Abstract
The transcription efficiency of an adhesion protein gene, ap65-1, in Trichomonas vaginalis varies with changes in the iron supply and with the growth stage. In the present study, two Myb recognition elements, MRE-1/MRE-2r and MRE-2f, were found to play antagonistic roles in regulating the iron-inducible activity of an ap65-1 reporter gene. Intriguingly, either of these elements was shown to be sufficient to repress basal activity, but together they were also shown to activate growth-related activity of the reporter gene in iron-depleted cells. A myb1 gene which encodes a 24-kDa protein containing a Myb-like R2R3 DNA binding domain was identified from Southwestern screening of MRE-2f-binding proteins. The Myb1 protein was detected as a major 35-kDa protein which exhibited variations in nuclear concentration with changes in the iron supply. A recombinant Myb1 protein was shown to differentially interact with MRE-1/MRE-2r and MRE-2f in vitro. Overexpression of hemagglutinin-tagged Myb1 in T. vaginalis resulted in repression or activation of ap65-1 transcription in iron-depleted cells at an early and a late stage of cell growth, respectively, while iron-inducible ap65-1 transcription was constitutively repressed. The hemagglutinin-tagged Myb1 protein was found to constantly occupy the chromosomal ap65-1 promoter at a proximal site, but it also selected two more distal sites only at the late growth stage. Together, these observations suggest that Myb1 critically regulates multifarious ap65-1 transcription, possibly via differential selection of multiple promoter sites upon environmental changes.
As the most common sexually transmitted disease of nonviral origin in humans, trichomoniasis caused by infection with the protozoan parasite Trichomonas vaginalis is an important risk factor for the transmission of human immunodeficiency virus (33). Along with increasing numbers of drug-resistant clinical T. vaginalis isolates (7, 9), trichomoniasis is emerging as a major threat to public health.
Iron availability, which periodically varies in the human vagina, where the parasite colonizes, regulates the cytoadherence of T. vaginalis, possibly through controlling the expression of several adhesion proteins in transcription initiation and protein trafficking steps (2, 11, 18, 35). Although only a few T. vaginalis genes have been characterized in detail, the most common example seems to suggest that T. vaginalis uses a conserved initiator (Inr) sequence as the sole core promoter element to regulate the basal transcription of protein-coding genes via interaction of the Inr with a unique Inr-binding protein, IBP39, to recruit α-ammanitin-resistant RNA polymerase II (17, 20, 22, 30). This basal transcription machinery significantly differs from that of higher eukaryotic systems, in which the α-ammanitin-sensitive RNA polymerase II machinery exhibits considerable diversity both in the core promoter context and in the components of the promoter-recognition TFIID complex (15, 24, 31). Thus, the transcription efficiency of a particular type II promoter in T. vaginalis may primarily be determined in cis by gene-specific distal promoter elements, such as those identified in the promoters of the α-scs and ap65-1 genes (21, 35).
The ap65-1 gene belongs to the ap65 (adhesion protein 65) multigene family encoding multiple homologous 65-kDa proteins (1, 25) whose protein sequences are identical to those of the hydrogenosomal malic enzymes (16). In addition to hydrogenosomal localization (4), these proteins have also been detected on plasma membranes as part of adhesion complexes during host-parasite encounters as well as when exposed to an ample iron supply (2, 11, 18). The transcription efficiency of the ap65-1 gene in T. vaginalis varies with changes in the iron supply as well as with the growth stage (27, 35). Multiple Myb recognition elements, namely, MRE-1/MRE-2r (which overlap) and MRE-2f, were found to regulate multifarious ap65-1 expression in cis (27, 35), implying the involvement of Myb-like transcription factors in the iron-inducible transcription machinery of the parasite.
Myb proteins that regulate myriad gene-specific transcription activities in a wide range of eukaryotic systems only share a consensus DNA binding domain. Vertebrates have three cellular Myb proteins, A-Myb, B-Myb, and c-Myb (38), each of which has a highly conserved N-terminal DNA binding domain (with ∼90% identity) consisting of three repeats (R1R2R3). They all recognize specific DNA contexts with a core pentanucleotide sequence, CNGTT, through three key base-contacting amino acid residues, i.e., K128 in the R2 domain and K182 and N183 in the R3 domain (26). In plants such as Arabidopsis thaliana, the Myb protein family is expanded and contains >100 distinct members, most of which contain an R2R3 DNA binding domain with only 40 to ∼80% identity (34). Although the key base-contacting amino acid residues are also conserved, the sequence contexts of the Myb recognition elements in plants are not limited to those with a CNGTT core (32). In T. vaginalis, neither MRE-1/MRE-2r (TAACGATA; MRE-1 sequence is in italics, and MRE-2r sequence is underlined) nor MRE-2f (TATCGT) conforms to the sequence context of the CNGTT core.
In the present study, MRE-1/MRE-2r and MRE-2f were further found to play multiple roles in controlling different phases of multifarious ap65-1 transcription. Through Southwestern screening and an in vitro DNA binding assay, an R2R3-type Myb1 protein encoded by a myb1 gene was found to bind MRE-1/MRE-2r and MRE-2f with differential activities. Further analyses showed that Myb1 plays distinct roles during different growth phases of ap65-1 transcription, possibly via differential selection of promoter entry sites. To the best of our knowledge, Myb1 is the first gene-specific transcription factor with defined roles in transcriptional regulation to be reported for T. vaginalis.
MATERIALS AND METHODS
Cultures.
T. vaginalis T1 cells were maintained as previously described (35), and the iron concentration in the growth medium was adjusted as previously described (18). The WT-13, m(MRE-1/MRE-2r), and m(MRE-2f) cell lines were obtained from a previous study (27).
Stable promoter assay.
Plasmid DNA was electroporated into T. vaginalis T1 cells, and transfectants were selected with paromomycin as previously described (35). Relative amounts of respective reporter plasmids in cells from individual cell lines were determined by a dot hybridization assay as previously described (27), and their promoter activities were normalized accordingly.
Oligonucleotides.
The sequences of the oligonucleotides used for the present study are listed in Table 1, unless otherwise specified in the text.
TABLE 1.
Sequences of oligonucleotides used for this study
| Primer name and purpose | Sequence (5′ to 3′) |
|---|---|
| RT-PCR | |
| ap65-1-5′ | AATGGCAAGGCCCTCTGCGCTAC |
| ap65-1-3′ | TAAATTAAGAAAGCTAAGTGTTTAAAAATCGCGC |
| tub-5′ | AAATCGTTCACATCCAAGCTGG |
| tub-3′ | TTGTATGGCTCGACGACTGTATCAG |
| ha-myb1-5′ | GATTACGCTCTTATGATGTTTG |
| myb1-5′ | TGCCTTTAATCGCATGATGTTTG |
| myb1-3′ | ATTATAATCAGTATCGACAGAGGCG |
| Southwestern screening | |
| mre-2sa | CAGAGCTGTATCGTCTAGCTGTATCGTCTAGCTGTATCGTCTAGC |
| mre-2as | GCTAGACGATACAGCTAGACGATACAGCTAGACGATACAGCT |
| Plasmid constructionb | |
| flp-5′-sac2 | TCCGCGGCTACTATAGGGCACGCGTGGTC |
| flp-3′-nco1 | ACCATGGATCCCATCATAAAAGTGATATGAA |
| flp-3′-bamh1-ha-hind3 | AGGATCCAAGAGCGTAATCTGGAACATCGTATGGGTAGGAAAGCTTCTCTTCTGTCGCC |
| ha-myb1-5′-bgl2 | CAGATCTTACCCATACGATGTTCCAGATTACGCTCTTATGATGTTTGACGGCCTTTCCG |
| myb1-5′-nde1 | TTCATATGATGTTTGACGGCCTTTCCG |
| myb1-3′-sal1 | TTGTCGACGGAAGCAACACTTAACTTAAAACG |
| ΔN(103)myb1-5′-nde1 | TTCATATGAAAGTCAAGTTCACGGAAGAGG |
| ChIP | |
| 1f | GGGCGAAATCACGGAAATCG |
| 1r | CCTTGCGGGAGAGTTCACGTGCTGG |
| 2f | TTCCAATCAACATTTATCTATACTATCGG |
| 2r | TGCCATTGCGATGATTATGGTTG |
| 3f | GCATTCAGTTTTGATCCAGGTTTCC |
| 3r | GATAGTATAGATAAATGTTGATTGGAAGC |
| 4f | GTTCTTATTCAGATGTTCCTGATTTG |
| 4r | GCCCTAGTATAGCATTCAAATGATCC |
The DNA sequence of MRE-2f is indicated in bold.
Sequences of restriction enzyme sites, as indicated in the names, are underlined.
Nucleic acid extraction.
RNAs were extracted from T. vaginalis T1 cells by use of the TRIZOL reagent (Invitrogen), and DNAs were extracted as previously described (5).
Construction and Southwestern screening of a cDNA library.
A T. vaginalis T1 cDNA library was constructed in a λ phage by using the OrientExpress cDNA system following the instructions of the supplier (Novagen). A 32P-labeled double-stranded DNA, mre-2, derived from annealing 20 pmol of a γ-32P-labeled antisense oligonucleotide, mre-2as, and 60 pmol of the sense oligonucleotide mre-2s, was ligated to produce a concatenated probe for screening of the expression library as previously described (37).
Cloning of genomic myb1 gene.
A T. vaginalis T1 genomic DNA library was constructed at the EcoRI site of pBluescript II SK (Stratagene). The 5′ sequence flanking the myb1 gene derived from a λc22 cDNA clone was amplified from the genomic DNA library by a PCR using the primer pair T3 and myb1-3′-2. The amplified DNA was then subcloned into pGEM-T Easy as described by the supplier (Promega).
Construction of plasmids.
The plasmids pAP65luc+ and pAP65-1luc+/TUBneo and the mutant plasmids pm(−95/−81) (with mutation of MRE-1/MRE-2r) and pm(−44/−39) (with mutation of MRE-2f) were obtained from a previous study (35). A DNA fragment from pm(−95/−81) was amplified by a PCR using the primer pair T7 and m(−44/−39)-3′ (35) and subcloned into pGEM-T Easy to generate pTA(−95/−81;−44/−39). A double mutant plasmid, pm(MRE-1/MRE-2r+MRE-2f), was obtained by replacing the SacII/BamHI fragment in pAP65-1luc+/TUBneo with the SacII/BamHI fragment from pTA(−95/−81;−44/−39).
A DNA fragment spanning the coding region of the myb1 gene was amplified by a PCR using the forward primer ha-myb1-5′-bgl2 or myb1-5′-nde1 along with the reverse primer myb1-3′-sal1 and was then subcloned into pGEM-T Easy to generate pTA-ha-myb1 and pTAmyb1, respectively.
A DNA fragment spanning the 5′ untranslated region of the flp-1 gene (6) was amplified by a PCR using the primer pair flp-5′-sac2 and flp-3′-bamh1-ha-hind3 and was then subcloned into pGEM-T Easy to generate pTA-FLP-ha. The SacII/BamHI fragment from pTA-FLP-ha and the BglII/SalI fragment from pTA-ha-myb1 were subsequently cloned into a plasmid backbone derived from pAP65-1luc+/TUBneo predigested with SacII and SalI to generate pFLP-ha-myb1/TUBneo.
The Myb1 expression plasmid, pET28/Myb1, was generated by subcloning of the NdeI/SalI fragment from pTAmyb1 into pET28a predigested with NdeI/SalI.
Northern hybridization.
An [α-32P]dCTP-labeled myb1 DNA probe was synthesized from the pTAmyb1 template by a random priming method using Redi-prime II, as described by the supplier (Amersham Biosciences). Ten micrograms of mRNA purified by oligo(dT) cellulose column chromatography was denatured in glyoxal prior to fractionation in a 1% agarose gel and then was blotted onto a Nytran membrane (Schleicher & Schuell) for Northern hybridization, using an ExpressHyb solution as suggested by the supplier (Clontech).
Reverse transcriptase PCR (RT-PCR).
Cellular RNAs were first treated with RNase-free DNase as described by the supplier (Promega). Ten micrograms of DNase-free RNA was then primed by oligo(dT) in a reaction to synthesize first-strand cDNA by the reverse transcriptase Superscript III (Invitrogen). The ap65-1 gene in the first-strand cDNA was amplified by a PCR using the primer pair ap65-1-5′ and ap65-1-3′, with an annealing temperature of 60°C. The β-tubulin gene was amplified by using the primer pair tub-5′ and tub-3′, with an annealing temperature of 56°C. The ha-myb1 or myb1 gene was amplified by using the forward primer ha-myb1-5′ or myb1-5′, respectively, along with the reverse primer myb1-3′, with an annealing temperature of 60°C. Multiple PCR amplifications were performed for each primer pair, using 1 μl of serially diluted (1×, 2×, 4×, or 8× dilution) first-strand cDNA as a template. PCR products were separated in 2% agarose gels by electrophoresis and stained with ethidium bromide. The intensity of each PCR product was analyzed by Meta Morp Offline, version 6.2r6 (Universal Imaging). Linear amplification of the mRNA signals was achieved by 24 to 26 PCR cycles for the β-tubulin gene signal, 28 to 30 cycles for the ha-myb1 and myb1 signals, and 21 to 23 cycles for the ap65-1 signal.
Expression of recombinant Myb1 protein.
pET28/Myb1 was transformed into Escherichia coli BL21-Codon Plus DE3-RIL (Stratagene) for recombinant Myb1 (rMyb1) production. E. coli transformed with pET28/Myb1 in shaking cultures was incubated at 30°C until the optical density at 600 nm reached 0.5. Induction was performed by the addition of 0.4 mM isopropyl-β-d-thiogalactopyranoside for 2 h. Under these conditions, the majority of rMyb1 was in inclusion bodies (see Fig. 4A). Inclusion bodies from 100 ml of E. coli culture were dissolved in 1 ml of 8 M urea and immediately refolded by the direct addition of 14 ml binding buffer (5 mM imidazole, 500 mM sodium chloride, and 20 mM Tris-HCl [pH 7.9]). After removal of the insoluble materials, rMyb1 in the supernatant was purified using a His-binding nickel column in the presence of 0.5 M urea as described by the supplier (Novagen).
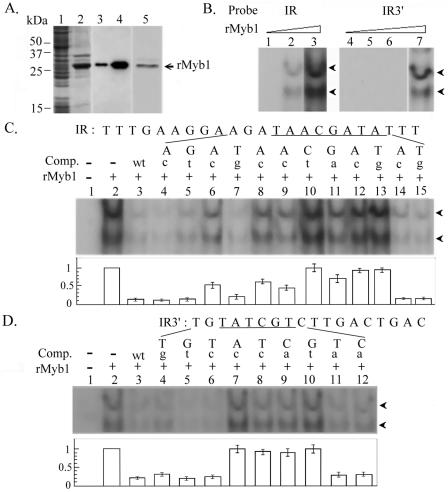
FIG. 4.
DNA binding specificity of rMyb1. (A) Soluble (lanes 1 and 3) and insoluble (lanes 2 and 4) fractions of a lysate from rMyb1-expressing E. coli cells and purified rMyb1 (lane 5) were separated by SDS-PAGE and stained with Coomassie blue (lanes 1, 2, and 5) or examined by Western blotting using an anti-six-His antibody (lanes 3 and 4). A prestained broad-range protein ladder was used as the size marker (Bio-Rad). (B) Five (lanes 1 and 4), 10 (lanes 2 and 5), 50 (lanes 3 and 6), or 100 ng (lane 7) of rMyb1 was incubated with γ-32P-labeled IR (lanes 1 to 3) or γ-32P-labeled IR3′ (lanes 4 to 7). (C and D) Fifty nanograms (C, lanes 2 to 15) or 100 ng (D, lanes 2 to 12) of rMyb1 was incubated with γ-32P-labeled IR (C, lanes 2 to 15) or γ-32P-labeled IR3′ (D, lanes 2 to 12). The DNA sequences of IR (C) and IR3′ (D) are shown at the top of each panel, with MRE-1/MRE-2r (C) and MRE-2f (D) underlined. A 1,000× molar excess of cold competitor (C and D, lanes 3) or a series of mutant competitors, each with a single point mutation, as indicated by a lowercase letter at the top of each lane, was included in the binding reactions (lanes 4 to 15 in panel C and lanes 4 to 12 in panel D). The reaction mixtures were separated in 10% polyacrylamide gels by electrophoresis. The DNA-protein complexes (arrowheads) in reactions with IR (B, left panel; C) or IR3′ (B, right panel; D) were detected in autoradiograms with 24 or 48 h of exposure, respectively, at room temperature. The average signal intensities in individual lanes from three separate experiments are depicted below each panel (C and D).
Antiserum production.
Soluble rMyb1 purified from inclusion bodies as described above was used for rabbit immunization by a standard protocol (14), and the antiserum was purified by protein A affinity chromatography as described by the supplier (Sigma).
Western blot assay.
Protein samples were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis in 12% gels, and the proteins were transferred to an Immobilon-P polyvinylidene difluoride membrane (Millipore) by use of a semidry electroblotter. Sequential immunoreactions were performed, and an ECL system was used for signal detection as instructed by the supplier (Pierce). Reaction conditions for antibodies from commercial sources, including a mouse monoclonal anti-α-tubulin antibody (DM1A; Sigma) (10,000×), a rat monoclonal anti-hemagglutinin (anti-HA) antibody (3F10; Roche), and a six-His monoclonal antibody (Clontech) (10,000×), were as described by the supplier. The expression of Myb1 and AP65 was assayed using a rabbit anti-Myb1 antiserum (1,000×) and a mouse monoclonal anti-malic enzyme antibody (15D7; 10,000×) (4), respectively. Serially diluted protein samples (1×, 2×, 4×, or 8× dilution) were assayed by Western blotting for semiquantification of protein expression levels in T. vaginalis. The signal intensities of individual bands were analyzed by Meta Morp Offline, version 6.2r6 (Universal Imaging), and the signal intensities of a particular protein from multiple samples were normalized to the signal intensity of α-tubulin in the same samples.
Immunofluorescence assay.
Cells were fixed on glass slides with cold methanol at −20°C for 5 min and sequentially stained using a mouse monoclonal anti-HA antibody (800×) (HA-7; Sigma) and a fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse immunoglobulin G antibody (500×) (Jackson). The fluorescent signal was recorded by confocal microscopy.
Electrophoretic mobility shift assay.
Probe labeling and an electrophoretic mobility shift assay were performed as previously described (35). The signal intensities of individual bands were analyzed by Meta Morp Offline, version 6.2r6 (Universal Imaging).
ChIP assay.
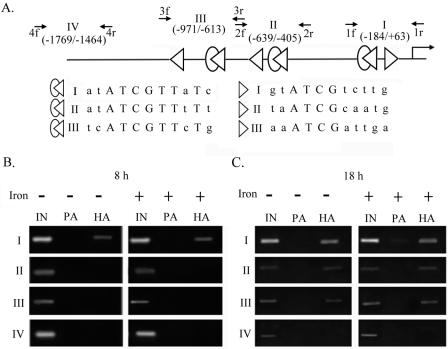
A chromatin immunoprecipitation (ChIP) assay was performed as previously described (23), except that 200 μg ml−1 of the protease inhibitor Nα-p-tosyl-l-lysine chloromethyl ketone was added to the reaction buffers used after the recovery of cells from formaldehyde cross-linking and before the elution of immunoprecipitated DNA. A detailed protocol is available upon request. About 6 × 107 cells were cross-linked by formaldehyde for each condition tested (see Fig. 7). The supernatant recovered from the DNA shearing step was divided into three parts: one was used as the input DNA control, another was incubated with 3 μl of anti-tubulin antibody plus 30 μl of protein A-agarose, and the other was incubated with 30 μl of anti-HA-agarose conjugate (Sigma). Purified DNAs were examined by PCR amplifications using the specific primer pairs shown in Fig. 7A. DNA spanning region I, II, or III of the ap65-1 promoter was amplified at an annealing temperature at 60°C, and DNA spanning region IV was amplified at an annealing temperature of 53°C.
FIG. 7.
Differential promoter selection of HA-Myb1 in transfected T. vaginalis cells. The high-affinity (open merged ovals and triangles) and low-affinity (open triangles) Myb1 binding sites in the entire ap65-1 promoter are depicted in panel A, and the consensus sequences are aligned in capital letters. Small arrows indicate primer pairs used for PCRs to amplify DNAs spanning regions I, II, III, and IV of the ap65-1 promoter. The boundaries of each region relative to the transcription start site are indicated in parentheses. Samples from pFLPha-myb1/TUBneo-transfected cells under iron-depleted (left panels) or iron-replete (right panels) conditions for 8 h (B) and 18 h (C) were evaluated by a ChIP assay. PCR amplifications were performed using 1 μl of 50×-diluted DNA from input (IN) samples or 4×-diluted DNA from protein A (PA)- or anti-HA-treated (HA) samples as a template for 30 cycles. The PCR products were assayed by agarose gel electrophoresis with ethidium bromide staining.
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this paper have been deposited in the GenBank database under accession number AY948337 for the myb2 gene and AY948338 for the myb1 gene.
RESULTS
Roles of two distinct Myb recognition elements in ap65-1 transcription.
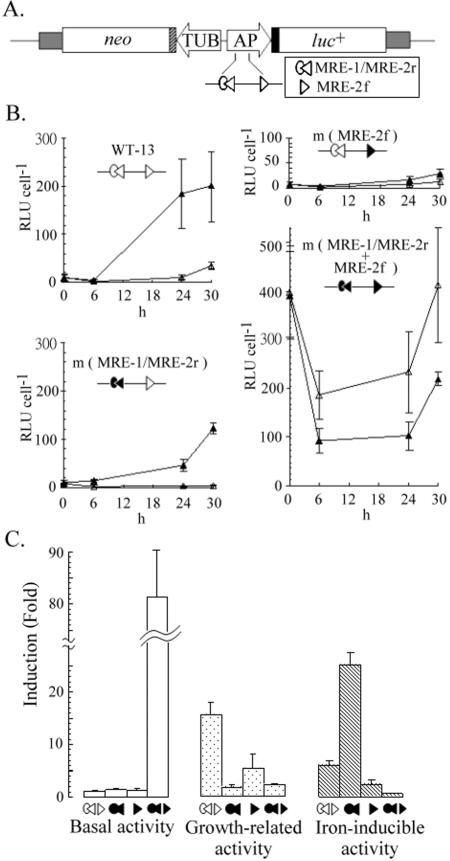
A stable reporter gene (pAPluc+/TUBneo)-based promoter assay was employed to define the roles of MRE-1/MRE-2r and MRE-2f in ap65-1 transcription (Fig. 1). Under iron depletion conditions, the lowest relative luciferase activity (referred to as basal activity) of pAPluc+/TUBneo-harboring WT-13 cells was detected at 6 h. This activity increased until 30 h, when the activity reached the highest level, which was 15-fold higher than the basal activity. At 6 h, the relative luciferase activity in cells with iron repletion was twofold higher than that in cells with iron depletion, and this difference increased to sixfold at 30 h. For the convenience of discussion in this paper, the ratio of the highest to the lowest activity in iron-depleted cells is referred to as growth-related activity, and the ratio of activity in iron-replete cells to that in iron-depleted cells at 30 h is referred to as iron-inducible activity.
FIG. 1.
Monitoring of iron-independent and -dependent transcriptional activities of ap65-1 promoter. Relative locations of MRE-1/MRE-2r and MRE-2f (see symbols in the box) in the reporter cassette, pAPluc+/TUBneo (27), are shown in panel A. In this plasmid, the ap65-1 promoter (AP) drives a luciferase gene (luc+), and the β-tubulin (TUB) promoter drives a selective marker, the neo gene. (B) Cell lines (as indicated at the top of each panel) harboring pAPluc+/TUBneo or one of the related mutant plasmids (open symbols indicate a wild-type MRE, and closed symbols indicate a mutant MRE in the plasmid) were established. Relative luciferase activities (relative light units [RLU] cell−1) of iron-depleted cells (open triangles) or iron-replete cells (closed triangles) were monitored 6 h, 24 h, and 30 h after treatment. The results are averages ± standard errors of duplicate samples from at least three separate experiments. (C) The basal activity (open bars) detected at 6 h in iron-depleted WT-13 cells was 2.3 ± 0.4 RLU per cell, and this value was taken as 1 for comparison. The ratio of the highest to the lowest activity in iron-depleted cells is defined as growth-related activity (stippled bars), and that of the highest activity detected at 30 h in iron-replete cells to that detected in iron-depleted cells is defined as iron-inducible activity (hatched bars).
The luciferase activity of iron-depleted m(MRE-1/MRE-2r) cells decreased for 24 h, when the lowest activity was comparable to that of WT-13 cells. The activity only increased slightly at 30 h. In contrast, the luciferase activity of iron-replete m(MRE-1/MRE-2r) cells increased for 30 h, when iron-inducible activity reached a 24-fold higher level. Basal activity of m(MRE-2f) cells was also detected at 6 h, at a level similar to that in WT-13 cells, whereas their growth-related and iron-inducible activities were repressed fivefold and twofold, respectively, at 30 h. Surprisingly, the basal activity of m(MRE-1/MRE-2r+MRE-2f) cells at 6 h was 80-fold higher than that of WT-13 cells. At 30 h, their growth-related activity was repressed to a twofold level, but these cells displayed a twofold iron-repressive activity.
Together, these results suggest that either MRE-1/MRE-2r or MRE-2f is sufficient to repress the basal activity of the ap65-1 promoter, whereas both are required for optimal growth-related activity in iron-depleted cells. In contrast, these DNA elements play antagonistic roles in iron-replete cells, as MRE-1/MRE-2r represses but MRE-2r activates iron-inducible ap65-1 transcription.
Molecular cloning of myb-like genes.
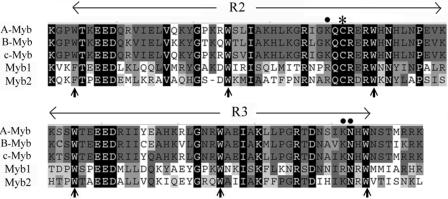
A T. vaginalis T1 cDNA expression library was screened using a 32P-labeled DNA probe, mre-2, which contains multiple copies of a concatenated MRE-2f sequence. Screening of nearly 8 × 105 plaques resulted in five partial cDNA clones, of which λc1, λc22, and λc24 overlap within a gene referred to as myb1 herein and λc20 and λc29 overlap within another gene, referred to as myb2. The protein sequences encoded by myb2 and myb1 share the consensus Myb R2R3 DNA binding domains, but with only 43% identity (Fig. 2). The recombinant protein derived from λc22 was found to specifically bind to MRE-2f in vitro (data not shown). The 5′ and 3′ ends of the λc22 cDNA were then extended by PCR from amplifications of respective DNA fragments from a genomic DNA library, and sequence analysis revealed a full-length myb1 gene which encodes an open reading frame of 206 amino acid residues, with a size estimated to be ∼24 kDa and a pI value of 8.84. It is notable that two of the key base-contacting amino acid residues, K128 and K182, of the vertebrate Myb proteins (26) are both replaced by R residues in T. vaginalis Myb1 (Fig. 2).
FIG. 2.
Identification of two myb-like genes in T. vaginalis. An alignment of protein sequences within the R2R3 domain of T. vaginalis Myb1 (GenBank accession no. AY948338) and Myb2 (AY948337) and human A-Myb (S03423), B-Myb (P10244), and c-Myb (CAF04477) is shown. The equally spaced tryptophan (W) residues, a signature of the Myb DNA binding domain, are indicated by upright arrows. Conserved amino acid residues are highlighted. The amino acid residues that provide strong base contacting for DNA recognition of vertebrate Mybs (26) are indicated by dots, and a redox-sensitive cysteine residue that needs to be reduced for DNA binding and transcriptional activity (3, 12, 13) is indicated by an asterisk.
Expression profile of myb1 gene.
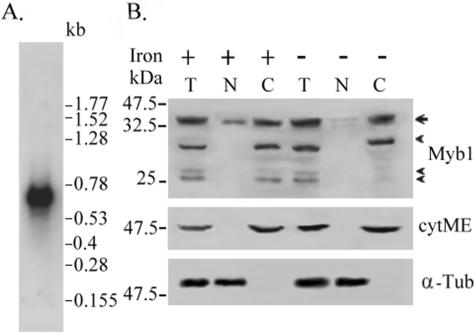
The myb1 gene in T. vaginalis T1 cells was expressed as a single 0.7-kilobase mRNA species, as revealed by Northern hybridization (Fig. 3A). Expression levels of myb1 mRNA in T. vaginalis T1 cells, as detected by RT-PCR, varied little in different growth phases as well as with a variable iron supply (data not shown).
FIG. 3.
Expression profile of myb1 gene in T. vaginalis. (A) Ten micrograms of mRNA purified from T. vaginalis T1 cells was examined by Northern hybridization with an [α-32P]dCTP-labeled DNA probe derived from pTAmyb1. A low-molecular-weight RNA ladder was used as the size marker (Invitrogen). (B) Total lysates (T) and nuclear (N) and cytoplasmic (C) fractions from T. vaginalis T1 cells with iron depletion (−) or iron repletion (+) for 18 h were assayed by Western blotting using anti-Myb1 (upper panel), anti-cytosolic malic enzyme (8) (middle panel), or anti-α-tubulin (bottom panel) antibody. A prestained protein ladder was used as the size marker (New England Biolabs). The arrow indicates the major Myb1 band, and arrowheads indicate minor bands. The cytosolic malic enzyme is indicated as cytME, and α-tubulin is indicated as α-Tub.
From a Western blot assay using anti-Myb1 antiserum, a major 35-kDa band and additional minor bands estimated to be 30-kDa and 25-kDa doublet bands were consistently detected in total lysates from iron-depleted or iron-replete cells (Fig. 3B). None of these protein bands was detected on a duplicate blot using preimmune serum (data not shown). The 35-kDa band was found to be more abundant in the cytoplasmic fractions than the nuclear fractions of cell lysates, but the minor and faster migrating bands were only detected in the cytoplasmic fractions. The purity of these cellular fractions was examined with an anti-cytosolic malic enzyme antibody (8), which detected a 50-kDa band in only the cytosolic fractions. In contrast, a constitutively expressed 55-kDa band was detected by anti-α-tubulin antibody in only the nuclear fractions. The results from three separate experiments revealed that the signal intensity of Myb1 in total cell lysates remained constant with changes in the iron supply; however, the signal intensity of the nuclear 35-kDa Myb1 protein was about threefold higher in samples from cells replete with iron than in those depleted of iron.
DNA binding specificity of recombinant Myb1 protein.
A recombinant His-tagged Myb1 protein, rMyb1 (Fig. 4A), was purified for use in an electrophoretic mobility shift assay. Ten nanograms of rMyb1 was sufficient to form two major complexes with a 32P-labeled IR double-stranded DNA probe (Fig. 4B, left panel) which contains the MRE-1/MRE-2r overlap (27). Although two major complexes were also formed in binding of rMyb1 to a 32P-labeled IR3′ double-stranded DNA probe (Fig. 4B, right panel) which contains the MRE-2f sequence (27), 100 ng of the protein was required to detect the signal. The DNA binding specificity of rMyb1 for 32P-labeled IR (Fig. 4C) or 32P-labeled IR3′ (Fig. 4D) was tested in competition assays using a 1,000× molar excess of mutated sequences of the mIR series or mIR3′ series, respectively, as previously described (27). The DNA-protein complexes were incompletely competed to various degrees by different mutant competitors, indicating that some nucleic acid residues are more important than others in the binding activities of these two DNA elements. The signal intensities of the DNA-protein complexes in individual reactions were measured, and the results from three separate competition assays indicated that ANAACGAT (or ATCGTTNT in reverse orientation), spanning MRE-1/MRE-2r, and ATCG,within MRE-2f, are the high-affinity and low-affinity binding sites of rMyb1, respectively.
Overexpression of HA-tagged Myb1 in T. vaginalis.
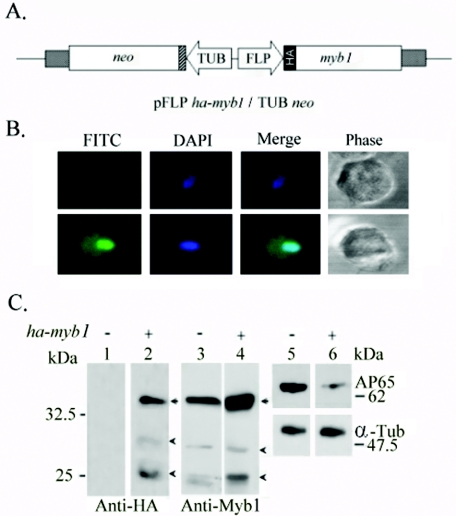
A DNA vector, pFLPha-myb1/TUBneo (Fig. 5A), was constructed to stably overexpress HA-tagged Myb1 in T. vaginalis T1 cells. The transfected cells were selected with 150 μg ml−1 of paromomycin. Freshly thawed transfected cells were used for the individual experiments described below. Higher concentrations of the drug or prolonged passages of transfected cells over a few weeks resulted in diminished Myb1 expression.
FIG. 5.
Overexpression of HA-Myb1 in T. vaginalis. An HA-Myb1 protein expression plasmid, pFLPha-myb1/TUBneo (A), was used to overexpress HA-Myb1 in T. vaginalis T1 cells. In this plasmid, the flp-1 promoter (FLP) drives an HA-tagged myb1 gene, and the β-tubulin (TUB) promoter drives a selective marker, the neo gene. (B) Subcellular localization of HA-Myb1 in pFLPha-myb1/TUBneo-transfected cells was detected using a mouse monoclonal anti-HA antibody and a FITC-conjugated secondary antibody. Images of cells labeled DAPI (4′,6′-diamidino-2-phenylindole), FITC, and phase contrast were recorded by confocal microscopy. (C) Lysates from nontransfected cells (lanes 1, 3, and 5) or cells transfected with pFLPha-myb1/TUBneo (lanes 2, 4, and 6) in normal medium for 8 h were examined by Western blotting using rat monoclonal anti-HA (lanes 1 and 2), anti-Myb1 (lanes 3 and 4), anti-malic enzyme (lanes 5 and 6, upper panels), or anti-α-tubulin (lanes 5 and 6, lower panels) antibody.
The HA-Myb1 signal was higher in the nuclei than in the cytoplasm of transfected cells, as determined by immunofluorescence microscopy using a mouse anti-HA monoclonal antibody (Fig. 5B, lower panels). All transfected cells exhibited positive signals, while a small fraction (<5%) of transfected cells exhibited stronger nuclear staining than others. No signal was detected in nontransfected T1 cells (Fig. 5B, upper panels). By Western blotting using a rat anti-HA monoclonal antibody, a major 35-kDa band and two barely detectable bands, of 30 and 25 kDa, were consistently detected in protein samples from transfected cells only (Fig. 5C). On a duplicate blot, the 35-kDa band and the two faster migrating minor bands were also detected by anti-Myb1 antiserum in samples from both transfected and nontransfected cells, and the signal intensity of the 35-kDa Myb1 protein detected in the protein sample from transfected cells was about 2.5-fold higher than that from nontransfected cells. In contrast, the signal intensity of AP65, as detected by an anti-malic enzyme 15D7 monoclonal antibody (4), in samples from transfected cells was only one-third that of nontransfected cells. The signal intensity of α-tubulin, as detected by an anti-α-tubulin monoclonal antibody, remained similar in these samples, implying that the expression of AP65 is repressed by overexpression of HA-Myb1.
Multiple roles of Myb1 in expression of the ap65-1 gene.
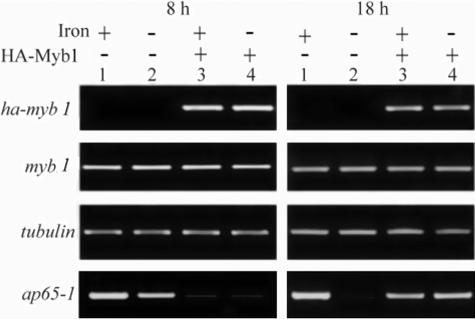
Although members of the ap65 gene family share nearly identical coding information (1, 16, 25), they display divergent 5′ intergenic regions, as revealed by a search of the database of the T. vaginalis Genome Sequencing Project (www.tigr.org/tdb/e2k1/tvg). To elucidate the roles of Myb1 in ap65-1 transcription, the expression profiles of several mRNA species in transfected cells versus nontransfected cells were examined by a semiquantitative RT-PCR assay in three separate experiments with consistent results, and the results from one representative experiment are shown. By 8 h (Fig. 6, left panel), the signal intensity of β-tubulin or myb1 mRNA detected in samples from transfected cells was similar to that from nontransfected cells, whereas ha-myb1 mRNA was only detected in samples from transfected cells, with a signal intensity slightly higher than that of myb1 mRNA in the same samples. Iron exerted little effect on the expression levels of these mRNA species. In contrast, the signal intensity of ap65-1 mRNA in samples from transfected cells was much lower than that in nontransfected cells exposed to iron depletion. Iron-inducible ap65-1 transcription, which was induced about twofold in nontransfected cells, was not apparent in transfected cells. The expression profiles of β-tubulin, myb1, and ha-myb1 mRNAs in transfected cells as well as nontransfected cells also varied little as growth progressed for 18 h (Fig. 6, right panel); however, the signal intensity of ap65-1 mRNA in samples from transfected cells was much higher than that in nontransfected cells exposed to iron depletion. Again, iron-inducible ap65-1 gene expression, which was apparent in nontransfected cells, was not observed in transfected cells. No signal was detected in total RNA if the reverse transcriptase was omitted from the RT-PCR amplifications described above (data not shown).
FIG. 6.
Effects of HA-Myb1 overexpression on ap65-1 transcription in T. vaginalis. T. vaginalis T1 cells (lanes 1 and 2) or pFLPha-myb1/TUBneo-transfected cells (lanes 3 and 4) were grown under iron-replete (lanes 1 and 3) or iron-depleted (lanes 2 and 4) conditions for 8 h (left panel) and 18 h (right panel). Expression levels of individual mRNA species were analyzed by RT-PCR. Using 1 microliter of 2×-diluted first-strand cDNA as a template, the ap65-1 signals at 8 h and 18 h were detected after 23 cycles and 21 cycles, respectively. Using 1 μl of 4×-diluted first-strand cDNA as a template, the β-tubulin signal was detected after 26 cycles, and myb1 and ha-myb1 signals were detected after 28 cycles. The PCR products were assayed by agarose gel electrophoresis with ethidium bromide staining.
Differential promoter selection by Myb1.
A search of the database from the T. vaginalis Genome Sequence Project (www.tigr.org/tdb/e2k1/tvg) revealed three Myb1 high-affinity binding sites, each of which is also adjacent to a low-affinity binding site, in the ap65-1 promoter in regions specified as I, II, or III (Fig. 7A). None of these DNA elements exists in distal region IV. Selection of the entry site(s) by HA-Myb1 in the ap65-1 promoter in transfected cells was then studied by a ChIP assay in three separate experiments with consistent results, and the results from one representative experiment are shown (Fig. 7B and C). A specific DNA fragment (referred to as DNA I to DNA IV) was amplified from each of these regions in samples from input DNA, but none of the fragments was amplified from samples pulled down by protein A under all tested conditions. A DNA fragment that spans two regions, either regions I and II or II and III, could not be amplified using primer pair 3f/2r or 2f/1r, respectively (data not shown). DNA I was detected at similar levels in anti-HA pull-down samples from both iron-depleted and iron-replete conditions at 8 h (Fig. 7B), but no DNA II, DNA III, or DNA IV was amplified from the same samples. In contrast, DNA I, DNA II, and DNA III, but not DNA IV, were detected to similar extents in anti-HA pull-down samples from iron-depleted and iron-replete conditions at 18 h (Fig. 7C). In conjunction with the roles of Myb1 in ap65-1 transcription, our results suggest that Myb1 may regulate multifarious transcription of the ap65-1 gene via differential promoter selection.
DISCUSSION
The transcription efficiency of the ap65-1 gene in T. vaginalis greatly varies with changes in the iron supply or the stage of cell growth in a closed culture system (27, 35). Such a gene expression profile is critically regulated in cis by multiple closely spaced DNA elements (27), among which two similar but oppositely oriented DNA sequences spanning the MRE-1/MRE-2r overlap and MRE-2f elements are shown herein to regulate the iron-independent activity of the ap65-1 promoter via repression of basal activity and gradual alleviation of this repressive activity during later growth stages. Moreover, they may also play antagonistic roles in the iron-inducible activity of the same promoter, implying that these two MREs act in concert to control various aspects of ap65-1 transcription in cis. Candidate genes that may regulate ap65-1 transcription in trans via interactions with MRE-2f were identified by Southwestern screening of a cDNA expression library, and a myb1 gene was found to encode a Myb1 protein differentially targeting multiple MRE sites in the ap65-1 promoter to regulate ap65-1 transcription in trans.
Unlike the ap65-1 gene, overall expression of the myb1 gene appears to be constant under our tested conditions (Fig. 3 and 6). Intriguingly, nuclear Myb1 in cells varied with changes in the iron supply (Fig. 3B), implying that nuclear translocation may be one of the key steps by which cells modulate Myb1's function. Given the size of His-tagged rMyb1, estimated to be ∼28 kDa (Fig. 4A), the estimated 35-kDa size of Myb1 (Fig. 3B) is most likely derived from posttranslational modifications. Since Myb1 was exclusively detected to be smaller than 35 kDa in the cytoplasmic fractions of cell lysates, a posttranslational modification such as protein phosphorylation, which regulates the nuclear translocation of some transcription factors in other eukaryotic systems (36), may play a critical role in the nuclear translocation of Myb1.
Although the DNA sequence spanning MRE-1/MRE-2r (ATCGTtaT) only differs from that spanning MRE-2f (ATCGTctT) by two residues (shown in lowercase), the difference in rMyb1 binding to these DNA elements was great (Fig. 4). Given the prevalence of the ATCG sequence in many promoter sites in the genome of T. vaginalis, secondary interactions between Myb1 and its low-affinity binding sites are probably determined by sequences flanking ATCG. As suggested by the DNA binding specificity assay, rMyb1 may simply recognize a 5′ core sequence, ATCG, for initial weak binding, and the binding affinity might be greatly enhanced in the presence of a 3′ accessory sequence, TNTT, on the DNA element. Such a DNA binding specificity is reminiscent of that of the vertebrate c-Myb protein, which recognizes a promoter context with a 5′ core sequence, c/tAACt/gG, for partial binding and a downstream stretch of T's for full activity (10). Intriguingly, either MRE-1/MRE-2r or MRE-2f was sufficient for repression of basal activity (Fig. 1B), implying that Myb1 may impartially target either of these MREs in vivo at the early stage of cell growth. Alternatively, MRE-2f may be a target for another Myb-like transcription factor which also represses basal transcription of the ap65-1 gene. In a previous study (29), a maize Myb protein, C1, was also found to bind two separate sites on an a1 promoter with different affinities in vitro, but either one of these sites was also sufficient for C1-mediated a1 gene activation in vivo.
The preference of Myb1 in selecting a target site(s) in vivo was further complicated by the presence of three potential Myb1 entry sites on the ap65-1 promoter (Fig. 7A). As revealed by RNA expression analysis (Fig. 6), Myb1 may regulate ap65-1 transcription via repression or activation of transcription in iron-depleted cells at different growth stages (Fig. 6). These antagonistic actions were correlated with differential promoter selection (Fig. 7), which in turn may allow Myb1 to alter local promoter structures and/or to interact with different transcription factors in performing distinct transcriptional regulation activities when the environment changes. This speculation remains to be studied. Nonetheless, Myb1's roles in ap65-1 transcription, as defined herein (Fig. 6), are consistent with the roles of its preferred targeting site, MRE-1/MRE-2r, in regulating the transcriptional activity of the ap65-1 promoter reporter gene (Fig. 1B). Temporary repression of basal transcription by Myb1 appears to be a key step by which the parasite controls expression of the ap65-1 gene in a new host environment or during periods of iron limitation. This activity may be sustained until the iron supply or the amount of another favorable factor(s) increases.
As predicted by our results, increased nuclear Myb1 as an effect of the action of iron (Fig. 3B) results in diminished iron-inducible ap65-1 transcription (Fig. 6); however, this prediction is contradictory to the fact that iron enhances ap65-1 transcription (35). It is likely that additional Myb proteins that also target MRE-1/MRE-2r or MRE-2f may counteract the actions of Myb1 to promote iron-inducible ap65-1 transcription. The myb2 gene, which encodes a Myb2 protein targeting the MRE-2r moiety of MRE-1/MRE-2r as well as MRE-2f (J. H. Tai, unpublished observation), is one of the candidates which can be used to test this hypothesis. The notion that multiple Myb proteins may participate in ap65-1 transcription in coordination or in competition is in contrast to the actions of vertebrate A-Myb, B-Myb, and c-Myb, which exhibit similar DNA binding specificities but transactivate distinct sets of endogenous genes in animal cells (19, 28).
In summary, we have identified and characterized a gene-specific transcription factor, Myb1, in T. vaginalis and showed that Myb1 probably critically regulates multifarious transcription of the ap65-1 gene. Information derived from this study will allow us to further investigate not only the signaling pathway leading to the activation of Myb1 but also the target genes regulated by Myb1 in this parasite.
Acknowledgments
We thank Ramarao Vepachedo, Hann Tsu, and Guan-Yi Wu for technical assistance, David Brooks and Yi-Juiang Chern for critical reviews of the manuscript, and Dan Chamberlin for English editing of the manuscript. The monoclonal anti-malic enzyme antibody 15D7 and the anti-cytosolic malic enzyme antibody were obtained from Guy Brugerolle at Universite Blaise Pascal, Clermont-Ferrand, France, and Ivan Hrdy at Charles University, Czech Republic, respectively. We also thank The Institute of Genomic Research (TIGR) for the use of preliminary T. vaginalis genome sequence data from the T. vaginalis Genome Sequence Project through the website at http://www.tigr.org.
This work was supported by grants from the National Science Council (NSC92-2314-B-001-009 and NSC93-2314-B-001-003) and IBMS, Academia Sinica.
REFERENCES
- 1.Alderete, J. F., J. L. O'Brien, R. Arroyo, J. A. Engbring, O. Musatovova, O. Lopez, C. Lauriano, and J. Nguyen. 1995. Cloning and molecular characterization of two genes encoding adhesion proteins involved in Trichomonas vaginalis cytoadherence. Mol. Microbiol. 17:69-83. [DOI] [PubMed] [Google Scholar]
- 2.Arroyo, R., A. Gonzalez-Robles, A. Martinez-Palomo, and J. F. Alderete. 1993. Signaling of Trichomonas vaginalis for amoeboid transformation and adhesion synthesis follows cytoadherence. Mol. Microbiol. 7:299-309. [DOI] [PubMed] [Google Scholar]
- 3.Bergholtz, S., T. O. Andersen, K. B. Andersson, J. Borrebaek, B. Luscher, and O. S. Gabrielsen. 2001. The highly conserved DNA-binding domains of A-, B- and c-Myb differ with respect to DNA-binding, phosphorylation and redox properties. Nucleic Acids Res. 29:3546-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brugerolle, G., G. Bricheux, and G. Coffe. 2000. Immunolocalization of two hydrogenosomal enzymes of Trichomonas vaginalis. Parasitol. Res. 86:30-35. [DOI] [PubMed] [Google Scholar]
- 5.Chou, C. F., and J. H. Tai. 1996. Simultaneous extraction of DNA and RNA from nuclease rich pathogenic protozoan Trichomonas vaginalis. BioTechniques 20:790-791. [DOI] [PubMed] [Google Scholar]
- 6.Crouch, M. V., and J. F. Alderete. 2001. Trichomonas vaginalis has two fibronectin-like iron-regulated genes. Arch. Med. Res. 32:102-107. [DOI] [PubMed] [Google Scholar]
- 7.Cudmore, S. L., K. L. Delgaty, S. F. Hayward-McClelland, D. P. Petrin, and G. E. Garber. 2004. Treatment of infections caused by metronidazole-resistant Trichomonas vaginalis. Clin. Microbiol. Rev. 17:783-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dolezal, P., S. Vanacova, J. Tachezy, and I. Hrdy. 2004. Malic enzymes of Trichomonas vaginalis: two enzyme families, two distinct origins. Genes 329:81-92. [DOI] [PubMed] [Google Scholar]
- 9.Dunne, R. L., L. A. Dunn, P. Upcroft, P. J. O'Donoghue, and J. A. Upcroft. 2003. Drug resistance in the sexually transmitted protozoan Trichomonas vaginalis. Cell Res. 13:239-249. [DOI] [PubMed] [Google Scholar]
- 10.Ganter, B., S. T. Chao, and J. S. Lipsick. 1999. Transcriptional activation by the Myb proteins requires a specific local promoter structure. FEBS Lett. 460:401-410. [DOI] [PubMed] [Google Scholar]
- 11.Garcia, A. F., T. H. Chang, M. Benchimol, D. J. Klumpp, M. W. Lehker, and J. F. Alderete. 2003. Iron and contact with cells induce expression of adhesions on surface of Trichomonas vaginalis. Mol. Microbiol. 47:1207-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grasser, F. A., K. LaMontagne, L. Whittaker, S. Stohr, and J. S. Lipsick. 1992. A highly conserved cysteine in the v-Myb DNA-binding domain is essential for transformation and transcriptional trans-activation. Oncogene 7:1005-1009. [PubMed] [Google Scholar]
- 13.Guehmann, S., G. Vorbrueggen, F. Kalkbrenner, and K. Moelling. 1992. Reduction of a conserved Cys is essential for Myb DNA-binding. Nucleic Acids Res. 20:2279-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harlow, E., and D. Land. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 15.Hochheimer, A., and R. Tjian. 2003. Diversified transcription initiation complexes expand promoter selectivity and tissue-specific gene expression. Genes Dev. 17:1309-1320. [DOI] [PubMed] [Google Scholar]
- 16.Hrdy, I., and M. Muller. 1995. Primary structure of the hydrogenosomal malic enzyme of Trichomonas vaginalis and its relationship to homologous enzymes. J. Eukaryot. Microbiol. 42:593-603. [DOI] [PubMed] [Google Scholar]
- 17.Lau, A., D. Liston, S. Vanacova, and P. Johnson. 2003. Trichomonas vaginalis initiator binding protein, IBP39, contains a novel DNA binding motif. Mol. Biochem. Parasitol. 130:167-171. [DOI] [PubMed] [Google Scholar]
- 18.Lehker, M. W., R. Arroyo, and J. F. Alderete. 1991. The regulation by iron of the synthesis of adhesins and cytoadherence levels in the protozoan parasite Trichomonas vaginalis. J. Exp. Med. 174:311-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lei, W., J. J. Rushton, L. M. Davis, F. Liu, and S. A. Ness. 2004. Positive and negative determinants of target gene specificity in myb transcription factors. J. Biol. Chem. 279:29519-29527. [DOI] [PubMed] [Google Scholar]
- 20.Liston, D. R., and P. J. Johnson. 1999. Analysis of a ubiquitous promoter element in a primitive eukaryote: early evolution of the initiator element. Mol. Cell. Biol. 19:2380-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liston, D. R., J. C. Carrero, and P. J. Johnson. 1999. Upstream regulatory sequences required for expression of the Trichomonas vaginalis α-succinyl CoA synthetase gene. Mol. Biochem. Parasitol. 104:323-329. [DOI] [PubMed] [Google Scholar]
- 22.Liston, D. R., A. O. T. Lau, D. Ortiz, S. T. Smale, and P. J. Johnson. 2001. Initiator recognition in a primitive eukaryote: IBP39, an initiator binding protein from Trichomonas vaginalis. Mol. Cell. Biol. 21:7872-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, Y., C. Kung, J. Fishburn, A. Z. Ansari, K. M. Shokat, and S. Hahn. 2004. Two cyclin-dependent kinases promote RNA polymerase II transcription and formation of the scaffold complex. Mol. Cell. Biol. 24:1721-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muller, F., and L. Tora. 2004. The multicolored world of promoter recognition complexes. EMBO J. 23:2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Brien, J. L., C. M. Lauriano, and J. F. Alderete. 1996. Molecular characterization of a third malic enzyme-like AP65 adhesin gene of Trichomonas vaginalis. Microb. Pathog. 20:335-349. [DOI] [PubMed] [Google Scholar]
- 26.Ogata, K., S. Morikawa, H. Nakamura, A. Sekikawa, T. Inoue, H. Kanai, A. Sarai, S. Ishii, and Y. Nishimura. 1994. Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell 79:639-648. [DOI] [PubMed] [Google Scholar]
- 27.Ong, S. J., S. C. Huang, H. W. Liu, and J. H. Tai. 2004. Involvement of multiple DNA elements in iron-inducible transcription of the ap65-1 gene in the protozoan parasite Trichomonas vaginalis. Mol. Microbiol. 52:1721-1730. [DOI] [PubMed] [Google Scholar]
- 28.Rushton, J. J., L. M. Davis, W. Lei, X. Mo, A. Leutz, and S. A. Ness. 2003. Distinct changes in gene expression induced by A-Myb, B-Myb and c-Myb proteins. Oncogene 22:308-313. [DOI] [PubMed] [Google Scholar]
- 29.Sainz, M. B., E. Grotewold, and V. L. Chandler. 1997. Evidence for direct activation of an anthocyanin promoter by the maize C1 protein and comparison of DNA binding by related Myb domain proteins. Plant Cell 9:611-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schumacher, M. A., A. O. Lau, and P. J. Johnson. 2003. Structural basis of core promoter recognition in a primitive eukaryote. Cell 115:413-424. [DOI] [PubMed] [Google Scholar]
- 31.Smale, S. T., and J. T. Kadonaga. 2003. The RNA polymerase II core promoter. Annu. Rev. Biochem. 72:449-479. [DOI] [PubMed] [Google Scholar]
- 32.Solano, R., A. Fuertes, L. Sanchez-Pulido, A. Valencia, and J. Paz-Ares. 1997. A single residue substitution causes a switch from the dual DNA binding specificity of plant transcription factor MYB.Ph3 to the animal c-MYB specificity. J. Biol. Chem. 272:2889-2895. [DOI] [PubMed] [Google Scholar]
- 33.Sorvillo, F., L. Smith, P. Kerndt, and L. Ash. 2001. Trichomonas vaginalis, HIV and African-Americans. Emerg. Infect. Dis. 7:927-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stracke, R., M. Werber, and B. Weisshaar. 2001. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4:447-456. [DOI] [PubMed] [Google Scholar]
- 35.Tsai, C. D., H. W. Liu, and J. H. Tai. 2002. Characterization of an iron-responsive promoter in the protozoan pathogen Trichomonas vaginalis. J. Biol. Chem. 277:5153-5162. [DOI] [PubMed] [Google Scholar]
- 36.Vandromme, M., C. Gauthier-Rouviere, N. Lamb, and A. Fernandez. 1996. Regulation of transcription factor localization: fine-tuning of gene expression. Trends Biochem. Sci. 21:59-64. [PubMed] [Google Scholar]
- 37.Walker, M. D. 1996. Screening expression cDNA libraries, p. 63-66. In K. Docherty (ed.), Gene transcription: DNA binding proteins. Wiley Interscience, New York, N.Y.
- 38.Weston, K. 1998. Myb proteins in life, death and differentiation. Curr. Opin. Genet. Dev. 8:76-81. [DOI] [PubMed] [Google Scholar]