Abstract
The movement of ammonium across biological membranes is mediated in both prokaryotes and eukaryotes by ammonium transport proteins (AMT/MEP) that constitute a family of related sequences. We have previously identified two ammonium permeases in Aspergillus nidulans, encoded by the meaA and mepA genes. Here we show that meaA is expressed in the presence of ammonium, consistent with the function of MeaA as the main ammonium transporter required for optimal growth on ammonium as a nitrogen source. In contrast, mepA, which encodes a high-affinity ammonium permease, is expressed only under nitrogen-limiting or starvation conditions. We have identified two additional AMT/MEP-like genes in A. nidulans, namely, mepB, which encodes a second high-affinity ammonium transporter expressed only in response to complete nitrogen starvation, and mepC, which is expressed at low levels under all nitrogen conditions. The MepC gene product is more divergent than the other A. nidulans AMT/MEP proteins and is not thought to significantly contribute to ammonium uptake under normal conditions. Remarkably, the expression of each AMT/MEP gene under all nitrogen conditions is regulated by the global nitrogen regulatory GATA factor AreA. Therefore, AreA is also active under nitrogen-sufficient conditions, along with its established role as a transcriptional activator in response to nitrogen limitation.
Ammonium transport proteins (AMT/MEP) have been identified in bacteria, fungi, plants, and animals and constitute a conserved family of polytopic membrane proteins (54). AMT/MEP proteins are predicted to contain 11 transmembrane (TM) helices with an Nout-Cin topology (26, 48). Bioinformatic topology predictions indicate that bacterial AMT/MEP proteins generally contain an additional N-terminal TM domain but that this region acts as a signal peptide which is removed from the mature protein (9). Aside from the small cytoplasmic loop between TM domains 3 and 4 that displays sequence similarity to a major facilitator superfamily motif (34), the AMT/MEP sequences represent a unique group of transport membrane proteins. Although over 300 members of the AMT/MEP/Rh family have currently been assigned based on amino acid sequence similarity (Pfam accession number PF00909), an ammonium transport function has not been confirmed for the vast majority of these sequences. Functional permeases have been described for bacteria, fungi, plants (reviewed in reference 54), and humans, where the rhesus blood group polypeptides, which display significant sequence identity to AMT/MEP proteins, have been shown to function as ammonium transporters in other systems (27, 53, 55, 59). The structure of Escherichia coli AmtB has been determined up to 1.35-Å resolution, and structural analysis revealed that the protein functions as a trimer that recruits ammonium which is then channeled as ammonia (18, 58). Genetic, molecular, and/or physiological evidence suggests that AMT/MEP proteins can function as homo- and/or heterocomplexes (6, 25, 29, 34).
The presence of multiple ammonium permeases with different kinetic properties within an organism is common (54). The yeast Saccharomyces cerevisiae has three AMT/MEP genes, MEP1, MEP2, and MEP3. Mep1 constitutes the majority of ammonium uptake for S. cerevisiae, with an affinity in the 5 to 10 μM range, Mep2 has the highest affinity for ammonium (1 to 2 μM), and Mep3 is a low-affinity (1 to 2 mM), high-capacity ammonium permease (28, 30). Certain fungal ammonium permeases, including S. cerevisiae Mep2, Candida albicans Mep2p, and Ustilago maydis UMP2, also function as ammonium sensors, generating a signal to regulate pseudohyphal or filamentous growth in response to nitrogen starvation (5, 23, 44). A MEP1 MEP2 MEP3 triple deletion mutant was unable to growon media containing <5 mM ammonium as the sole nitrogen source, whereas single deletion strains displayed normal growth (28). Each of the MEP genes displays an expression profile typical of genes subjected to nitrogen catabolite repression. The expression of these genes is low on good nitrogen sources, such as asparagine, glutamine, and ammonium, that support optimal yeast growth, and levels are elevated on low-ammonium or suboptimal nitrogen sources such as proline (28). The GATA factors Gln3p and Nil1p, which mediate nitrogen catabolite repression in S. cerevisiae, control the expression of all three MEP genes (28).
Two Aspergillus nidulans AMT/MEP genes, meaA and mepA, have been characterized (33). A meaAΔ mutant displays reduced growth on ammonium as the sole nitrogen source, whereas a mepAΔ mutant exhibits normal growth under these conditions. A mepAΔ meaAΔ mutant is unable to grow with low ammonium concentrations at pH 4.5, and the residual growth at pH 6.5 has been attributed to the passive diffusion of ammonium (24, 33, 45). The MepA permease displays a higher affinity for methylammonium than does MeaA (Km, 44.3 μM and 3.04 mM, respectively). MeaA serves as the main ammonium transporter, whereas the higher-affinity MepA permease is likely to have a scavenging role in ammonium uptake (33).
Here we present a further characterization of the A. nidulans ammonium transport system and its regulation. We have identified two additional AMT/MEP-like sequences in the A. nidulans genome and present a functional analysis of these genes, designated mepB and mepC. We show that even though the four A. nidulans AMT/MEP genes are differentially regulated in response to the nitrogen status of the cell, their full expression requires the function of the GATA transcription factor AreA (3, 20, 31).
MATERIALS AND METHODS
A. nidulans strains, media, and transformation.
The A. nidulans strains used for this study are shown in Table 1. A. nidulans media (ANM) and growth conditions were as described by Cove (10), except that the pH of the media was adjusted to either pH 6.5 or pH 4.5. Genetic analysis was carried out using previously described techniques (8). Strains of A. nidulans were transformed according to the method of Andrianopoulos and Hynes (1).
TABLE 1.
A. nidulans strains used for this study
| Strain | Genotype |
|---|---|
| MH1 | biA1 |
| MH5699 | yA1 areA::riboB pyroA4 |
| MH8694 | biA1 tamAΔ riboB2 |
| MH9102 | biA1 nmrA::Bleor areAUTRΔ pyroA4 riboB2 |
| MH9233 | biA1 wA3 argB::trpCΔB pyroA4 riboB2 |
| MH9828 | biA1 wA3 argB::trpCΔB meaA::argB pyroA4 mepA::riboB riboB2 |
| MH9829 | biA1 meaA::argB pyroA4 mepA::riboB riboB2 |
| MH9830 | biA1 amdS::lacZ meaA::argB pyroA4 riboB2 |
| MH9861 | biA1 wA3 meaA::argB pyroA4 riboB2 |
| MH9924 | biA1 amdS::lacZ pyroA4 riboB2 |
| MH9961 | biA1 wA3 argB::trpCΔB meaA::argB pyroA4 riboB2 |
| MH9965 | biA1 meaA::argB pyroA4 riboB2 |
| MH10311 | yA1 areA::riboB meaA::argB pyroA4 |
| MH10313 | biA1 areA::riboB mepA::riboB |
| MH10314 | biA1 areA::riboB meaA::argB pyroA4 mepA::riboB |
| MH10315 | biA1 meaA::argB pyroA4 tamAΔ riboB2 |
| MH10316 | wA3 pyroA4 tamAΔ mepA::riboB riboB2 |
| MH10317 | biA1 meaA::argB pyroA4 tamAΔ mepA::riboB riboB2 |
| MH10319 | biA1 amdS::lacZ meaA::argB pyroA4 mepA::riboB riboB2 mepB::Bleor (pBJM5639) |
| MH10321 | biA1 meaA::argB pyroA4 mepA::riboB mepB::Bleor |
| MH10322 | biA1 amdS::lacZ pyroA4 mepA::riboB mepB::Bleor |
| MH10323 | biA1 amdS::lacZ meaA::argB pyroA4 riboB2 mepB::Bleor |
| MH10324 | biA1 pyroA4 mepB::Bleor |
| MH10325 | biA1 amdS::lacZ meaA::argB pyroA4 mepA::riboB mepB::Bleor |
| MH10326 | wA3 amdS::lacZ pyroA4 mepB::Bleor |
| MH10328 | biA1 amdS::lacZ meaA::argB pyroA4 mepA::riboB |
| MH11056 | nkuA:argB argB2 pabaB22 riboB2 |
| MH11136 | nkuA:argB argB2 pabaB22 riboB2 mepC:BAR |
Molecular techniques.
Standard methods for the manipulation of E. coli cells and DNA were done as described by Sambrook and Russell (39). The E. coli strain used for this study was NM522. Restriction enzymes (Promega) were used according to the manufacturer's recommendations. DNA fragments were recovered from agarose gels using a BresaClean DNA purification kit (Geneworks). DNA fragments were subcloned into the pBluescript SK(+) (Stratagene) plasmid vector. A. nidulans genomic DNA was isolated by the method of Lee and Taylor (22). DNA gels were transferred to Hybond N+ membranes (Amersham) using 0.4 M NaOH. [α-32P]dATP (Bresatec)-labeled DNA probes for hybridization were created using the random hexanucleotide priming method (39). Taq DNA polymerase (Promega) was used for PCR. Automated DNA sequencing was performed by the Australian Genome Research Facility (Brisbane, Queensland, Australia), using plasmid DNA prepared with a High Pure plasmid kit (Roche).
Cloning of mepB and mepC.
Based on the partial A. nidulans genome sequence available at the Monsanto Microbial Sequence Database (http://microbial.cereon.com/), mepB-specific primers were designed (Table 2). An 1,129-bp mepB product was amplified using primers mepB-F and mepB-R and 100 ng genomic DNA at an annealing temperature of 58°C with 1.5 mM MgCl2. Hybridization of the mepB PCR product to an A. nidulans bacterial artificial chromosome (BAC) library (kindly provided by Ralph Dean, Department of Plant Pathology and Physiology, Clemson University) identified seven positive clones (3O2, 11P5, 15F8, 19C1, 27E10, 31H17, and 7P14). A 5.3-kb BamHI-XbaI fragment from BAC 7P14 was subcloned into pBluescript SK(+) (Stratagene) to create pBJM5377 and was sequenced. Based on the A. nidulans genome sequence available at the time via the Aspergillus Sequencing Project, Whitehead Institute/MIT Center for Genome Research (http://www-genome.wi.mit.edu), the mepC-specific primers mepC-1 and mepC-2 were designed and used to amplify a 5-kb product containing the coding region of the gene. The PCR product was ligated into pGEM-T Easy (Promega), creating pMA5741.
TABLE 2.
Oligonucleotides used for this study
| Primer name | Primer sequence (5′-3′) |
|---|---|
| Btub2 | AGTTGTTACCAGCACCGGAC |
| Btub3 | GCTCCGGTGTTTACAATGG |
| meaA2 | GACGGTACCGAGCAGAATGAG |
| meaA-RT | GGAAACTCAAGGGTGGAAAAC |
| mepA-RT | GCAATGTAGTCAGCTGCGAAG |
| MEPg1 | ACTTTGGGTTCAAGGGTGTCC |
| mepC-F | GCGCATTGACGGTACCATCTC |
| mepC-R | ACTCATTGCTGTTGCAGTGGC |
| mepC-RT | GCTCCAGAGGAAGATAAATGG |
| mepB1 | TTCTGCAGCGTCCCAATGTAG |
| mepB-F | GTATTCACTCACCTACTCGCG |
| mepB-R | CAGTCCGACCATCTCAGCTTC |
| mepB-RT | CTCATCATGTCTAGTCCCGAA |
| mepC-1 | TACCCGTCACTATCCGGAAC |
| mepC-2 | CTTTGAGCGGTGGTGATTCT |
| mepCxlp-1 | ATGGCCAAGATGTCAATCCA |
| mepCxlp-2 | ATGGCGCATTTCGTCACAAG |
Creation of mepBΔ and mepCΔ mutants by homologous gene replacement.
The mepB deletion construct, pBJM5639, was made by inserting a 1.45-kb BamHI bleomycin resistance cassette (Bleor) from pAmPh520 (4) into the BglII sites of pBJM5377. pBJM5639 was linearized by digestion with KpnI and NotI and then transformed into the mepAΔ meaAΔ mutant MH10328. Transformants were selected for resistance to 1 mg/ml bleomycin, and 66 transformants were screened by Southern blot analysis. MH10319 was identified as a mepB deletion mutant that contained additional ectopic copies of the knockout construct (data not shown). To remove these extra copies containing the Bleor cassette, MH10319 was crossed with MH9828 (meaAΔ mepAΔ wA3), and the progeny were screened for decreased resistance to 1 and 5 μg/ml bleomycin compared to that of MH10319. Southern blot analysis of genomic DNA confirmed MH10321 as a mepB deletion mutant that contained no extra copies of the knockout construct (data not shown). Genetic crosses were performed with MH10321 to create the following strains (see Table 1 for exact genotypes): mepBΔ (MH10324 and MH10326), mepBΔ meaAΔ (MH10323), mepBΔ mepAΔ (MH10322), and mepBΔ meaAΔ mepAΔ (MH10325).
A mepC gene inactivation construct was made by inserting a 1.44-kb EcoRV-HindIII fragment of pMT1612 into the EcoICR1-HindIII sites of pMA5741 to create pMA6403. pMT1612 contains the glufosinate resistance (bar) gene from the plasmid pBP1T (47) flanked by the amdS I9I66 promoter and the Aspergillus niger glucoamylase terminator and was constructed by Mogens Hansen, Novo Nordisk A/S, Bagsvaerd, Denmark. A linear version of the gene inactivation construct was generated using the mepC-specific primers and transformed into MH11056. Transformants were selected for resistance to 25 μl/ml glufosinate. Two transformants were screened by Southern blot analysis, both of which had the correct restriction pattern for a gene inactivation. MH11136 was selected for further analysis.
Construction of xylp::mepC fusion.
Primers mepCxyl-1 and mepCxyl-2 were designed to introduce one-half of an NcoI site 5′ and 3′ of the coding region of mepC. The 1.72-kb product was ligated into the SmaI site of pBluescript SK(+), reconstructing complete NcoI sites at either end. Using an NcoI digest, a 1.72-kb band containing the mepC coding region was gel purified and ligated into the NcoI site of pMH6102, resulting in a translational fusion of the xylP promoter (57) and the mepC coding region, creating pMA6382. This plasmid was cotransformed with pI4 carrying the pyroA gene (37) into the triple deletion strain MH10325, and PyroA+ transformants were selected for growth on medium lacking pyridoxine.
Northern blot analysis.
Total A. nidulans RNA was obtained by using an RNA Red Fast Prep kit (BIO 101). Eight-microgram RNA samples were mixed with 2 volumes of denaturing solution (50% formamide, 30% formaldehyde, 2× MOPS [morpholinepropanesulfonic acid], 1% ethidium bromide), incubated at 68°C for 15 min, and then placed on ice for 5 min. Samples were run in a 20% formaldehyde-1.2% agarose gel at 80 V in 1× MOPS buffer and then transferred to a Hybond N+ membrane (Amersham) using 0.04 M NaOH. The 2.2-kb meaA cDNA insert from pBJM5280 (33) and the 2-kb mepA cDNA from pBJM5281 (33) were amplified with standard M13 forward and reverse primers at an annealing temperature of 55°C with 1.5 mM MgCl2. mepB-specific (1.1 kb) and mepC-specific (0.6 kb) PCR products were used as probes for Northern analysis.
RT-PCR analysis.
The primers used for reverse transcription-PCRs (RT-PCRs) are shown in Table 2. The constitutively expressed β-tubulin gene benA (32) was used as an internal loading control within multiplex RT-PCRs. Reactions were performed by using a Superscript One-Step RT-PCR kit (Invitrogen) with AMT/MEP gene-specific and β-tubulin gene-specific (Btub2 and Btub3; predicted size of product, 209 bp) primers. The AMT/MEP gene-specific primers and the predicted sizes of their products were as follows: for meaA, meaA-RT and meaA2 (646 bp); for mepA, mepA-RT and MEPg1 (784 bp); for mepB, mepB-RT and mepB1 (308 bp); and for mepC, mepC-RT and mepC-F. The cDNA synthesis step was performed at 50°C, and the annealing temperature for all reactions was 58°C. One hundred nanograms of A. nidulans total RNA was used per reaction. Primers with the RT notation spanned an intron and therefore would only prime from RNA and cDNA targets. Control reactions using Taq DNA polymerase alone (no reverse transcriptase activity) and no-RNA control reactions were also performed for each experiment (data not shown). To confirm that amplification of both products was in the exponential phase for both primer sets (AMT/MEP gene specific and β-tubulin gene specific), cycle titration (15, 18, 21, 24, 27, 30, and 35 cycles) was performed for each combination (data not shown). From this analysis, 27 cycles was determined to be optimal for mepC with β-tubulin and 24 cycles was optimal for meaA, mepA, and mepB with β-tubulin.
[14C]methylammonium uptake assays.
[14C]methylammonium uptake assays were performed as described previously (33), except that cultures were grown in 250-ml Erlenmeyer flasks containing 100 ml ANM medium. The methylammonium concentration used for each assay is indicated in the text. To determine the apparent Km of MepB, the multiple-copy mepB cotransformant D (the transformant with the highest mepB copy number) was used in [14C]methylammonium uptake studies using nitrogen-starved mycelia.
Bioinformatic analysis.
Genomic data for Aspergillus fumigatus were provided by The Institute for Genomic Research (www.tigr.org/tbd/e2kl/aful/) and The Wellcome Trust Sanger Institute (www.sanger.ac.uk/Projects/A_fumigatus). Genomic data were provided by the Broad Institute for Aspergillus nidulans (http://www.broad.mit.edu/annotation/fungi/aspergillus/), Neurospora crassa (http://www.broad.mit.edu/annotation/fungi/neurospora_crassa_7/), Magnaporthe grisea (http://www.broad.mit.edu/annotation/fungi/magnaporthe/), and Fusarium graminearum (http://www.broad.mit.edu/annotation/fungi/fusarium/index.html), and Aspergillus oryzae genomic data were provided by the National Institute of Advanced Industrial Science and Technology (www.bio.nite.go.jp/dogan/Top). Coordination of analyses of these data was enabled by an international collaboration involving more than 50 institutions from 10 countries coordinated from Manchester, United Kingdom (www.cadre.man.ac.uk and www.aspergillus.man.ac.uk).
RESULTS
Aspergillus nidulans contains four AMT/MEP genes.
Two A. nidulans AMT/MEP genes, meaA and mepA, have been described previously (33). An analysis of the draft A. nidulans genome sequence identified partial sequences of two additional AMT/MEP sequences, designated mepB and mepC. A mepB-specific PCR product was used to screen an A. nidulans genomic BAC library, and a mepC-specific PCR product was generated from genomic DNA (see Materials and Methods). Sequence analysis indicated that mepB is comprised of five exons and is predicted to encode a 472-amino-acid protein andthat the mepC open reading frame of 1,452 bp contains threeexons encoding a predicted product of 453 amino acids. Subsequent annotation of the A. nidulans genome (http://www.broad.mit.edu/annotation/fungi/aspergillus/) has confirmed the sequences for all four genes, i.e., meaA (AN7463.2), mepA (AN1181.2), mepB (AN0209.2), and mepC (AN0496.2).
Alignment of the four predicted A. nidulans AMT/MEP protein sequences reveals substantial amino acid conservation throughout the proteins (Fig. 1A). While MeaA and MepA share 55% amino acid sequence identity (75% similarity), MepB displays 46% identity (65% and 68% similarity, respectively) with MeaA and MepA. In contrast, MepC shares only 31% identity (53% similarity) with MeaA, 35% identity (56% similarity) with MepA, and 31% identity (53% similarity) with MepB. Like MeaA and MepA, MepB and MepC contain both ammonium transporter signature motifs (PF00909) (54), although MepC has a total of seven mismatches across both signature sequences (Fig. 1B). Eleven putative transmembrane helices with an Nout-Cin topology were identified for MepB, using the prediction program HMMTOP (52), which is in agreement with predictions made for other AMT/MEP transporters, including MeaA and MepA (33). Although it is likely that the structure of MepC resembles that of other AMT/MEP transporters, topology predictions for MepC using the HMMTOP and TMHMM (46) programs were not as convincing. Together, these comparisons suggest that the MepC protein is more divergent than MeaA, MepA, and MepB.
FIG. 1.
Four proteins constitute the A. nidulans AMT/MEP family. (A) Alignment of A. nidulans AMT/MEP predicted protein sequences. Identical residues present in at least half of the sequences are indicated by black boxes, whereas gray shading represents similar residues. The alignment was performed using ClustalX (49) and shaded using MacBoxshade 2.15. Large boxes indicate the positions of the AMT/MEP signature sequences. (B) Alignment of the ammonium permease signature sequences, SS 1 and SS 2 (54). Bold residues represent the MepC residues that do not match the signature sequence.
The A. nidulans mepB gene encodes a high-affinity ammonium permease.
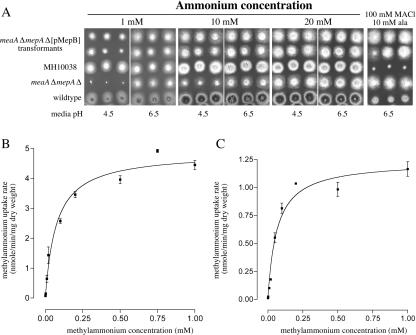
The mepAΔ meaAΔ double mutant MH9828 is unable to grow on 1 mM ammonium at pH 4.5 and is resistant to methylammonium, a toxic analogue of ammonium (33). To assess whether mepB encoded a functional ammonium permease, multiple copies of mepB (pBJM5377) were cotransformed with the pyroA+-carrying plasmid pI4 into MH9828, and transformants were screened for suppression of the double mutant phenotype. Thirty-seven of the 83 PyroA+ transformants tested displayed increased growth on 1 mM ammonium (pH 4.5), whereas no growth was observed for either pI4-only transformants or the recipient strain (Fig. 2A). The phenotypes observed were dependent on the mepB copy number (an approximate range of 4 to 10 copies per cell), and transformants with higher mepB copy numbers displayed increased growth on 1 mM ammonium and reduced methylammonium resistance (Southern blot analysis; data not shown). It should be noted that the extent of growth of the mepB cotransformants on any ammonium concentration or the degree of methylammonium sensitivity never reached wild-type levels (Fig. 2A). Overall, these results indicate that mepB encodes a functional ammonium permease and suggest that the capacity of this permease is limiting.
FIG. 2.
The A. nidulans mepB gene encodes a high-affinity (methyl)ammonium permease. (A) Suppression of mepA meaA double deletion mutant's ammonium growth defect by multiple copies of mepB. The growth of six different mepB transformants (meaAΔ mepAΔ [pMepB {pBJM5377}]) compared to that of MH10038, the meaAΔ mepAΔ recipient strain (MH9828), and the wild type (MH1) is shown. MH10038 is a wA3 (white conidia) strain that displays wild-type ammonium growth and was included to allow growth comparisons between the wA3 strains. Growth was tested on 1, 10, and 20 mM ammonium (NH4+) at either pH 4.5 or 6.5, as indicated. Resistance to 100 mM methylammonium chloride (MACl) on medium containing 10 mM alanine is also shown. Cotransformant D, which was used for panel B, is the first colony in the second row. (B and C) Plots of velocity versus substrate concentration for MepB and MepA activity. [14C]methylammonium uptake rates for nitrogen-starved mycelia were determined at methylammonium concentrations of 1, 2, 10, 20, 50, 100, 200, 500, 750, and 1,000 μM. (B) [14C]methylammonium uptake rates for multiple-copy mepB transformant D, assessing MepB activity. (C) MepA methylammonium transport activity in the mepBΔ meaAΔ (mepA+) mutant (MH10323).
The transformant with the highest mepB copy number (cotransformant D) was used in [14C]methylammonium uptake studies (Fig. 2B). The apparent Km (methylammonium) for MepB was determined to be 73.5 (±12.14) μM, with an apparent Vmax of 4.87 (± 0.176) nmoles/min/mg dry weight. Since mepB was overexpressed in cotransformant D, the MepB Vmax value is not comparable to that for a wild-type strain, whereas the difference in the actual relative amounts of active MepB does not affect Km determination. These results indicate that mepB encodes a high-affinity (methyl)ammonium permease.
The Km calculated for MepB was very similar to that calculated previously for MepA (44.3 μM), which was determined for a meaAΔ mepB+ strain (33). A mepBΔ mutant was created by homologous gene replacement (see Materials and Methods), and the MepA kinetic parameters were reassessed in the mepBΔ meaAΔ (mepA+) strain MH10323 (Fig. 2C). This analysis yielded an apparent Km (methylammonium) for MepA of 69.11 (± 9.46) μM and a Vmax value of 1.28 (± 0.052) nmoles/min/mg (Fig. 2C). The Km value for MepA presented here is within the 95% confidence intervals for the value estimated previously and confirms that A. nidulans contains two high-affinity ammonium permease genes, mepA and mepB.
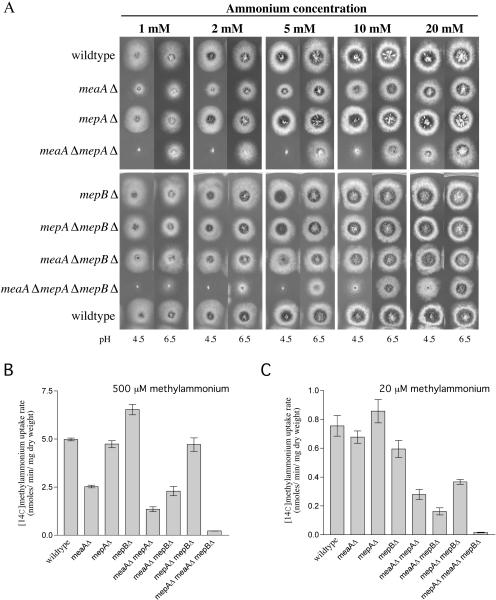
Analysis of A. nidulans mepBΔ mutant.
Deletion of the two high-affinity permease genes, mepA and mepB, either singly or combined, had no effect on growth for all ammonium concentrations tested, at either pH 6.5 or 4.5 (Fig. 3A). Indeed, the [14C]methylammonium uptake rates of the mutants with 500 μM methylammonium showed that the presence of MeaA alone was sufficient for wild-type levels of methylammonium uptake under these conditions (Fig. 3B). The methylammonium transport activity of the meaAΔ mepBΔ double mutant was similar to that of the meaAΔ single mutant, indicating that transport in the absence of MeaA was mediated by MepA. However, a comparison of the transport rates of the meaAΔ mepAΔ double mutant and the meaAΔ mepAΔ mepBΔ triple mutant indicated that MepB is able to contribute about 27% of the wild-type transport activity under these conditions. The [14C]methylammonium uptake assays performed with 20 μM methylammonium showed that the transport rate for the mepBΔ mutant was slightly lower than that for the meaA or mepA single deletion mutant (Fig. 3C). The combined deletion of the high-affinity ammonium transport genes mepA and mepB only reduced the transport activity of the cell to 48% that of the wild type, compared to 37% and 21% for the mepAΔ meaAΔ and mepBΔ meaAΔ mutants, respectively (Fig. 3). This indicates that the MeaA permease has a greater capacity than either MepA or MepB, so cells relying on only MepA or MepB activity have low methylammonium transport rates.
FIG. 3.
Analysis of A. nidulans meaA, mepA, and mepB deletion mutants. (A) Growth of AMT/MEP deletion mutants on a range of ammonium concentrations. At each ammonium concentration, tests were performed at pH 4.5 and pH 6.5, as indicated. (B and C) [14C]methylammonium uptake analyses of meaA, mepA, and mepB single, double, and triple deletion mutants. Assays were performed as described in Materials and Methods with nitrogen-starved mycelia at final methylammonium concentrations of 500 μM (B) and 20 μM (C). Error bars represent standard errors calculated for the results from at least two independent experiments. The strains used for this analysis were the wild type (MH1) and mepAΔ (MH9831), meaAΔ (MH9830), meaAΔ mepAΔ (MH9829), mepBΔ (MH10324), mepBΔ meaAΔ (MH10323), mepBΔ mepAΔ (MH10322), and mepBΔ meaAΔ mepAΔ (MH10325) mutants.
Across all ammonium concentrations tested at pH 6.5, the meaAΔ mepAΔ mepBΔ mutant displayed reduced growth compared to the meaAΔ mepAΔ strain (Fig. 3). This was particularly notable at 1 mM ammonium, where no growth was observed for the meaAΔ mepAΔ mepBΔ mutant, indicating that the growth seen for the meaAΔ mepAΔ mutant on this medium was due to MepB activity. Interestingly, no growth of the meaAΔ mepAΔ mepBΔ or meaAΔ mepAΔ mutant was observed at pH 4.5 on ammonium concentrations of 10 mM or less, indicating that MepB activity appeared to be absent on media at pH 4.5. As shown by the ammonium growth phenotypes of the multiple-copy mepB transformants (Fig. 2A), the MepB protein is able to function at pH 4.5, suggesting that differences in MepB activity at pH 4.5 and 6.5 may be regulated at the transcriptional level. Ambient pH regulation of genes in A. nidulans is controlled by the zinc finger transcription factor PacC, which activates the expression of alkaline-expressed genes and represses the transcription of acid-expressed genes (50). Two potential PacC binding sites (GCCARG) are present in the mepB promoter.
MepC does not normally contribute to ammonium uptake.
To analyze the function of MepC, a mepCΔ mutant was created by homologous gene replacement (see Materials and Methods). The phenotype of the mepCΔ mutant was indistinguishable from that of the wild type, and the deletion of mepC in all possible combinations with meaA, mepA, and mepB deletions did not alter the growth phenotypes of the various single, double, and triple mutants at any tested ammonium concentration or pH (data not shown). Furthermore, no significant [14C]methylammonium uptake activity was detected for the meaAΔ mepAΔ mepBΔ strain MH10325 (Fig. 3). For example, with 20 μM methylammonium, the meaAΔ mepAΔ mepBΔ mutant had an uptake rate of 0.0161 (±0.0023) nmoles methylammonium/min/mg dry weight, which is only 2% that of the wild type and less than the standard error values for all of the other strains. Therefore, no methylammonium transport attributable to MepC activity was present under the conditions tested.
To assess whether the structurally different MepC protein could function as an ammonium transporter, the effect of overexpression was determined. Multiple copies of mepC (pMA5741) were introduced into the mepAΔ meaAΔ meaBΔ MH10321 recipient by cotransformation with the pyroA+ plasmid pI4. No PyroA+ transformants showed stronger growth than the recipient strain when tested on 1 mM or 10 mM ammonium (pH 6.5 or pH 4.5). The presence of additional copies of mepC (approximate range of 2 to 20 copies) was confirmed by Southern blot analysis (data not shown). Therefore, multiple copies of mepC were unable to suppress the ammonium growth phenotype of the triple mutant. Since the expression of mepC from its native promoter is low (see below), it was possible that even with multiple copies, the expression levels were not sufficiently high to allow suppression. To further elevate the levels of MepC expression, the mepC coding region was fused to the highly inducible xylP promoter (see Materials and Methods). The xylP::mepC construct pMA6382 was cotransformed into MH10325 with pI4, and PyroA+ transformants were screened on 1 mM and 10 mM ammonium at pH 4.5. In the presence of glucose, where the xylP promoter is repressed, all transformants retained the phenotype of the recipient strain. On xylose medium, where xylP expression is induced, approximately 20% of the transformants showed partial restoration of growth on ammonium at pH 4.5. The copy numbers of the xylP::mepC construct in these complementing cotransformants (approximate range of 3 to 20 copies) were determined by Southern blot analysis (data not shown). The extent of growth of the complementing xylP::mepC cotransformants on any ammonium concentration or the degree of methylammonium sensitivity never reached wild-type levels and was comparable to that of transformants overexpressing mepB (Fig. 2A). These results indicate that MepC can function as an ammonium permease, but only when highly overexpressed from multiple copies of a xylP-driven gene.
Differential expression of A. nidulans ammonium permease genes.
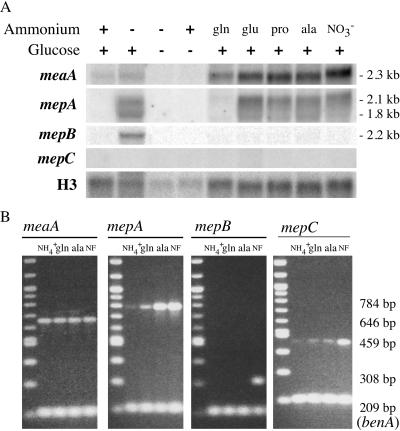
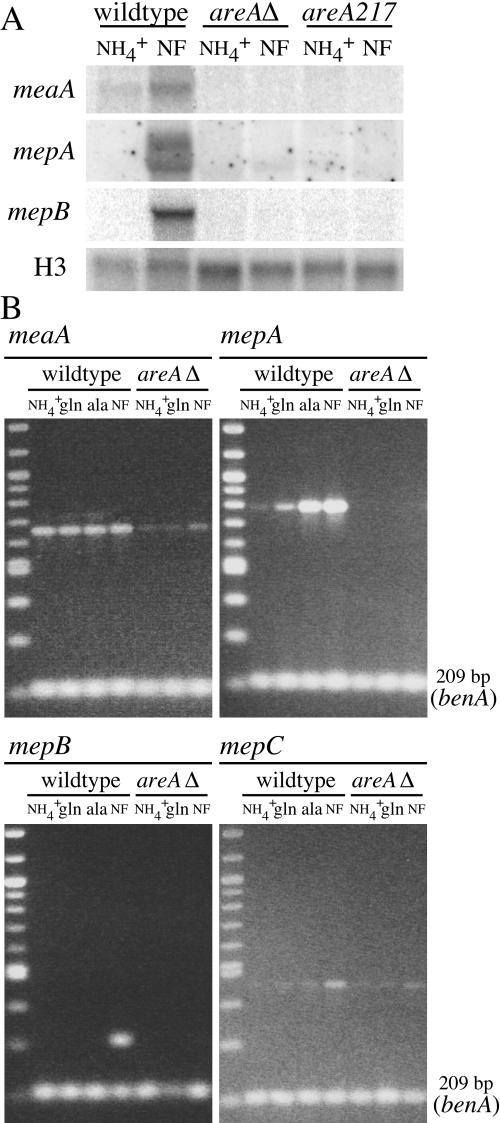
The expression of the A. nidulans ammonium permease genes under different nitrogen conditions was investigated by Northern blotting and RT-PCR analysis (Fig. 4A and B). The expression of meaA was readily detected for cells grown in ammonium and also glutamine, indicating that the expression of meaA occurs under nitrogen-sufficient conditions. In contrast, the expression of mepA was repressed under these nitrogen-sufficient conditions but was readily observed for cells grown in nonrepressing nitrogen sources (glutamate, proline, alanine, or nitrate) or nitrogen-starved cultures (Fig. 4). The expression of meaA or mepA was not detected under carbon starvation conditions. The expression profile for mepB was unique, with a single transcript detected only under conditions of complete nitrogen starvation and not under conditions of nitrogen limitation (Fig. 4). This nitrogen starvation response of mepB expression was not detected in nitrogen-starved cultures that were simultaneously starved of carbon or in carbon-starved and ammonium-sufficient cultures. The expression of mepC was not detected by Northern analysis under any conditions tested. However, RT-PCR analysis showed low levels of mepC expression on ammonium, glutamine, and alanine that increased slightly in response to nitrogen starvation (Fig. 4). These results show that each of the A. nidulans ammonium permease genes displays a distinct expression pattern.
FIG. 4.
Expression analysis of A. nidulans ammonium permease genes in wild-type background. (A) Northern analysis of ammonium permease gene expression from the wild-type (MH1) strain. RNAs were isolated from mycelia grown in 1% glucose-ANM medium, pH 6.5, with 20 mM ammonium (NH4+) at 37°C for 16 h and then transferred to fresh medium that was nitrogen- and/or carbon-free, as indicated, for 4 hours. Alternatively, RNAs were isolated from mycelia grown for 16 h at 37°C in 1% glucose medium, pH 6.5, with 10 mM glutamine (gln), glutamate (glu), proline (pro), alanine (ala), or nitrate (NO3−), as indicated. Northern blots were hybridized with probes specific for meaA, mepA, mepB, and mepC (see Materials and Methods) or A. nidulans histone H3 as a loading control (13), as indicated. Sizes to the right (in kb) represent the approximate sizes of the respective transcripts. (B) Multiplex RT-PCR analysis of meaA, mepA, mepB, and mepC gene expression. The sizes of the AMT/MEP products (upper bands) and the benA loading control (lower bands) are indicated, and the name of the respective AMT/MEP gene is shown at the top of each gel picture. The growth conditions were the same as those for panel A. RT-PCR conditions and primer details are described in Materials and Methods.
Full expression of A. nidulans AMT/MEP genes requires the global nitrogen activator AreA.
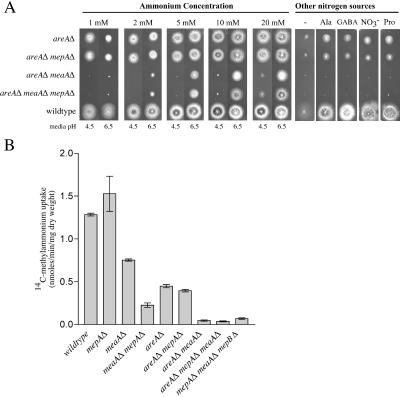
To investigate the role of AreA in the regulation of the A. nidulans AMT/MEP genes, areAΔ meaAΔ and areAΔ meaAΔ mepAΔ double and triple mutants were created by genetic crosses and assessed for growth on a range of ammonium concentrations (Fig. 5A). The areAΔ mutant MH5699 displayed poorer-than-wild-type growth for all ammonium concentrations tested. The growth of the areAΔ mepAΔ mutant MH10313 was indistinguishable from that of the areAΔ single mutant. At 1 mM ammonium, the areAΔ meaAΔ mutant MH10311 displayed no growth, indicating that the residual growth observed for the areAΔ mutant was due to MeaA function (Fig. 5A). The lack of growth of the areAΔ meaAΔ mutant at 1 mM ammonium (pH 6.5) also suggested that AreA is absolutely required for the expression of MepA and MepB activity. [14C]methylammonium uptake assays on nitrogen-starved mycelia were in agreement with the ammonium growth phenotypes (Fig. 5B). The areAΔ mutant and the areAΔ mepAΔ strain displayed an appreciably reduced methylammonium transport activity (35% of the wild-type activity). The areAΔ meaAΔ mepAΔ mutant MH10312 had a methylammonium uptake rate similar to that of the meaAΔ mepAΔ mepBΔ mutant MH10321 (approximately 3% that of the wild type). These results showed the AreA is required for MepA, MepB, and MeaA transport activity, although there is a level of AreA-independent MeaA function.
FIG. 5.
Functional analysis of A. nidulans ammonium permeases in areA mutant backgrounds. (A) Growth on a range of ammonium concentrations of areAΔ (MH5699), areAΔ mepAΔ (MH10313), areAΔ meaAΔ (MH10311), and areAΔ meaAΔ mepAΔ (MH10314) mutant and wild-type (MH1) strains. For each ammonium concentration, the pH of the medium was either 4.5 or 6.5 (normal growth pH), as indicated. Growth on nitrogen-free (−), 10 mM alanine (ala), GABA, nitrate (NO3−), and proline (pro) media is also shown. (B) [14C]methylammonium transport rates for the wild-type strain and mepAΔ, meaAΔ, meaAΔ mepAΔ, areAΔ, areAΔ mepAΔ, areAΔ meaAΔ, areAΔ meaAΔ mepAΔ, and meaAΔ mepAΔ mepBΔ mutant strains. Assays were performed with nitrogen-starved mycelia at a final methylammonium concentration of 200 μM. Error bars represent standard errors calculated for the results from at least two independent experiments.
Consistent with the established function of AreA as an activator of catabolic gene expression, the single areAΔ mutant showed very poor growth on nonpreferred nitrogen sources (Fig. 5A). The slight level of growth observed is presumed to be due to basal levels of catabolic gene transcription. Surprisingly, the growth observed for the areAΔ and areAΔ mepAΔ strains on nitrogen sources such as alanine and nitrate was absent for the areAΔ meaAΔ mutant. This is likely to reflect a requirement for retention and/or uptake of trace amounts of ammonium released by the catabolism of alternative nitrogen sources (34).
Northern blot analysis and RT-PCR were performed to assess the steady-state transcript levels of the AMT/MEP genes for the areAΔ mutant and the loss-of-function areA217 mutant, which contains a mutation within the DNA-binding domain (20). The expression of meaA was detected by RT-PCR but not by Northern blot analysis for the areAΔ or areA217 strain from either ammonium-grown or nitrogen-starved mycelia (Fig. 6). The significant but not total reduction of meaA expression in the areAΔ mutant is consistent with the growth phenotypes and [14C]methylammonium uptake data and indicates that the full expression of meaA under nitrogen-sufficient conditions requires AreA function.
FIG. 6.
Expression of A. nidulans AMT/MEP genes in areA mutant background. (A) Northern analysis of meaA, mepA, and mepB gene expression in the wild-type (MH1) and areAΔ (MH5699) and areA217 (MH341) mutant strains. RNAs were isolated from mycelia grown for 16 h at 37°C in 1% glucose medium, pH 6.5, with 20 mM ammonium (NH4+) and then transferred to nitrogen-free (NF) medium at pH 6.5 for 4 hours. Northern blots were hybridized with probes specific for meaA, mepA, mepB (see Materials and Methods), and A. nidulans histone H3 as a loading control (13), as indicated. (B) Multiplex RT-PCR analysis of meaA, mepA, mepB, and mepC expression in wild-type and areAΔ mutant strains. The name of the respective gene is shown at the top of each picture, and in all cases, the lower band is the internal loading control benA. RT-PCR conditions and primer details are described in Materials and Methods.
AreA-dependent expression of mepA was shown by Northern blotting and RT-PCR analysis, as no mepA mRNA was detected for the areAΔ or areA217 strain for all growth conditions tested (Fig. 6). Furthermore, the nitrogen starvation-specific expression of mepB was shown to be absolutely dependent on AreA. The increase of mepC expression in response to nitrogen starvation was reduced in the areAΔ mutant, but expression levels on ammonium and glutamine were unaffected (Fig. 6). Since Northern analysis showed identical results for the areA217 point mutant and the areAΔ mutant, AreA regulation of the AMT/MEP genes is at the level of DNA binding.
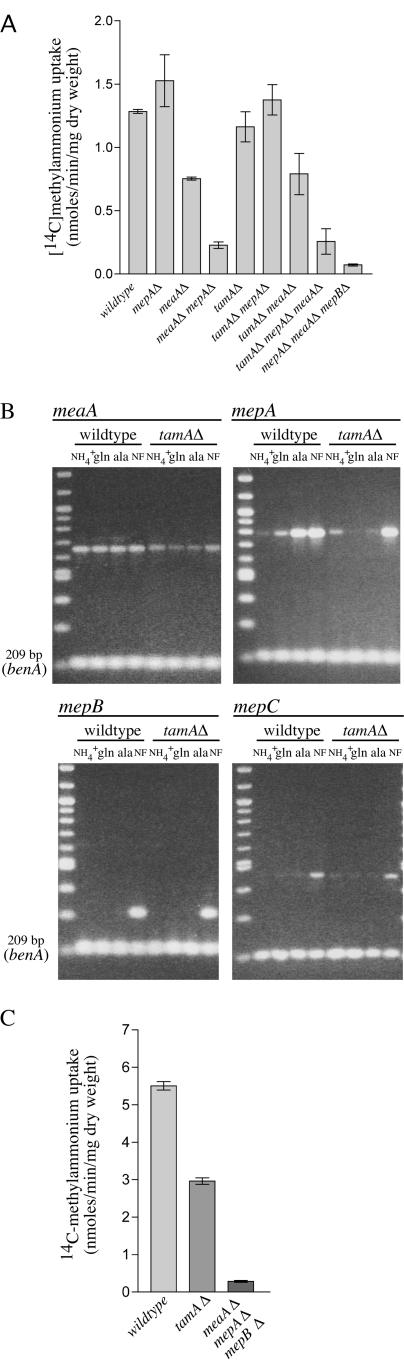
Effects of TamA on AMT/MEP activity and gene expression.
TamA has been shown to be a coactivator required for the optimal expression of certain AreA-regulated genes (42, 43). The tamAΔ mutation had very little effect on [14C]methylammonium transport, other than a slight reduction attributable to a reduction in MeaA-mediated transport (Fig. 7A). Consistent with this, RT-PCR results for meaA, mepA, mepB, and mepC expression in the tamAΔ strain under ammonium growth or nitrogen-starved conditions indicated no effect of the tamAΔ mutation other than a slight reduction in meaA expression compared to the wild type (Fig. 7B). However, clear reductions in meaA and mepA expression were noted for the tamAΔ strain grown in glutamine or alanine, and [14C]methylammonium uptake analysis performed on alanine-grown mycelia confirmed that the tamAΔ mutant had reduced methylammonium transport activity under these conditions (Fig. 7B and C). Such a reduction is consistent with the methylammonium-resistant phenotype observed for tamA mutants which were originally identified as methylammonium resistant on media containing alanine as the nitrogen source (12, 19). Northern analysis of meaA, mepA, and mepB expression on a range of nitrogen sources (ammonium, glutamine, glutamate, alanine, proline, and nitrate) indicated a clear reduction of meaA and mepA expression in a tamAΔ strain compared to the wild-type level for all nonammonium nitrogen sources tested (data not shown). These results show that the full expression of meaA and mepA on nonammonium nitrogen sources requires the combined activities of AreA and the transcriptional coactivator TamA.
FIG.7.
Analysis of A. nidulans AMT/MEP genes in tamAΔ mutant background. (A) [14C]methylammonium transport rates forwild-type (MH1) and mepAΔ (MH9831), meaAΔ (MH9830), meaAΔ mepAΔ (MH9829), tamAΔ (MH8694), tamAΔ mepAΔ (MH10316), tamAΔ meaAΔ (MH10315), tamAΔ meaAΔ mepAΔ (MH10317), and meaAΔ mepAΔ mepBΔ (MH10325) mutant strains. Assays were performed with nitrogen-starved mycelia at a final methylammonium concentration of 200 μM. (B) RT-PCR analysis of meaA, mepA, mepB, and mepC expression in wild-type and tamAΔ mutant strains. RNAs were isolated from mycelia grown for 16 h at 37°C in 1% glucose medium, pH 6.5, with 20 mM ammonium (NH4+) and then transferred to nitrogen-free (NF) medium, pH 6.5, for 4 hours or from mycelia grown for 16 h at 37°C in 1% glucose medium, pH 6.5, with 10 mM glutamine (gln) or alanine (ala), as indicated. The name of the respective gene is shown at the top of each picture, and in all cases, the lower band is benA. (C) [14C]methylammonium transport rates for the wild-type and tamAΔ and meaAΔ mepAΔ mepBΔ mutant strains for mycelia grown for 16 h with 10 mM alanine. A final methylammonium concentration of 200 μM was used. Error bars represent standard errors from at least two independent experiments.
DISCUSSION
The A. nidulans genome contains a family of four AMT/MEP genes, namely, meaA, mepA, mepB, and mepC. These genes are differentially expressed in response to nitrogen availability, and the full expression of each AMT/MEP gene requires the global nitrogen regulator AreA. AreA functions as a transcriptional activator in response to nitrogen limitation, and its known targets are GATA sites in the promoters of genes involved in nitrogen acquisition (3, 17, 40, 56). The transcription of genes subjected to nitrogen metabolite repression is reduced under nitrogen-sufficient conditions by changes in the level and activity of AreA. Regulated areA mRNA degradation (35, 36) and the interaction of AreA with the negative regulator NmrA (2, 21) reduce or prevent AreA-dependent activation of these genes. Nitrogen limitation leads to stabilization of areA mRNA and a loss of NmrA-associated inhibition of activity. Together, these factors account for the influence of the quality of nitrogen source availability on AreA activity and levels of catabolic gene expression. Recent studies have shown that the complete absence of a nitrogen source results in enhanced AreA activity and increased expression of AreA-regulated genes through additional mechanisms that are independent of mRNA stability and NmrA and are correlated with modification of AreA and its hyperaccumulation inside the nucleus (51).
AreA-mediated regulation of mepA represents the pattern observed for the amdS, gmdA, and fmdS genes, which are regulated by nitrogen metabolite repression but do not require induction (11, 14-16). mepA is repressed under conditions of nitrogen sufficiency, activated in response to nitrogen limitation, and increased further in response to nitrogen starvation. AreA is absolutely required for these responses of mepA expression. Like gmdA and fmdS, mepA is not expressed under carbon starvation conditions, where AreA is thought to be inactive (14). MepA is a high-affinity ammonium transporter and is likely to serve an ammonium-scavenging function when nitrogen availability is limiting to growth.
The mepB gene has been shown to encode a second high-affinity ammonium permease. mepB expression has a novel expression profile whereby this gene is not expressed in the presence of any nitrogen source tested, whether it is repressing or limiting. This is thought to be the first example of an AreA-regulated gene in A. nidulans that is expressed specifically in response to complete nitrogen starvation. This highlights the observation that nitrogen starvation conditions result in enhanced AreA activity above that observed for nitrogen limitation. The mepB gene appears to be exquisitely sensitive to AreA, such that its expression is observed only under conditions where AreA is highly active. This pattern of regulation is consistent with MepB having a scavenging role under extreme nitrogen deprivation conditions.
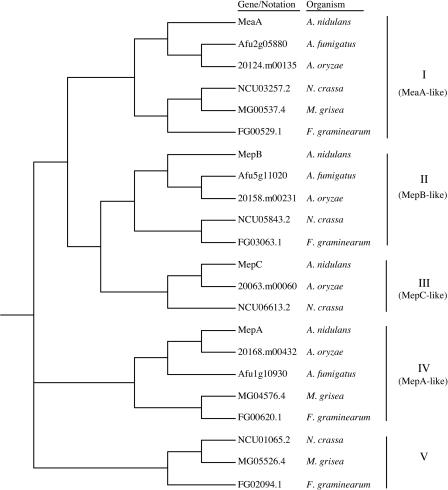
A fourth potential member of the A. nidulans AMT/MEP family has been identified. However, we suggest that mepC, which is more poorly expressed than the other AMT/MEP genes, does not normally contribute to ammonium acquisition in A. nidulans. Deletion of mepC resulted in no detectable phenotypic effect, and furthermore, no significant methylammonium uptake activity was detected in a meaAΔ mepAΔ mepBΔ triple mutant retaining only MepC function. In a recent study, Candida albicans was shown to contain two MEP genes, mep1 and mep2, that are similar in structure and function to the S. cerevisiae MEP1/MEP3 and MEP2 genes (5). A third, more divergent MEP-like gene identified in C. albicans was considered nonfunctional based on the phenotype of the Δmep1/2 double mutant (5). The predicted MepC product is also the most divergent of the A. nidulans family of AMT/MEP permeases, including differences in the ammonium signature sequences. Despite these differences, MepC expressed at high levels can partially compensate for the loss of the other three AMT/MEP permeases. Database searches revealed that orthologs of the A. nidulans AMT/MEP genes meaA, mepA, and mepB are present in the genomes of A. fumigatus, A. oryzae, Neurospora crassa, Magnaportha grisea, and Fusarium graminearum (Fig. 8). In contrast, orthologs of mepC can be identified in the N. crassa genome but not in the related M. grisea genome, and among the Aspergillus spp., mepC is present in A. oryzae but not in A. fumigatus. The additional amino acid sequences apparent in the MepC amino acid sequence compared to the MeaA, MepA, and MepB sequences (Fig. 1) are present in all MepC orthologs. Therefore, mepC may be the result of an early duplication that has been independently lost in several fungal lineages. This further argues that mepC does not have a critical physiological or regulatory function in A. nidulans, although a subtle function in nitrogen signaling cannot be excluded.
FIG. 8.
Relatedness of AMT/MEP protein sequences from A. nidulans, A. fumigatus, A. oryzae, N. crassa, F. graminearum, and M. grisea. The dendrogram was constructed by the neighbor-joining method, using ClustalX (49) with default settings. Organisms and gene names or sequence notations are indicated. Each cluster is numbered with a roman numeral.
The expression of meaA, encoding the major ammonium transporter, is both AreA dependent and AreA independent. The factors that promote AreA-independent expression are unknown, but it is clear that AreA plays an active role in the expression of meaA under all nitrogen conditions. It is paradoxical that AreA activity is required under ammonium-sufficient conditions, in which it has been assumed to be inactive. It is apparent that AreA does retain the capacity to activate gene expression from certain promoters under repressed conditions. The gdhA gene, encoding NADP-linked glutamate dehydrogenase, the major enzyme of ammonium assimilation in A. nidulans, is also regulated by AreA under nitrogen-sufficient conditions (7, 38, 41). However, the mechanisms that underlie the expression of meaA and gdhA on ammonium appear to be different. The full expression of gdhA under ammonium-sufficient conditions is dependent on TamA acting as a coactivator of AreA and an additional transcriptional activator, LeuB (38). In contrast, TamA has a relatively minor role in the activation of meaA expression on ammonium. Instead, TamA appears to act with AreA to activate meaA (and mepA) expression on nonammonium nitrogen sources, similar to the role that it plays in contributing to the expression of other nitrogen-regulated genes (12, 42, 43). Furthermore, LeuB is not required for meaA expression, consistent with the lack of predicted LeuB binding sites in the meaA promoter (B. J. Monahan, unpublished results). There is no indication that NmrA, which acts to inhibit AreA function at the promoters of genes subject to nitrogen metabolite repression, has a role in modulating meaA expression (Monahan, unpublished data). The factors that facilitate AreA function at the meaA promoter under nitrogen-sufficient conditions are unknown but are of considerable interest. By analogy with the complex interactions of AreA, TamA, and LeuB at the gdhA promoter, it is possible that interactions between AreA and other transcription factors at the meaA promoter are involved.
The contrasting expression profiles of the four AMT/MEP genes in A. nidulans have revealed that nitrogen sufficiency, limitation, and starvation can be differentiated as distinct physiological states by the organism. Furthermore, this study has highlighted the finding that promoter-specific contexts must be an important factor in determining the activation capacity of AreA under different nitrogen conditions. The AMT/MEP genes in A. nidulans provide an excellent system with which to further investigate these complexities of AreA function.
Acknowledgments
This work was supported by the Australian Research Council and the award of Melbourne Research Scholarships to B.J.M. and M.C.A.
REFERENCES
- 1.Andrianopoulos, A., and M. J. Hynes. 1988. Cloning and analysis of the positively acting regulatory gene amdR from Aspergillus nidulans. Mol. Cell. Biol. 8:3532-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrianopoulos, A., S. Kourambas, J. A. Sharp, M. A. Davis, and M. J. Hynes. 1998. Characterization of the Aspergillus nidulans nmrA gene involved in nitrogen metabolite repression. J. Bacteriol. 180:1973-1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arst, H. N., Jr., and D. J. Cove. 1973. Nitrogen metabolite repression in Aspergillus nidulans. Mol. Gen. Genet. 126:111-141. [DOI] [PubMed] [Google Scholar]
- 4.Austin, B., R. M. Hall, and B. M. Tyler. 1990. Optimized vectors and selection for transformation of Neurospora crassa and Aspergillus nidulans to bleomycin and phleomycin resistance. Gene 93:157-162. [DOI] [PubMed] [Google Scholar]
- 5.Biswas, K., and J. Morschhauser. 2005. The Mep2p ammonium permease controls nitrogen starvation-induced filamentous growth in Candida albicans. Mol. Microbiol. 56:649-669. [DOI] [PubMed] [Google Scholar]
- 6.Blakey, D., A. Leech, G. H. Thomas, G. Coutts, K. Findlay, and M. Merrick. 2002. Purification of the Escherichia coli ammonium transporter AmtB reveals a trimeric stoichiometry. Biochem. J. 364:527-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christensen, T., M. J. Hynes, and M. A. Davis. 1998. Role of the regulatory gene areA of Aspergillus oryzae in nitrogen metabolism. Appl. Environ. Microbiol. 64:3232-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clutterbuck, A. J. 1974. Aspergillus nidulans genetics, p. 447-510. In R. C. King (ed.), Handbook of genetics, vol. 1. Plenum Publishing Corp., New York, N.Y. [Google Scholar]
- 9.Conroy, M. J., S. J. Jamieson, D. Blakey, T. Kaufmann, A. Engel, D. Fotiadis, M. Merrick, and P. A. Bullough. 2004. Electron and atomic force microscopy of the trimeric ammonium transporter AmtB. EMBO Rep. 5:1153-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cove, D. J. 1966. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim. Biophys. Acta 113:51-56. [DOI] [PubMed] [Google Scholar]
- 11.Davis, M. A., J. M. Kelly, and M. J. Hynes. 1993. Fungal catabolic gene regulation: molecular genetic analysis of the amdS gene of Aspergillus nidulans. Genetica 90:133-145. [DOI] [PubMed] [Google Scholar]
- 12.Davis, M. A., A. J. Small, S. Kourambas, and M. J. Hynes. 1996. The tamA gene of Aspergillus nidulans contains a putative zinc cluster motif which is not required for gene function. J. Bacteriol. 178:3406-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ehinger, A., S. H. Denison, and G. S. May. 1990. Sequence, organization and expression of the core histone genes of Aspergillus nidulans. Mol. Gen. Genet. 222:416-424. [DOI] [PubMed] [Google Scholar]
- 14.Fraser, J. A., M. A. Davis, and M. J. Hynes. 2001. The formamidase gene of Aspergillus nidulans: regulation by nitrogen metabolite repression and transcriptional interference by an overlapping upstream gene. Genetics 157:119-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraser, J. A., M. A. Davis, and M. J. Hynes. 2002. The genes gmdA, encoding an amidase, and bzuA, encoding a cytochrome P450, are required for benzamide utilization in Aspergillus nidulans. Fungal Genet. Biol. 35:135-146. [DOI] [PubMed] [Google Scholar]
- 16.Hynes, M. J. 1994. Regulatory circuits of the amdS gene of Aspergillus nidulans. Antonie Leeuwenhoek 65:179-182. [DOI] [PubMed] [Google Scholar]
- 17.Hynes, M. J. 1975. Studies on the role of the areA gene in the regulation of nitrogen catabolism in Aspergillus nidulans. Aust. J. Biol. Sci. 28:301-313. [DOI] [PubMed] [Google Scholar]
- 18.Khademi, S., J. O'Connell III, J. Remis, Y. Robles-Colmenares, L. J. Miercke, and R. M. Stroud. 2004. Mechanism of ammonia transport by Amt/MEP/Rh: structure of AmtB at 1.35 A. Science 305:1587-1594. [DOI] [PubMed] [Google Scholar]
- 19.Kinghorn, J. R., and J. A. Pateman. 1975. Studies of partially repressed mutants at the tamA and areA loci in Aspergillus nidulans. Mol. Gen. Genet. 140:137-147. [DOI] [PubMed] [Google Scholar]
- 20.Kudla, B., M. X. Caddick, T. Langdon, N. M. Martinez-Rossi, C. F. Bennett, S. Sibley, R. W. Davies, and H. N. Arst, Jr. 1990. The regulatory gene areA mediating nitrogen metabolite repression in Aspergillus nidulans. Mutations affecting specificity of gene activation alter a loop residue of a putative zinc finger. EMBO J. 9:1355-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamb, H. K., J. Ren, A. Park, C. Johnson, K. Leslie, S. Cocklin, P. Thompson, C. Mee, A. Cooper, D. K. Stammers, and A. R. Hawkins. 2004. Modulation of the ligand binding properties of the transcription repressor NmrA by GATA-containing DNA and site-directed mutagenesis. Protein Sci. 13:3127-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, S., and J. Taylor. 1990. Isolation of DNA from fungal mycelia and single spores, p. 282-287. In M. A. Innis, D. H. Gelfand, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, San Diego, Calif.
- 23.Lorenz, M. C., and J. Heitman. 1998. The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J. 17:1236-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ludewig, U., N. von Wiren, and W. B. Frommer. 2002. Uniport of NH4+ by the root hair plasma membrane ammonium transporter LeAMT1;1. J. Biol. Chem. 277:13548-13555. [DOI] [PubMed] [Google Scholar]
- 25.Ludewig, U., S. Wilken, B. Wu, W. Jost, P. Obrdlik, M. El Bakkoury, A. M. Marini, B. Andre, T. Hamacher, E. Boles, N. von Wiren, and W. B. Frommer. 2003. Homo- and hetero-oligomerization of ammonium transporter-1 NH4+ uniporters. J. Biol. Chem. 278:45603-45610. [DOI] [PubMed] [Google Scholar]
- 26.Marini, A. M., and B. Andre. 2000. In vivo N-glycosylation of the Mep2 high-affinity ammonium transporter of Saccharomyces cerevisiae reveals an extracytosolic N-terminus. Mol. Microbiol. 38:552-564. [DOI] [PubMed] [Google Scholar]
- 27.Marini, A. M., G. Matassi, V. Raynal, B. Andre, J. P. Cartron, and B. Cherif-Zahar. 2000. The human rhesus-associated RhAG protein and a kidney homologue promote ammonium transport in yeast. Nat. Genet. 26:341-344. [DOI] [PubMed] [Google Scholar]
- 28.Marini, A. M., S. Soussi-Boudekou, S. Vissers, and B. Andre. 1997. A family of ammonium transporters in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:4282-4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marini, A. M., J. Y. Springael, W. B. Frommer, and B. Andre. 2000. Cross-talk between ammonium transporters in yeast and interference by the soybean Sat1 protein. Mol. Microbiol. 35:378-385. [DOI] [PubMed] [Google Scholar]
- 30.Marini, A. M., S. Vissers, A. Urrestarazu, and B. Andre. 1994. Cloning and expression of the MEP1 gene encoding an ammonium transporter in Saccharomyces cerevisiae. EMBO J. 13:3456-3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marzluf, G. A. 1997. Genetic regulation of nitrogen metabolism in the fungi. Microbiol. Mol. Biol. Rev. 61:17-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.May, G. S., J. Gambino, J. A. Weatherbee, and N. R. Morris. 1985. Identification and functional analysis of beta-tubulin genes by site specific integrative transformation in Aspergillus nidulans. J. Cell Biol. 101:712-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monahan, B. J., J. A. Fraser, M. J. Hynes, and M. A. Davis. 2002. Isolation and characterization of two ammonium permease genes, meaA and mepA, from Aspergillus nidulans. Eukaryot. Cell 1:85-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monahan, B. J., S. E. Unkles, I. T. Tsing, J. R. Kinghorn, M. J. Hynes, and M. A. Davis. 2002. Mutation and functional analysis of the Aspergillus nidulans ammonium permease MeaA and evidence for interaction with itself and MepA. Fungal Genet. Biol. 36:35-46. [DOI] [PubMed] [Google Scholar]
- 35.Morozov, I. Y., M. Galbis-Martinez, M. G. Jones, and M. X. Caddick. 2001. Characterization of nitrogen metabolite signalling in Aspergillus via the regulated degradation of areA mRNA. Mol. Microbiol. 42:269-277. [DOI] [PubMed] [Google Scholar]
- 36.Morozov, I. Y., M. G. Martinez, M. G. Jones, and M. X. Caddick. 2000. A defined sequence within the 3′ UTR of the areA transcript is sufficient to mediate nitrogen metabolite signalling via accelerated deadenylation. Mol. Microbiol. 37:1248-1257. [DOI] [PubMed] [Google Scholar]
- 37.Osmani, A. H., G. S. May, and S. A. Osmani. 1999. The extremely conserved pyroA gene of Aspergillus nidulans is required for pyridoxine synthesis and is required indirectly for resistance to photosensitizers. J. Biol. Chem. 274:23565-23569. [DOI] [PubMed] [Google Scholar]
- 38.Polotnianka, R., B. J. Monahan, M. J. Hynes, and M. A. Davis. 2004. TamA interacts with LeuB, the homologue of Saccharomyces cerevisiae Leu3p, to regulate gdhA expression in Aspergillus nidulans. Mol. Genet. Genomics 272:452-459. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Scazzocchio, C. 2000. The fungal GATA factors. Curr. Opin. Microbiol. 3:126-131. [DOI] [PubMed] [Google Scholar]
- 41.Small, A. J. 2000. Characterisation of the tamA gene of Aspergillus nidulans. Ph.D. thesis. University of Melbourne, Melbourne, Australia.
- 42.Small, A. J., M. J. Hynes, and M. A. Davis. 1999. The TamA protein fused to a DNA-binding domain can recruit AreA, the major nitrogen regulatory protein, to activate gene expression in Aspergillus nidulans. Genetics 153:95-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Small, A. J., R. B. Todd, M. C. Zanker, S. Delimitrou, M. J. Hynes, and M. A. Davis. 2001. Functional analysis of TamA, a coactivator of nitrogen-regulated gene expression in Aspergillus nidulans. Mol. Genet. Genomics 265:636-646. [DOI] [PubMed] [Google Scholar]
- 44.Smith, D. G., M. D. Garcia-Pedrajas, S. E. Gold, and M. H. Perlin. 2003. Isolation and characterization from pathogenic fungi of genes encoding ammonium permeases and their roles in dimorphism. Mol. Microbiol. 50:259-275. [DOI] [PubMed] [Google Scholar]
- 45.Sohlenkamp, C., C. C. Wood, G. W. Roeb, and M. K. Udvardi. 2002. Characterization of Arabidopsis AtAMT2, a high-affinity ammonium transporter of the plasma membrane. Plant Physiol. 130:1788-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sonnhammer, E. L., G. von Heijne, and A. Krogh. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6:175-182. [PubMed] [Google Scholar]
- 47.Straubinger, B., E. Straubinger, S. Wirsel, G. Turgeon, and O. Yoder. 1992. Versatile fungal transformation vectors carrying the selectable bar gene of Streptomyces hygroscopicus. Fungal Genet. Newsl. 39:82-83. [Google Scholar]
- 48.Thomas, G. H., J. G. Mullins, and M. Merrick. 2000. Membrane topology of the Mep/Amt family of ammonium transporters. Mol. Microbiol. 37:331-344. [DOI] [PubMed] [Google Scholar]
- 49.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tilburn, J., S. Sarkar, D. A. Widdick, E. A. Espeso, M. Orejas, J. Mungroo, M. A. Penalva, and H. N. Arst, Jr. 1995. The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. EMBO J. 14:779-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Todd, R. B., J. A. Fraser, K. H. Wong, M. A. Davis, and M. J. Hynes. 2005. Nuclear accumulation of the GATA factor AreA in response to complete nitrogen starvation by regulation of nuclear export. Eukaryot. Cell 4:1646-1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tusnady, G. E., and I. Simon. 1998. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J. Mol. Biol. 283:489-506. [DOI] [PubMed] [Google Scholar]
- 53.Van Kim, C. L., Y. Colin, and J. P. Cartron. 2005. Rh proteins: key structural and functional components of the red cell membrane. Blood Rev. [Epub ahead of print.] doi: 10.1016/j.bire.2005.04.002. [DOI] [PubMed]
- 54.von Wiren, N., and M. Merrick. 2004. Regulation and function of ammonium carriers in bacteria, fungi, and plants. Top. Curr. Genet. 9:95-120.
- 55.Westhoff, C. M., D. L. Siegel, C. G. Burd, and J. K. Foskett. 2004. Mechanism of genetic complementation of ammonium transport in yeast by human erythrocyte Rh-associated glycoprotein. J. Biol. Chem. 279:17443-17448. [DOI] [PubMed] [Google Scholar]
- 56.Wilson, R. A., and H. N. Arst, Jr. 1998. Mutational analysis of AreA, a transcriptional activator mediating nitrogen metabolite repression in Aspergillus nidulans and a member of the “streetwise” GATA family of transcription factors. Microbiol. Mol. Biol. Rev. 62:586-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zadra, I., B. Abt, W. Parson, and H. Haas. 2000. xylP promoter-based expression system and its use for antisense downregulation of the Penicillium chrysogenum nitrogen regulator NRE. Appl. Environ. Microbiol. 66:4810-4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng, L., D. Kostrewa, S. Berneche, F. K. Winkler, and X. D. Li. 2004. The mechanism of ammonia transport based on the crystal structure of AmtB of Escherichia coli. Proc. Natl. Acad. Sci. USA 101:17090-17095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zidi-Yahiaoui, N., I. Mouro-Chanteloup, A. M. D'Ambrosio, C. Lopez, P. Gane, C. le van Kim, J. P. Cartron, Y. Colin, and P. Ripoche. 2005. Human rhesus B and rhesus C glycoproteins: properties of facilitated ammonium transport in recombinant kidney cells. Biochem. J. 391:33-40. [DOI] [PMC free article] [PubMed] [Google Scholar]