Abstract
Azf1 activates CLN3 transcription in Saccharomyces cerevisiae cells growing in glucose. Paradoxically, other studies have shown Azf1 to be almost undetectable in glucose-grown cells. Microarray experiments showed that Azf1 activates nonoverlapping gene sets in different carbon sources: in glucose, Azf1 activates carbon and energy metabolism genes, and in glycerol-lactate, Azf1 activates genes needed for cell wall maintenance. Consistent with the decreased expression of cell wall maintenance genes observed with azf1Δ mutants, we observed a marked growth defect in the azf1Δ cells at 37°C in nonfermentable medium. Cell wall integrity assays, such as sensitivity to calcofluor white, sodium dodecyl sulfate, or caffeine, confirmed cell wall defects in azf1Δ mutants in nonfermentable medium. Gel shift experiments show that Azf1 binds to DNA elements with the sequence AAAAGAAA (A4GA3), a motif enriched in the promoters of Azf1-sensitive genes and predicted by whole-genome studies. This suggests that many of the Azf1-dependent transcripts may be regulated directly by Azf1 binding. We found that the levels of Azf1 protein in glucose-grown cells were comparable to Azf1 levels in cells grown in glycerol-lactate; however, this could only be demonstrated with a cell extraction procedure that minimizes proteolysis. Glucose produces conditions that destabilize the Azf1 protein, a finding that may reflect a glucose-induced change in Azf1 tertiary or quaternary structure.
Free-living organisms, such as the yeast Saccharomyces cerevisiae, have evolved in a continuously changing environment, switching between fermentative and respiratory metabolism. Glucose is catabolized by fermentation. As glucose is exhausted, cells enter the diauxic shift, during which proliferation ceases and the cell switches from fermentative to oxidative metabolism. This is followed by a slow post-log phase of oxidative growth. As the remaining carbon is exhausted, this gradually merges into a G0-like state, stationary phase. Progression toward stationary phase brings about major reconfigurations of yeast cell structure and metabolism (5). Stationary-phase cells have thicker cell walls and are more resistant to heat and other stress than log-phase cells growing by fermentation (5, 7, 18). Although still slowly growing, cells in nonfermentable carbon sources share many of the properties of stationary-phase cells (18).
Carbon source has a considerable effect on the transcription of yeast genes. Microarray work by DeRisi et al. demonstrated that nearly a third of the yeast genome is regulated by the change from growth on glucose to growth on ethanol at the diauxic shift (4). Many of the expected changes in gene expression are observed. Transcripts necessary for glycolysis are downregulated as glucose is exhausted, while genes involved in oxidative growth and carbohydrate storage are induced (4). Although the regulation of some of these genes has been well studied, for a great many genes the mechanism for carbon source regulation remains unknown.
Azf1 is an asparagine-rich zinc finger protein first identified as a high-copy suppressor of a mutation in the mitochondrial RNA polymerase gene RPO41. Azf1 was identified as a transcription factor based on its zinc finger domain and nuclear localization (2, 19). We confirmed the transcriptional role, showing that Azf1 binds to the CLN3 promoter to activate CLN3 in response to glucose (16). Glucose induction of CLN3 helps propel cells through G1 in response to positive growth conditions (22). Our demonstration of an Azf1 function in glucose produces a paradox because Stein et al. reported that Azf1 levels are almost undetectable in glucose and are expressed at much higher levels in nonfermentable medium (19).
In this study we report that Azf1 regulates two distinct sets of genes: a set of genes in glucose that is involved in growth and metabolism and a set in glycerol-lactate that maintains cell wall integrity. As predicted by these data, Azf1 is required for proper cell wall integrity during growth on nonfermentable medium. A search of the promoter regions of both sets of Azf1-dependent genes showed enrichment in the DNA sequence AAAAGAAA (A4GA3) that comprises an Azf1 binding motif. Biochemical experiments show that Azf1 protein levels in glucose are similar to those in nonfermentable medium but that the stability of Azf1 is dramatically decreased in glucose. This suggests a model in which a nutrient-sensitive process affects Azf1, switching its function from controlling growth and metabolism to activating cell wall maintenance genes.
MATERIALS AND METHODS
Yeast strains and growth conditions.
The media used were yeast extract-peptone (YEP) (1% yeast extract, 2% peptone) and synthetic (S) (0.67% yeast nitrogen base) with the indicated carbon source at 2%. Strains used in this study were BY4741 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0), TTL9 (azf1Δ::URA3 in BY4741), DS10 (MATα his3-11,15 leu2-3,112 lys1 lys2 ura3-52 trp1Δ) (1), and TLN81 (azf1Δ::KAN in DS10) (16). BY4741 was obtained from Research Genetics. AZF1 was deleted in TTL9 by PCR amplification of the region spanning the AZF1 deletion in strain TLN60 (16) and by use of this fragment to delete AZF1 in BY4741.
Labeled-cRNA preparation and microarray hybridization.
Total yeast RNA was isolated as described previously (6). cDNA and labeled cRNA were generated from total yeast RNA according to the Affymetrix protocol. Briefly, first-strand cDNA was generated using a T7-oligo(dT)24 primer and SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA). Second-strand cDNA synthesis was performed using Escherichia coli DNA ligase, E. coli DNA polymerase I, and RNase H, followed by incubation with T4 DNA polymerase. After phenol-chloroform cleanup and ethanol precipitation of cDNA, biotin-labeled antisense cRNA was generated using an Enzo BioArray high-yield RNA transcript labeling kit. Labeled cRNA was purified using a QIAGEN RNeasy mini kit, concentrated by ethanol precipitation, and fragmented in RNA fragmentation buffer (40 mM Tris-acetate, pH 8.1, 100 mM potassium acetate, 30 mM magnesium acetate). Fragmented cRNA was then mixed with hybridization controls and hybridized to a yeast genome S98 array (Affymetrix) at 45°C with rotation (60 rpm) for 16 h. Microarrays were then washed and stained with streptavidin-phycoerythrin in a GeneChip Fluidics Station 400.
Microarray data analysis.
Affymetrix yeast genome S98 arrays were scanned using an Agilent gene array scanner and Microarray Suite 5.0. The Microarray Suite-generated .CEL files were then analyzed using dChip 1.3 (12). Intensity values were normalized across 12 independent microarrays by using dChip's invariant set normalization method (11). Model-based analysis, including log2 transformation of expression indexes, using the perfect match-mismatch model was performed using values from three independent microarrays for each strain/condition. Probe sets called absent greater than 80% of the time within both the experimental and the baseline group were excluded from further analysis. Only probe sets representing mRNAs were analyzed, as these are the only transcripts efficiently converted to cDNA during the first step of labeled-cRNA preparation (see Table S1 in the supplemental material). Probe sets were considered upregulated in azf1Δ if the mean azf1Δ expression value minus the mean wild-type expression value was greater than or equal to 1 (twofold induction) or downregulated in azf1Δ if the mean wild-type expression value minus the mean azf1Δ expression value was greater than or equal to 1 (twofold repression). The genes YBR178W and YBR277C were found to be Azf1 dependent in glucose and glycerol-lactate, respectively, but were not included in the analysis (see Fig. 2) because they were both dubious open reading frames (as classified by the Saccharomyces Genome Database [SGD]) and their upstream DNA regions completely overlapped another open reading frame. Overrepresented DNA motifs were extracted using the oligonucleotide analysis pattern discovery program in Regulatory Sequence Analysis Tools (20).
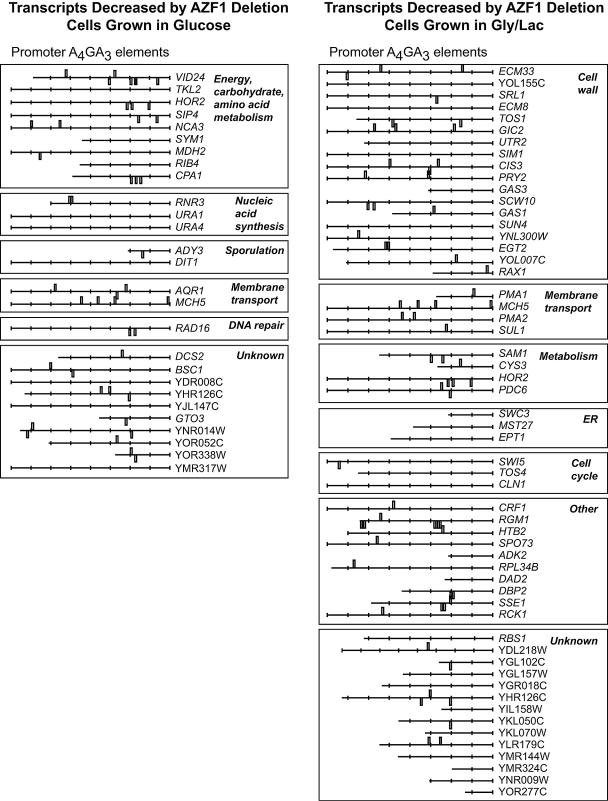
FIG. 2.
Azf1 regulates distinct gene sets depending on carbon source. Azf1-dependent genes were determined as described in Materials and Methods and are grouped by functional categories based on gene ontology information at SGD (http://www.yeastgenome.org/). Each Azf1-dependent group was examined for enrichment of promoter sequences by use of Regulatory Sequence Analysis Tools (RSAT, http://rsat.scmbb.ulb.ac.be/rsat/). AAAAGAAA was the most significant enriched sequence in both the glucose and the glycerol-lactate (Gly/Lac) gene set, with significance indexes of 3.72 and 2.88, respectively. The significance index is a −log10 conversion of the expected number of patterns that would be returned at random for the motif's P value. Each gene is accompanied by a representation of its promoter (up to 800 bp upstream, with each tick marking 100 bp). Rectangles represent the positions of A4GA3 elements. ER, endoplasmic reticulum.
Cell wall integrity assays.
For cell wall integrity assays, cells were grown in YEP-dextrose (YEPD) to log phase and then the indicated numbers of cells were spotted onto the plates in 10-μl drops. The plates used were YEP medium with 2% glycerol and 2% lactate added as the carbon source, containing 0.1 mg/ml calcofluor white, 15 mM caffeine, or 0.01% sodium dodecyl sulfate (SDS), as indicated. To test for zymolyase resistance, wild-type and azf1Δ cultures were grown to mid-log phase in the indicated medium at 30°C, and aliquots (7.5 × 106 cells) were harvested, washed with 1.5 ml water, and resuspended in 0.5 ml 10 mM Tris, pH 7.4, containing 0.05 U/μl zymolyase (Zymo Research, Orange, CA). The optical density at 600 nm (OD600) was measured immediately, and the cells were incubated at 37°C with optical density measurements at the indicated intervals. The OD600 at each time point for each sample was divided by the initial OD600; error bars represent the standard deviations of this ratio across triplicate experiments.
Gel shift assays.
Gel shift assays were performed as described previously (17). Binding reactions were carried out in a 20-μl reaction volume containing 20 mM HEPES (pH 8.0), 0.1% Nonidet P-40, 8% glycerol, 2 mM dithiothreitol, 65 mM KCl, 2.5 mM MgCl2, 2 μg poly(dI-dC), 10 fmol of labeled DNA probe, and 10 μg of yeast extract. The protein extract was first incubated on ice with a molar excess of the cold competitor (when added) for 5 min followed by a 15-min incubation at room temperature in the presence of labeled probe. The labeled DNA probe was made by annealing two complementary oligonucleotides corresponding to the region of DBP2 from −225 to −181 with 5′ overhanging G residues at each end. These overhangs were then filled in with [α-32P]dCTP and Klenow fragment. Antibodies against Azf1, where present, were included at 1 μl per lane.
Fluorescence microscopy.
Fluorescence microscopy was performed with a Zeiss Axioplan 2 microscope with a 63× oil immersion objective as described previously (8).
Western blots.
Protein extracts were prepared as described for the gel shift experiments (protocol A) or by a simpler method (protocol B). For protocol B, yeast samples were collected, washed, and resuspended in ice cold extraction buffer (200 mM Tris [pH 8], 150 mM ammonium sulfate, 10% glycerol, 1 mM EDTA, 200 nM dithiothreitol) with protease inhibitors (200 μg/ml phenylmethylsulfonyl fluoride, 300 ng/ml leupeptin, 1 μg/ml pepstatin A, and 1 μg/ml TPCK [tosylsulfonyl phenylalanyl chloromethyl ketone]). Two-thirds volume of glass beads was added and samples were vortexed for 2.5 min, placed on ice for 1 min, and then vortexed for another 2.5 min. Cellular debris was removed by 3 min of centrifugation at 16,000 × g. Additional insoluble material was removed from the supernatant by centrifugation for 10 min at 16,000 × g. The supernatant was immediately frozen and stored at −70°C. For both protocols, protein concentration was determined using a Bio-Rad protein assay kit. Western blotting was performed as described previously (6) by using antibodies against Azf1 provided as a generous gift from Thomas Lisowsky (19). For each experiment, an equal amount of protein was loaded onto each lane, and equal gel loading and transfer to nitrocellulose was confirmed by Ponceau S staining.
RESULTS
Deletion of AZF1 produces a stress-sensitive phenotype in nonfermentable medium.
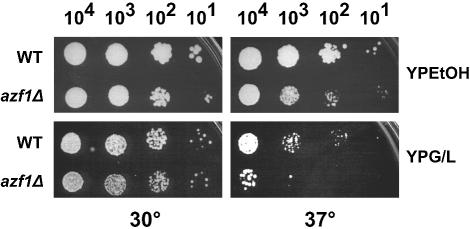
We were unable to find any marked phenotype for azf1Δ mutations in glucose medium. However, in nonfermentable carbon sources, deletion of AZF1 had a modest effect on growth at 30°C and produced a pronounced growth defect at 37°C (Fig. 1). This indicates that in addition to upregulating CLN3 in response to glucose, AZF1 has a role to play during growth on poor carbon sources.
FIG. 1.
Deletion of AZF1 produces a growth defect on nonfermentable medium at 37°C. Wild-type (WT) (BY4741) and azf1Δ (TTL9) mutant strains were plated on the media indicated in serial 10-μl dilutions. Plates were incubated at 30°C or 37°C as indicated. YPEtOH, YEP-ethanol; YPG/L, YEP-glycerol-lactate.
Carbon source determines the gene sets affected by Azf1.
Since we have characterized Azf1 as a glucose-dependent transcriptional activator, a reasonable approach to understanding function was to determine what transcripts are affected by AZF1 deletion in glucose. The phenotype in nonfermentable medium also made it important to determine the effect of AZF1 deletion in nonfermentable media. We used Affymetrix DNA microarrays and triplicate independent experiments to compare azf1Δ mutant and wild-type transcripts from cells at mid-log phase in either glycerol-lactate or glucose medium. Transcripts downregulated twofold or greater in the azf1Δ mutant were considered AZF1 dependent. A total of 28 genes were AZF1 dependent in cells grown in glucose, while 55 were AZF1 dependent in glycerol-lactate. Surprisingly, only three genes were dependent on AZF1 under both conditions (Fig. 2; see Table S2 in the supplemental material).
We used SGD gene ontology annotations to assign functions to the genes in these sets (Fig. 2). For cells growing in glucose medium, the largest group of Azf1-dependent genes was involved in carbon metabolism and/or energy production. This group included SIP4, involved in regulating gluconeogenic genes (10, 21), and VID24, encoding a regulator of fructose-1,6-bisphosphatase degradation (3). This is consistent with a role for Azf1 in regulating processes related to growth.
In glycerol-lactate, AZF1 deletion affected genes involved in a completely different function. The majority of Azf1-dependent genes in glycerol-lactate medium encode proteins involved in cell wall organization and biogenesis or proteins found at the cell surface. For example, expression of GAS1 and GAS3, encoding β-1,3-glucanosyl transferases (14, 15), was decreased in azf1Δ cells growing in glycerol-lactate but was not decreased in cells growing in glucose. These results reinforce the idea that Azf1 responds to the carbon source and suggest a role in regulating metabolism during growth in glucose and in cell wall buildup as glucose is depleted and cells switch to growth in nonfermentable medium.
Azf1 is necessary for cell wall integrity in poor carbon sources.
The reduction in metabolism-linked transcripts in azf1Δ mutants in glucose is apparently not enough to impact growth, since these cells appear to grow normally in glucose. However, the reduction in cell wall transcripts observed with growth in glycerol-lactate suggested the possibility that loss of Azf1 could lead to cell wall defects. Such defects tend to have more-severe consequences at higher temperatures, due to increased intracellular pressure associated with higher temperature (7). This hypothesis is consistent with the temperature-sensitive phenotype of azf1Δ mutant cells in poor medium.
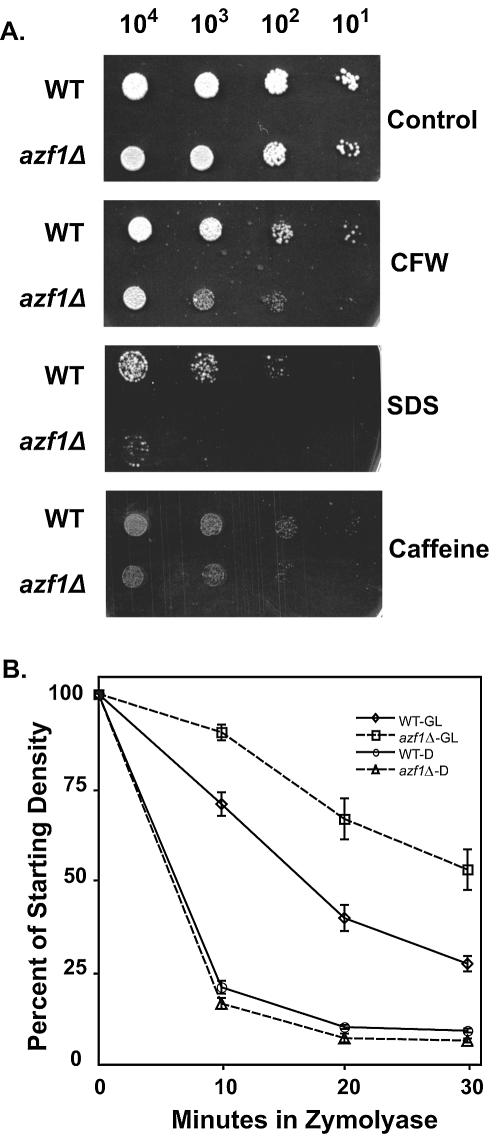
To test this hypothesis, we examined the ability of azf1Δ cells to grow in the presence of inhibitors of cell wall formation and maintenance: calcofluor white, SDS, and caffeine. Cells lacking AZF1 were more sensitive to calcofluor white than were the wild-type controls, regardless of carbon source. In addition, we observed the predicted sensitivity to SDS and caffeine; this was limited to cells grown on poor carbon sources (Fig. 3).
FIG. 3.
Deletion of AZF1 produces cell wall defects. (A) Wild-type (WT) (BY4741) and azf1Δ (TTL9) mutants were serially diluted and plated in 5-μl drops on YEP-glycerol-lactate media with the indicated cell wall disruptors as described in Materials and Methods. Plates were incubated for 2 days at 30°C. CFW, calcofluor white. (B) WT (BY4741) and azf1Δ (TTL9) mutants were grown to mid-log phase in either YEP-glycerol-lactate (GL) or YEPD (D) medium. Aliquots were resuspended in zymolyase (0.05 U/μl) as described in Materials and Methods, and cell disruption by zymolyase was measured by monitoring the OD600 at the indicated intervals. Error bars represent the standard deviations of this ratio across triplicate experiments.
Loss of β-1,3-glucan linkages generally weakens the cell wall but also produces a seemingly paradoxical resistance to the cell wall-degrading enzyme zymolyase, a β-1,3-glucanase. This is likely because a cell without these linkages lacks the targets for zymolyase (13). Our observation that cells in glycerol-lactate are dependent on Azf1 for proper GAS1 and GAS3 expression, encoding β-1,3-glucanosyltransferases, suggests that these mutants should have fewer β-1,3-glucan linkages and should be zymolyase resistant. This zymolyase resistance in poor carbon medium was confirmed using experiments monitoring cell wall degradation by zymolyase in both glucose-grown and glycerol-lactate-grown cells (Fig. 3). As expected, this phenotype was not evident with glucose-grown azf1Δ cells.
Taken together, our results indicate that Azf1 shifts roles as cells change carbon source. During log-phase growth in glucose, Azf1 supports growth processes. As glucose is exhausted, Azf1 then changes roles to strengthening and maintaining the cell wall.
Azf1 recognizes the A4GA3 motif upstream of its target genes.
We examined the upstream regions of the genes that are coregulated by Azf1 to determine whether these genes have short oligonucleotide sequences in common. Computational analysis of the promoter regions by using Regulatory Sequence Analysis Tools (RSAT, http://rsat.scmbb.ulb.ac.be/rsat/) revealed enrichment of a motif with the sequence AAAAGAAA (A4GA3). This site was found in both the glucose and the glycerol-lactate group at frequencies much higher than expected by random chance (Fig. 2). We have previously reported that the Azf1 binding site in the CLN3 promoter is the related sequence AAGAAAAA (A2GA5) (16). However, a large-scale chromatin immunoprecipitation experiment determined the Azf1 consensus site to be GA5GMA7 (where M is A or C) (9). This matches the A4GA3 sequence enriched in the Azf1-dependent genes. A closer look at the Azf1 binding region of the CLN3 promoter revealed that A4GA3 motifs are embedded within the tandem A2GA5 repeats. Therefore, Azf1 binding to either motif can explain our previous results.
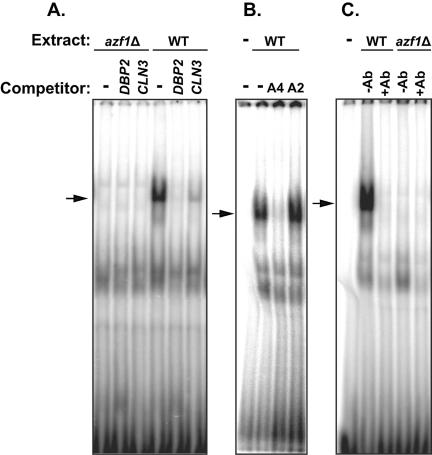
To determine whether the identified A4GA3 motifs provide a binding site for Azf1, we selected a region from the DBP2 promoter containing a pair of A4GA3 motifs as a gel shift probe. DBP2 expression is dependent on Azf1 in glycerol-lactate but not in glucose, and the DBP2 promoter contains no A2GA5 sequences. As demonstrated in Fig. 4A, this A4GA3 probe produced a strong, specific protein/DNA complex that was completely absent with azf1Δ extracts. The gel shift band could be competed with the unlabeled A4GA3 oligonucleotide from DBP2 and with the Azf1 binding region of CLN3 used in the previous report that contains both A4GA3 and A2GA5 motifs. We also found that while a synthetic oligonucleotide containing A4GA3 sequences could effectively compete for Azf1 binding, the same oligonucleotide rearranged to produce A2GA5 sequences could not (Fig. 4B). To demonstrate that Azf1 protein is a part of the protein/DNA complex observed with our assays, we performed gel shift experiments using the antibody against the Azf1 protein. This antibody abolished the complex, consistent with disruption of a complex between Azf1 and the DNA probe (Fig. 4C). We conclude that Azf1 is bound directly to the DNA and that the antibody either alters the conformation of Azf1 or binds at the DNA binding site of Azf1, preventing formation of the Azf1/A4GA3 DNA complex.
FIG. 4.
Azf1 binds to A4GA3 motifs. Cell extracts for gel shift assays were prepared from wild-type (WT) (DS10) and isogenic azf1Δ (TLN81) cells grown in YEP-glycerol-lactate as described in Materials and Methods. A double-stranded DNA fragment corresponding to the −225 to −181 region of the DBP2 promoter, carrying a pair of A4GA3 motifs (antiparallel on the opposite strand), was used as a gel shift probe. Unlabeled competitor was added at a 100-fold excess as indicated. The competitor was the unaltered probe sequence (labeled DBP2 in panel A or A4 in panel B), the Azf1 binding −626 to −569 region of the CLN3 promoter (labeled CLN3 in panel A), or the −225 to −181 DBP2 fragment in which the G residues were moved so as to change the A4GA3 motifs into A2GA5 elements (competitor A2 in panel B). The arrow indicates a specific band lost in the azf1Δ extract lanes and not competed by the A2GA5 oligonucleotide. (C) Gel shift in which Azf1 antibodies were included (+Ab).
Our experiments demonstrate the ability of the A4GA3 motifs found enriched in Azf1-dependent genes to interact with Azf1. Despite the finding that Azf1 regulates very distinct groups of genes under different conditions, the same potential Azf1 binding sites are found in the promoters for both gene sets.
Azf1 is located in the nucleus during growth.
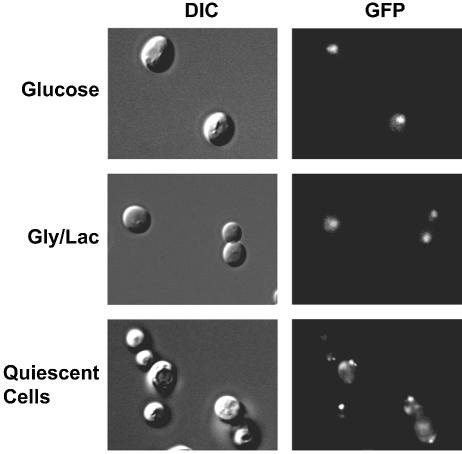
We have not detected glucose-dependent changes in the ability of Azf1 to bind DNA (17). However, this may not be significant because DNA binding can be regulated by changes in nuclear localization. This would not be detected in gel shift experiments in which protein compartmentalization is destroyed. To address this, we used green fluorescent protein (GFP)-tagged Azf1 to monitor localization. This chromosomal GFP-tagged Azf1 appears to be functional in that this allele does not produce the growth defects in poor medium at 37°C normally observed with AZF1 deletion. Consistent with a transcriptional role in both carbon sources, we found Azf1 to be predominantly nuclear in cells growing in either glucose or glycerol-lactate (Fig. 5). However, the pattern changed in quiescent, nutrient-depleted cells. After 2 days in culture, we observed Azf1 throughout the cell, often with punctate fluorescence in the cytoplasm. This is not simply the result of a shift to oxidative metabolism of ethanol after glucose is exhausted, as Azf1 was nuclear in cells growing logarithmically on YEP-ethanol (not shown). These results show that Azf1 is present in the nucleus under both types of growth conditions examined.
FIG. 5.
Azf1 is nuclear in growing cells. Cells expressing GFP-tagged Azf1 (0423) were examined using a 63× objective with differential interference contrast (DIC) (left) and fluorescence microscopy (right) in log phase in S-glucose (SD) medium, log phase in S-glycerol-lactate (Gly/Lac) medium, or after 2 days of growth in SD medium (quiescent cells).
The Azf1 protein is affected by glucose.
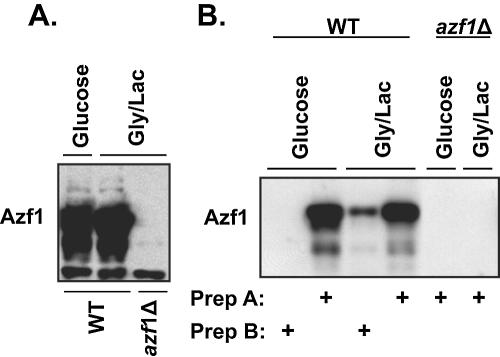
Azf1 plays divergent roles depending on the carbon source, yet Azf1 is in the nucleus and is able to bind DNA in both fermentable and nonfermentable media. We hypothesized that nutrient conditions affect some other property of Azf1. Stein et al. (19) reported that Azf1 levels are almost undetectable in glucose, yet our results point to Azf1-dependent genes in glucose-grown cells. We used Western blotting to examine this paradox more carefully. In contrast to the previous report, we found that Azf1 protein was present at approximately equal levels in both rich and poor media. However, the observed level of Azf1 was strongly dependent on extraction technique. When we used a technique that we have used for isolating nuclear DNA binding proteins (17) (protocol A), we consistently found Azf1 protein present at roughly equal levels in either glucose-grown cells or glycerol-lactate-grown cells (Fig. 6A). However, this result was observed only with this protein extraction protocol, involving cell disruption in liquid nitrogen by use of a cocktail of protease inhibitors and ammonium sulfate as a stabilizer. A simpler extraction method (protocol B) produced results that matched those of Stein and colleagues, in which Azf1 was found only in glycerol-lactate-grown cells (Fig. 6B). We conclude that Azf1 is present in cells growing in both fermentable and nonfermentable carbon sources but that glucose changes the state of the protein or its surroundings so that Azf1 becomes very unstable or less extractable.
FIG. 6.
Azf1 protein is present at equal levels in fermentable and nonfermentable media. Proteins were isolated from wild-type (WT) (BY4741) and isogenic azf1Δ (TTL9) cells grown in YEPD or YEP-glycerol-lactate (Gly/Lac). (A) Proteins were isolated using protocol A as described in Materials and Methods and loaded onto an SDS-polyacrylamide gel (20 μg/lane) for Western blotting. (B) Proteins were isolated using protocol A or B as indicated for Western blotting (20 μg/lane). Prep, preparation.
Azf1 protein stability.
A time course experiment showed that shifting cells from glycerol-lactate to glucose medium produced a rapid reduction in the recovery of Azf1 protein with protocol B (Fig. 7). We hypothesized that glucose destabilizes Azf1 and that protocol A prevents loss of Azf1 because of the very low temperatures used and ammonium sulfate fractionation at the later stages of the preparation. Azf1 was relatively stable in the extracts prepared using protocol A, and we saw little degradation in extracts from either glycerol-lactate-grown or glucose-grown cells incubated for more than an hour at 30°C (Fig. 8A). This indicates that there is no difference in intrinsic stability of Azf1 obtained from cells grown in the two carbon sources.
FIG. 7.
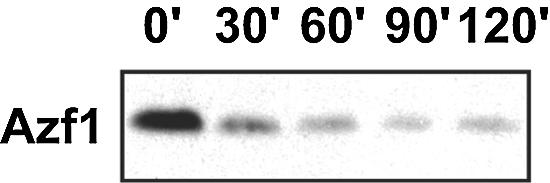
Azf1 protein recovery is altered by carbon source. Wild-type (BY4741) cells were grown to mid-log phase in YEP-glycerol-lactate and resuspended in YEPD. Samples were collected at the indicated times (in minutes) after the medium change for protein isolation (protocol B) and Western blotting (20 μg/lane).
FIG. 8.
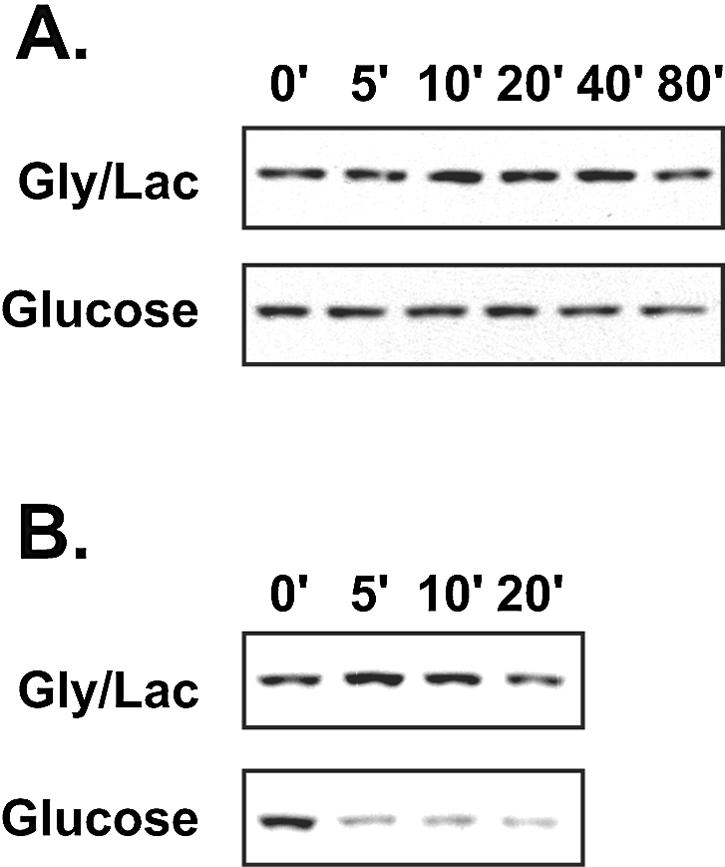
Azf1 stability is affected by carbon source and extraction conditions. (A) Extracts were isolated from wild-type (BY4741) cells grown in YEP-glycerol-lactate (Gly/Lac) or YEPD by use of protocol A as described in Materials and Methods. The extract was incubated at 30°C, and aliquots (20 μg) were placed on ice at the indicated times (in minutes) prior to Western blotting. (B) The YEP-glycerol-lactate extract mentioned above (20 μg/lane) was mixed with extract isolated via protocol B (20 μg/lane) from either YEP-glycerol-lactate-grown or YEPD-grown azf1Δ mutants and incubated as described above prior to Western blotting.
One possibility is that glucose increases the activity of an Azf1 protease. In this model, the protease is removed or inactivated with protocol A but not with the simpler extraction process. To test this, we added some extract made using protocol B to the stable Azf1 prepared with protocol A (Fig. 8B). To avoid the complication of multiple sources of Azf1, the protocol B extracts were made from azf1Δ cells. Consistent with the model, the protocol B extract rapidly destroyed the Azf1, but only when it was prepared from cells grown in glucose.
DISCUSSION
Previous work identified Azf1 as a DNA binding protein involved in the positive regulation of CLN3 transcription by glucose. Despite this role in CLN3 regulation, cells lacking AZF1 displayed no growth defects in glucose-containing medium. Perhaps not surprisingly, microarray results suggest that Azf1 is responsible for efficient transcription of a relatively small set of genes in cells growing logarithmically in rich medium. Many of these genes appear to play a role in carbohydrate metabolism, but no single metabolic pathway is especially affected. While it is possible that Azf1 simply plays a minor role in cells growing on glucose, we think that it is likely that the functional significance of Azf1 may be masked by the presence of redundant transcriptional activators. Further experiments will be necessary to define the role of Azf1 in cells growing in glucose.
In light of the concordance between the phenotype of azf1Δ mutants and the microarray data, conclusions regarding the role of Azf1 in nonfermentable carbon sources are more straightforward. When grown in glycerol-lactate medium, cells lacking AZF1 are unable to efficiently transcribe a number of genes involved in maintaining cell wall integrity. As would be predicted, azf1Δ mutants are sensitive to cell wall perturbing agents when grown in glycerol-lactate medium. The zymolyase resistance in nonfermentable medium also matches the microarray data showing downregulation of GAS1 and GAS3 in azf1Δ cells grown in nonfermentable medium. These two genes encode β-1,3-glucanosyltransferases, producing the β-1,3-glucan linkages that are the target of zymolyase (14, 15). Furthermore, the array data show a decrease in TOS1 expression. Deletion of TOS1 also leads to zymolyase resistance (23). Thus, the predictions of the microarray experiments were born out experimentally.
HOR2 stands out as a gene that is affected by Azf1 in both carbon sources. Since HOR2, encoding glycerol 3-phosphatase (GPP2), is required for proper responses to salt stress, it would be predicted that azf1Δ cells might be salt stress sensitive. This proved to be the case: azf1Δ cells were clearly more sensitive to 0.75 M NaCl than were wild-type cells in glycerol-lactate (not shown). The mutants showed no increased sensitivity to NaCl in glucose. However, this might be expected because the related phosphatase GPP1, encoded by RHR2, is expressed in glucose but is downregulated in poor carbon sources (16a). Thus, the presence of GPP1 in the glucose-grown cells would be expected to at least partially mask the decrease in GPP2.
While we previously identified the Azf1 binding motif as A2GA5, it now appears that Azf1 does not recognize A2GA5 but rather the related sequence A4GA3. These A4GA3 elements were embedded in all of the test sequences used to reach the original conclusion. The fact that this binding motif is found enriched in the promoters of Azf1-dependent genes suggests that Azf1 may regulate many of these genes directly. While these sequences appear to be very closely related, the failure of A2GA5 oligonucleotides to compete for binding to Azf1 suggests that more than two adenine residues are needed 5′ of the guanine to make an Azf1 binding motif; we have not determined whether longer stretches of A residues 5′ or 3′ affect binding.
If both of the distinct nonoverlapping sets of Azf1-regulated genes have Azf1 binding sites, how then does Azf1 induce one set of genes in glucose and the other set in glycerol-lactate? Perhaps the simplest model is one in which the state of Azf1 changes as cells move between fermentable and nonfermentable carbon sources.
It does not appear that movement in and out of the nucleus is a mechanism for regulating the change in Azf1 activity between cells growing in different carbon sources. However, we find Azf1 excluded from the nucleus in cells in stationary phase. This might be expected to downregulate all of the Azf1-regulated transcripts. However, examination of public gene expression databases (4a) does not show any significant trend for these genes in stationary-phase cells. We conclude that many other regulatory pathways come into play as cells enter stationary phase, obscuring the effect of Azf1 export.
We emphasize the fact that we can isolate as much Azf1 from cells grown in glucose as from cells grown in glycerol-lactate. This tells us that lower Azf1 recovery from cells in glucose is an artifact of extract preparation: the steady-state level of Azf1 protein in the cell appears the same regardless of carbon source. Our results suggest that protocol A somehow separates Azf1 from a protease that is present in protocol B extracts from glucose-grown cells. It is tempting to link the glucose-induced change in Azf1 function with the glucose-induced difference in Azf1 stability seen in vitro. However, we have no evidence for this and the result does not clearly point to a regulatory mechanism. In this regard, identification of Azf1-interacting proteins would be very valuable.
In summary, while loss of Azf1 produces a subtle phenotype, our results indicate that Azf1 fits an important niche, activating genes needed for growth and division when the cell is surrounded by glucose and strengthening the cell wall as the quality of the medium declines. These are important processes for the yeast cell, and it is likely that Azf1 does not act alone in regulating them.
Supplementary Material
Acknowledgments
We acknowledge D. Porcaro, B. Bagley, M. Dapp, P. Floyd, and S. Eloundou for assistance.
This work was supported by National Science Foundation grant 0235379 (W.H.) and a predoctoral fellowship from the American Foundation for Pharmaceutical Education (M.G.S.).
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Boorstein, W. R., and E. A. Craig. 1990. Transcriptional regulation of SSA3, an HSP70 gene from Saccharomyces cerevisiae. Mol. Cell. Biol. 10:3262-3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brohl, S., T. Lisowsky, G. Riemen, and G. Michaelis. 1994. A new nuclear suppressor system for a mitochondrial RNA polymerase mutant identifies an unusual zinc-finger protein and a polyglutamine domain protein in Saccharomyces cerevisiae. Yeast 10:719-731. [DOI] [PubMed] [Google Scholar]
- 3.Chiang, M. C., and H. L. Chiang. 1998. Vid24p, a novel protein localized to the fructose-1,6-bisphosphatase-containing vesicles, regulates targeting of fructose-1,6-bisphosphatase from the vesicles to the vacuole for degradation. J. Cell Biol. 140:1347-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. [DOI] [PubMed] [Google Scholar]
- 4a.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gray, J. V., G. A. Petsko, G. C. Johnston, D. Ringe, R. A. Singer, and M. Werner-Washburne. 2004. “Sleeping beauty”: quiescence in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 68:187-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall, D. D., D. D. Markwardt, F. Parviz, and W. Heideman. 1998. Regulation of the Cln3-Cdc28 kinase by cAMP in Saccharomyces cerevisiae. EMBO J. 17:4370-4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klis, F. M., P. Mol, K. Hellingwerf, and S. Brul. 2002. Dynamics of cell wall structure in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 26:239-256. [DOI] [PubMed] [Google Scholar]
- 8.Laabs, T. L., D. D. Markwardt, M. G. Slattery, L. L. Newcomb, D. J. Stillman, and W. Heideman. 2003. ACE2 is required for daughter cell-specific G1 delay in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 100:10275-10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee, T. I., N. J. Rinaldi, F. Robert, D. T. Odom, Z. Bar-Joseph, G. K. Gerber, N. M. Hannett, C. T. Harbison, C. M. Thompson, I. Simon, J. Zeitlinger, E. G. Jennings, H. L. Murray, D. B. Gordon, B. Ren, J. J. Wyrick, J. B. Tagne, T. L. Volkert, E. Fraenkel, D. K. Gifford, and R. A. Young. 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298:799-804. [DOI] [PubMed] [Google Scholar]
- 10.Lesage, P., X. Yang, and M. Carlson. 1996. Yeast SNF1 protein kinase interacts with SIP4, a C6 zinc cluster transcriptional activator: a new role for SNF1 in the glucose response. Mol. Cell. Biol. 16:1921-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li, C., and W. H. Wong. 2003. DNA-Chip analyzer (d-Chip), p. 120-141. In G. Parmigiani, E. S. Garrett, R. A. Irizarry, and S. L. Zeger (ed.), The analysis of gene expression data: methods and software, Springer-Verlag KG, Berlin, Germany.
- 12.Li, C., and W. H. Wong. 2001. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA 98:31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lussier, M., A.-M. White, J. Sheraton, T. di Paolo, J. Treadwell, S. B. Southard, C. I. Horenstein, J. Chen-Weiner, A. F. J. Ram, J. C. Kapteyn, T. W. Roemer, D. H. Vo, D. C. Bondoc, J. Hall, W. W. Zhong, A.-M. Sdicu, J. Davies, F. M. Klis, P. W. Robbins, and H. Bussey. 1997. Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics 147:435-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mouyna, I., T. Fontaine, M. Vai, M. Monod, W. A. Fonzi, M. Diaquin, L. Popolo, R. P. Hartland, and J. P. Latge. 2000. Glycosylphosphatidylinositol-anchored glucanosyltransferases play an active role in the biosynthesis of the fungal cell wall. J. Biol. Chem. 275:14882-14889. [DOI] [PubMed] [Google Scholar]
- 15.Mouyna, I., M. Monod, T. Fontaine, B. Henrissat, B. Lechenne, and J. P. Latge. 2000. Identification of the catalytic residues of the first family of beta(1-3)glucanosyltransferases identified in fungi. Biochem. J. 347:741-747. [PMC free article] [PubMed] [Google Scholar]
- 16.Newcomb, L. L., D. D. Hall, and W. Heideman. 2002. AZF1 is a glucose-dependent positive regulator of CLN3 transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 22:1607-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a.Pahlman, A. K., K. Granath, R. Ansell, S. Hohmann, and L. Adler. 2001. The yeast glycerol 3-phosphatases Gpp1p and Gpp2p are required for glycerol biosynthesis and differentially involved in the cellular responses to osmotic, anaerobic, and oxidative stress. J. Biol. Chem. 276:3555-3563. [DOI] [PubMed] [Google Scholar]
- 17.Parviz, F., D. D. Hall, D. D. Markwardt, and W. Heideman. 1998. Transcriptional regulation of CLN3 expression by glucose in Saccharomyces cerevisiae. J. Bacteriol. 180:4508-4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rolland, F., J. Winderickx, and J. M. Thevelein. 2002. Glucose-sensing and-signalling mechanisms in yeast. FEMS Yeast Res. 2:183-201. [DOI] [PubMed] [Google Scholar]
- 19.Stein, T., J. Kricke, D. Becher, and T. Lisowsky. 1998. Azf1p is a nuclear-localized zinc-finger protein that is preferentially expressed under non-fermentative growth conditions in Saccharomyces cerevisiae. Curr. Genet. 34:287-296. [DOI] [PubMed] [Google Scholar]
- 20.van Helden, J. 2003. Regulatory Sequence Analysis Tools. Nucleic Acids Res. 31:3593-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vincent, O., and M. Carlson. 1998. Sip4, a Snf1 kinase-dependent transcriptional activator, binds to the carbon source-responsive element of gluconeogenic genes. EMBO J. 17:7002-7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu, M., L. Newcomb, and W. Heideman. 1999. Regulation of gene expression by glucose in Saccharomyces cerevisiae: a role for ADA2 and ADA3/NGG1. J. Bacteriol. 181:4755-4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin, Q. Y., P. W. de Groot, H. L. Dekker, L. de Jong, F. M. Klis, and C. G. de Koster. 2005. Comprehensive proteomic analysis of Saccharomyces cerevisiae cell walls: identification of proteins covalently attached via glycosylphosphatidylinositol remnants or mild alkali-sensitive linkages. J. Biol. Chem. 280:20894-20901. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.