Merz, K. M. Collins, and W. Wickner. 2003. Hierarchy of protein assembly at the vertex ring domain for yeast vacuole docking and fusion. J. Cell Biol. 160:365-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, L., E. S. Seeley, W. Wickner, and A. J. Merz. 2002. Vacuole fusion at a ring of vertex docking sites leaves membrane fragments within the organelle. Cell 108:357-369. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.