Abstract
This paper describes the first physiological response at the translational level towards heterologous protein production in Saccharomyces cerevisiae. In yeast, the phosphorylation of eukaryotic initiation factor 2α (eIF-2α) by Gcn2p protein kinase mediates derepression of GCN4 mRNA translation. Gcn4p is a transcription factor initially found to be required for transcriptional induction of genes responsible for amino acid or purine biosynthesis. Using various GCN4-lacZ fusions, knockout yeast strains, and anti-eIF-2α-P/anti-eIF-2α antibodies, we observed that heterologous expression of the membrane-bound α1β1 Na,K-ATPase from pig kidney, the rat pituitary adenylate cyclase seven-transmembrane-domain receptor, or a 401-residue soluble part of the Na,K-ATPase α1 subunit derepressed GCN4 mRNA translation up to 70-fold. GCN4 translation was very sensitive to the presence of heterologous protein, as a density of 1‰ of heterologous membrane protein derepressed translation maximally. Translational derepression of GCN4 was not triggered by misfolding in the endoplasmic reticulum, as expression of the wild type or temperature-sensitive folding mutants of the Na,K-ATPase increased GCN4 translation to the same extent. In situ activity of the heterologously expressed protein was not required for derepression of GCN4 mRNA translation, as illustrated by the expression of an enzymatically inactive Na,K-ATPase. Two- to threefold overexpression of the highly abundant and plasma membrane-located endogenous H-ATPase also induced GCN4 translation. Derepression of GCN4 translation required phosphorylation of eIF-2α, the tRNA binding domain of Gcn2p, and the ribosome-associated proteins Gcn1p and Gcn20p. The increase in Gcn4p density in response to heterologous expression did not induce transcription from the HIS4 promoter, a traditional Gcn4p target.
Heterologous expression is used intensively to produce large amounts of active proteins that are difficult or impossible to purify from native tissue. This approach has been very successful, and many soluble proteins are expressed to high levels by standard protocols, typically in microbial hosts (5, 33) or baculovirus-infected insect cells (37). The tremendous impact of heterologous expression on protein chemistry is evident from the increase in the number of solved three-dimensional protein structures, from about 1,000 in 1992 to almost 16,000 in March 2002 (4). However, the number of known membrane protein structures is extremely small compared to the coding capacities of presently sequenced genomes. Bioinformatic analysis of eubacterial, archaeal, yeast, worm, and human genomes predicts that about one-third of all genes encode membrane proteins (39). This is in contrast to the approximately 50 membrane protein structures of predominantly prokaryotic origin found in the Protein Data Bank (http://www.pdb.org/). Only five structures are known for higher eukaryotic membrane proteins (http://www.mpibp-frankfurt.mpg.de/michel/public/memprotstruct.html). All of these are based on proteins purified from native tissue. The high-resolution structure of recombinant Ca-ATPase produced in Saccharomyces cerevisiae (23) was recently shown to be identical to the structure previously determined for the native protein (19). The essential role of membrane proteins in the transport of solutes and information across membranes and the fact that approximately 60% (26) of all approved drugs target members of the seven-transmembrane-domain (7TM) superfamily underscore the importance of developing methods to produce, purify, and crystallize membrane proteins on a routine basis. Almost all membrane proteins are found in minute amounts in specialized cells, preventing purification from natural tissue on the milligram scale required for crystallization attempts. Unfortunately, heterologous expression of membrane proteins is very difficult and by no way routine (9). The density of heterologous protein in the membrane is grossly independent of the applied host, pointing to a general failure of cells to cope with high-level expression of membrane proteins. In fact, very few examples are found in the literature on the heterologous expression of membrane proteins to a level where large-scale purification is achievable. The molecular reasons for this inherent difficulty have not been identified, as the physiological responses to heterologous membrane protein production have not been characterized.
Gcn4p is a transcription factor initially identified as being responsible for the induction of amino acid biosynthesis genes in response to starvation for any of several amino acids (14, 15). GCN4 expression is regulated at the translational level through four short open reading frames (ORFs) positioned in the 5′ noncoding part of GCN4 mRNA. The presence of the four open reading frames blocks translation of the GCN4 coding sequence in cells not suffering from amino acid starvation. The eukaryotic initiation factor 2α (eIF-2α)-specific kinase Gcn2p is activated by starvation for one or more amino acids. Phosphorylated eIF-2α works as an inhibitor of the GTP-GDP exchange protein eIF-2B and causes a reduction in the eIF-2α-GTP concentration. This circumvents the inhibitory effect of the four open reading frames and allows translation of the GCN4 open reading frame.
Further studies have shown that Gcn4p biosynthesis is also induced by starvation for purines (31), glucose limitation (41), growth on ethanol (41), high salinity in the growth medium (8), alkylation with methyl methanesulfonate (31), or treatment with the Tor1p and Tor2p kinase inhibitor rapamycin (36). Furthermore, GCN4 mRNA translation is repressed by nitrogen starvation (10). Microarray studies have recently shown that 10% of all yeast genes are induced by Gcn4p (18, 27). Gcn4p therefore seems to be a master regulator of gene expression in yeast and particularly involved in responses to various kinds of stress (15).
The focus of the present study was to examine if GCN4 plays a role in the physiological response to heterologous protein production in the yeast Saccharomyces cerevisiae. Using various GCN4-lacZ fusions, knockout and mutant yeast strains, eIF-2α-specific antibodies, three different membrane proteins, and one soluble protein, we have extended the list of stress factors able to induce GCN4 production to include heterologous protein production.
MATERIALS AND METHODS
Plasmid construction.
pPAP1553 was constructed by insertion of a 12-bp EcoRI linker into SalI- and Klenow polymerase-treated YDp-H (3). pPAP1555 was generated by insertion of an EcoRI-HIS3-EcoRI fragment from pPAP1553 into pPAP1486 (29). pPAP2231 was constructed by inserting a SalI-HindIII PCR fragment, generated with pPAP1666 (29) as a template and with primers alfaup (5′ CACACAGTCGACCAGAACTGCCTTGTGAAGAAC 3′) and alfado (5′ GTCGACAAGCTTAGAAGTTGTCATCCAGGAG 3′), into pEMBLyex4 (6). pPAP2915 and pPAP3608 were generated by insertion of the PstI-SalI fragment from p180 (13) into similarly digested YDp-K and YDp-W, respectively (3). pPAP2983 and pPAP3626 carry the PstI-SalI fragment from p227 (13) in similarly digested YDp-K and YDp-W, respectively (3). pPAP3277 was created by insertion of a PstI-SalI HIS4-lacZ fragment, generated by PCR with p929 (24) as a template, into PstI- and SalI-digested YDp-K (3). The PMA1 coding region was amplified by PCR, digested with XhoI and HindIII, and inserted into SalI- and HindIII-restricted pEMBLyex4, generating pPAP3437. Overlap extension PCR was used to generate a DNA fragment encompassing nucleotides −478 to −10 relative to the adenosine in the GCN2 initiation codon and nucleotides +10 to +485 relative to the thymine in the GCN2 termination codon, using yeast chromosomal DNA as a template. This fragment was blunt-end cloned into EcoRV-digested pBluescript KS II (Stratagene), generating plasmid pPAP3038. The GCN2 flanking sequences were transferred to pQE40 (QIAGEN Inc.) after XhoI and HindIII digestion, generating pPAP3054. An EcoRI-G418r-BamHI fragment from pPAP1511 was inserted into similarly digested pPAP3054, giving rise to pPAP3056. An EcoRI-G418r-BamHI fragment from pPAP1511 was inserted into similarly digested pPAP3140, giving rise to pPAP3161. A YIp352 (12) derivative carrying an sui2S51A mutant was constructed by overlap extension PCR and designated pPAP4159. pPAP4692 was generated by inserting a SmaI-XhoI fragment from p332 (40) into similarly digested pRS403 (Stratagene). Plasmid pPAP4126, allowing doxycycline-inducible expression of the pig Na,K-ATPase α1 subunit was created by inserting an α1 PCR fragment into pCM252 (2). A similar plasmid, pPAP4144, was generated by cloning the doxycycline-inducible promoter and tetR from pCM252 and a β1 PCR fragment into Yep352 (12).
Yeast strains.
The genotypes of strains used for the present study are shown in Table 1. NheI-digested reporter plasmid pPAP2915 or pPAP2983 was inserted into the yeast lys2 allele of PAP1503 by homologous recombination. A GCN2 knockout strain was generated by the one-step gene disruption procedure after transformation with a gcn2-G418r-gcn2 fragment generated by a PCR with pPAP3056 as the template. GCN1 and GCN20 knockout strains were generated after transformation with gcn1-G418r-gcn1 and gcn20-G418r-gcn20 PCR fragments, respectively, generated with pFA6a-KanMX4 (38) as the template. Strain BMA64-1B (W303) was obtained from Euroscarf and made to carry the ADE2 allele by transformation with an ADE2 PCR fragment. pPAP3608 was targeted to the trp1 locus, and BsaBI-digested pPAP1555 was targeted to the his3 locus, by homologous recombination. A prb1Δ derivative was constructed by transformation with a prb1-GCN4-prb1 PCR fragment. A strain producing only the SUI2S51A mutant protein was generated by transforming PAP2861 with ClaI-digested pPAP4159, followed by selection of 5-fluoroorotic acid-resistant cells and screening for the presence of the correct sui2 allele by PCR. A strain carrying only the gcn2m2 allele was created in a similar way, using plasmid pPAP4692. A W303 strain allowing maximal repression of doxycycline-inducible promoters was generated by targeting pCM242 (2) to the leu2 locus of BMA64-1B from Euroscarf, generating PAP4284. The StuI-digested GCN4-lacZ reporter pPAP3639 was subsequently targeted to the ade2 locus of PAP4284, creating PAP4287.
TABLE 1.
Yeast strains used for the present study
| Straina | Genotype | Reference or source |
|---|---|---|
| PAP1503 | α ura3-52 trp::GAL10-GAL4 lys2-801 leu2Δ1 his3Δ200 pep4::HIS3 prb1Δ1.6R can1 GAL | 29 |
| PAP2571 | α ura3-52 trp::GAL10-GAL4 lys2-801::UPR-lacZ leu2Δ1 his3Δ200 pep4::HIS3 prb1Δ1.6R can1 GAL | 21 |
| PAP2861 | α ura3-52 trp::GAL10-GAL4 lys2-801::GCN4-lacZ leu2Δ1 his3Δ200 pep4::HIS3 prb1Δ1.6R can1 GAL | This study |
| PAP3059 | α ura3-52 trp::GAL10-GAL4 lys2-801::GCN4ΔuORF-lacZ leu2Δ1 his3Δ200 pep4::HIS3 prb1Δ1.6R can1 GAL | This study |
| PAP3096 | α ura3-52 trp::GAL10-GAL4 lys2-801::GCN4-lacZ leu2Δ1 his3Δ200 pep4::HIS3 prb1Δ1.6R can1 GAL gcn2Δ | This study |
| PAP3258 | α ura3-52 trp::GAL10-GAL4 lys2-801::GCN4ΔuORF-lacZ leu2Δ1 his3Δ200 pep4::HIS3 prb1Δ1.6R can1 GAL gcn2Δ | This study |
| PAP3313 | α ura3-52 trp::GAL10-GAL4 lys2-801::HIS4-lacZ leu2Δ1 his3Δ200 pep4::HIS3 prb1Δ1.6R can1 GAL | This study |
| PAP4069 | α ura3-52 trp::GAL10-GAL4 lys2-801::GCN4-lacZ leu2Δ1 his3Δ200 pep4::HIS3 prb1Δ1.6R can1 GAL gcn1Δ | This study |
| PAP4072 | α ura3-52 trp::GAL10-GAL4 lys2-801::GCN4-lacZ leu2Δ1 his3Δ200 pep4::HIS3 prb1Δ1.6R can1 GAL gcn20Δ | This study |
| PAP4185 | α ura3-52 trp::GAL10-GAL4 lys2-801::GCN4-lacZ leu2Δ1 his3Δ200 pep4::HIS3 prb1Δ1.6R can1 GAL sui2S51A | This study |
| PAP4284* | α ura3-52 trp1Δ2 leu2-3-112::tetR-Snn6 his3-11 ade2-1 can1-100 | This study |
| PAP4713 | α ura3-52 trp::GAL10-GAL4 lys2-801::GCN4-lacZ leu2Δ1 his3Δ200 pep4::HIS3 prb1Δ1.6R can1 GAL gcn2m2 | This study |
| BMA64-1B | α ura3-52 trp1Δ2 leu2-3-112 his3-11 ade2-1 can1-100 | Euroscarf |
| PAP3975* | α ura3-52 trp1Δ2::GCN4-lacZ TRP1 leu2-3-112 his3-11::GAL10-GAL4 HIS3 ADE2 can1-100 prb1Δ | This study |
All strains are isogenic to the parental S288C derivative PAP1503, a derivative of BJ5457 (20), except for those marked with an asterisk, which were derived from W303.
Heterologous expression.
For experiments using galactose-regulated promoters, yeast cells were grown in shake flasks in minimal medium containing 0.5% glucose or 2% glucose and 3% glycerol as carbon sources and supplemented with all amino acids except leucine, isoleucine, glutamine, asparagine, and glycine. Cultures were grown at 30°C until the optical density at 450 nm (OD450) reached 1.0 and then induced with 2% galactose dissolved in growth medium with or without glucose but including all amino acids except glutamine, asparagine, and glycine. For experiments involving the tetracycline-inducible promoters, yeast cells were grown in minimal medium with 2% glucose and supplemented with all amino acids except leucine, tryptophan, glutamine, asparagine, and glycine. Cultures were grown at 30°C until the OD450 reached 1.0 and then induced with doxycycline.
Cytosolic and membrane fractions were prepared as described previously (21), except for the inclusion of 37 mM β-glycerophosphate, 48 mM NaF, and 1 mM dithiothreitol to inhibit yeast phosphatases when required.
RNA slot blotting, sodium dodecyl sulfate-polyacrylamide gel electrophoresis, Western blotting, β-galactosidase activity measurements, and [3H]ouabain binding were performed as described previously (21). Western blots were quantified using UN-SCAN-IT software from Silk Scientific.
Antibodies against eIF-2α-P were purchased from New England Biolabs, and anti-yeast eIF-2α antibodies were generous gifts from A. G. Hinnebusch, National Institutes of Health, and R. C. Wek, Indiana University School of Medicine. Anti-PMA1 antibodies were kindly provided by R. Serrano, Technical University of Valencia, Valencia, Spain.
Cytoplasmic and vacuolar pools of amino acids were determined according to the method of Ohsumi et al. (28).
Polysomes were prepared and analyzed according to the method of Marton et al. (25).
RESULTS
Generation of yeast strains reporting GCN4 mRNA translation, GCN4 transcription, and HIS4 transcription.
The expression plasmids used for the present study are diagrammed in Fig. 1. The lacZ reporter fusions were transferred from previously described plasmids (13, 24) to integrative yeast vectors carrying other selective markers and targeted to the yeast chromosome. All fusions were transferred to PAP1503, an S288C derivative, and the most important were transferred to PAP3975, of W303 origin. The data in Table 2 verify that the generated reporter strains correctly monitor GCN4 mRNA translation, GCN4 transcription, or HIS4 transcription under derepressing and repressing conditions. Derepression was induced by the addition of 3-aminotriazole (3-AT), which inhibits histidine biosynthesis and consequently causes the histidine starvation that induces translational derepression of GCN4 mRNA and increased HIS4 transcription.
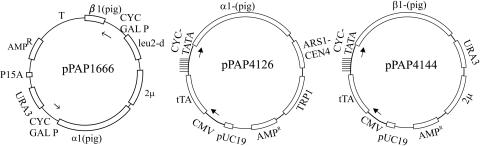
FIG. 1.
Structural map of expression plasmids pPAP1666, pPAP4126, and pPAP4144. Plasmid pPAP1666 allows galactose-inducible transcription of α1 and β1 cDNAs encoding pig kidney Na,K-ATPase subunits. Plasmid pPAP2231 is identical to pPAP1666 except that the α1(TM4/TM5) DNA (encoding amino acids 348 to 748 from the pig Na,K-ATPase α1 subunit) has replaced the α1 cDNA, and the β1 cDNA has been removed. In pPAP3437 and pPAP2902, the α1(TM4/TM5) DNA has been replaced by the PMA1 coding region and rat PACR1 receptor cDNA, respectively. Expression from plasmids pPAP4126 and pPAP4144 is induced by doxycycline. Ampr, β-lactamase gene; p15A, p15A origin of replication; pUC19, origin of replication from pUC19; 2μ, 2μm origin of replication; ARS1-CEN4, autonomous replicating sequence and centromere region from yeast; URA3, yeast orotinin-5′-P decarboxylase gene; leu2-d, a poorly expressed allele of the yeast β-isopropylmalate dehydrogenase gene; TRP1, phosphoribosylanthranilate isomerase gene; CYC-GALP, a galactose-inducible promoter; CYC1-TATA, a crippled promoter from the cytochrome c gene; CMV, cytomegalovirus promoter; |, operator site from tetracycline resistance gene; tTA, reverse transactivator; α1 (pig) and β1 (pig), cDNAs encoding the α1 and β1 Na,K-ATPase subunits from the pig kidney.
TABLE 2.
Verification of reporter strains used in the present study
| Reporter gene | β-Galactosidase activity (nmol/min mg of protein)a
|
|||
|---|---|---|---|---|
| S288C mutant
|
W303
|
|||
| R | DR | R | DR | |
| GCN4-lacZ | 20 | 270 | 16 | 140 |
| GCN4Δuorf-lacZ | 710 | 680 | ND | ND |
| HIS4-lacZ | 579 | 1726 | ND | ND |
Reporter strains were grown in amino acid-supplemented minimal medium to an OD450 of 1, supplemented (derepressed [DR]) or not (repressed [R]) with 50 mM 3-AT, and harvested 24 hours later. β-Galactosidase activities were measured in purified cytosolic preparations. ND, not determined; GCN4-lacZ, a translational fusion between GCN4 at codon 56 and lacZ; GCN4Δuorf-lacZ, identical to the GCN4-lacZ fusion except that the initiation codons of the four upstream open reading frames were altered to GTG; HIS4-lacZ, a translational fusion between codon 10 of HIS4 and codon 8 of lacZ.
Transcription and translation are unaffected by heterologous expression.
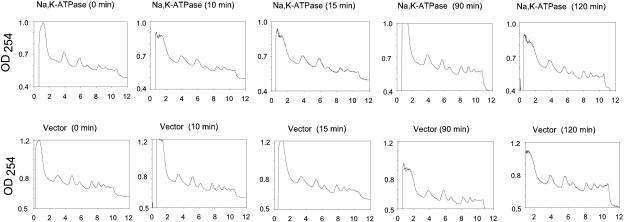
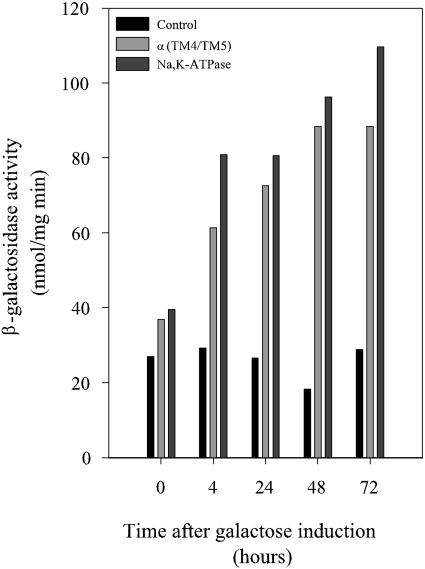
For initial characterization of how the induction of heterologous protein production affects macromolecular synthesis in S. cerevisiae, we chose the membrane-bound pig kidney Na,K-ATPase and a soluble part of this huge protein complex. The Na,K-ATPase is an α1β1 heterodimer containing a 1,019-amino-acid α1 subunit with 10 transmembrane segments and an N-glycosylated β1 subunit with a single transmembrane segment and three disulfide bonds. The enzyme produced in yeast has been shown to be targeted to the plasma membrane and to have the same activity as the native enzyme purified from the kidneys (29, 30) The soluble part, α1(TM4/TM5), contains 401 residues located between transmembrane segments 4 and 5 of the α1 subunit. cDNAs encoding each of the three model proteins were expressed from galactose-inducible promoters (Fig. 1). We determined the time-dependent accumulation of functional membrane-bound Na,K-ATPase protein by ouabain binding, soluble α1(TM4/TM5) protein by Western blotting, and α1 mRNA, actin mRNA, and the short-lived PAB1 mRNA (t1/2, ∼11 min) (11) by slot blotting. In addition, we compared the polysome distributions in cells expressing either the intact Na,K-ATPase or no heterologous protein. The data in Fig. 2A show that the density of ouabain sites in yeast membranes increased until 48 h after galactose induction. Accumulation of the soluble α1(TM4/TM5) protein was faster than membrane accumulation of the Na,K-ATPase, and the density peaked approximately 20 h after induction (Fig. 2A). The results of Northern slot blots in Fig. 2B show that α1 mRNA increased for 48 h. Transcription initiation in general was unaffected by heterologous expression, as the level of the short-lived PAB1 mRNA was fairly constant for 72 h after induction. The data in Fig. 3 show that translation in general was unaffected until 2 hours after induction of heterologous expression, as polysome distributions in cells carrying the α1β1 expression plasmid or the empty vector were similar. The polysome analysis was restricted to the first 2 hours for the following two reasons: other conditions known to affect translation initiation are effective within this time span (1, 35), and the data in Fig. 2A and B demonstrate that expression was present prior to induction.
FIG. 2.
Time-dependent accumulation of ouabain sites (A), α1(TM4/TM5) loop protein (A), α1 mRNA (B), and PAB1 mRNA (B). The Na,K-ATPase or the α1(TM4/TM5) loop was expressed from galactose-inducible promoters in the S288C mutant strain, as described in Materials and Methods. Membranes, cytosolic fractions, and total RNA were isolated from cells harvested at the indicated time points. Data represent one of two or three independent growth experiments with each yeast strain. Ouabain binding to crude membranes was performed with 15 nM [3H]ouabain. The amount of α1(TM4/TM5) was determined by quantitative Western blotting. PAB1 mRNA, α1 mRNA, or actin mRNA was detected by Northern slot blot analysis. Blots were quantified on a STORM PhosphorImager. RNA data were normalized to actin mRNA levels. The maximum value for each experiment was defined as 100%.
FIG. 3.
Effect of heterologous expression of Na,K-ATPase on polysome distribution. S288C mutant yeast cells expressing either Na,K-ATPase or no heterologous protein were grown as described in Materials and Methods. At an OD450 of 0.3, each culture was induced with 2% galactose dissolved in growth medium lacking glucose but including amino acids. Polysomes were prepared from cells harvested at the indicated time points and analyzed as described in Materials and Methods.
Production of recombinant protein derepresses translation of GCN4 mRNA.
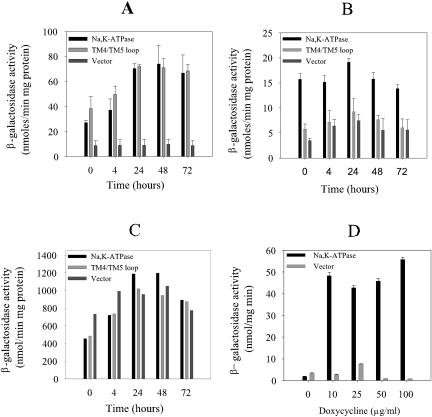
To determine whether heterologous expression of membrane-bound pig Na,K-ATPase or the soluble α1(TM4/TM5) loop affects the level of GCN4 mRNA or Gcn4p protein, we measured the kinetics of β-galactosidase accumulation in the S288C mutant and W303 strains carrying a GCN4-lacZ fusion and in the S288C mutant strain carrying the GCN4Δuorf-lacZ fusion. The former construct reports translation of GCN4 mRNA, while the latter measures GCN4 transcription. The data in Fig. 4A and B show that heterologous membrane protein production derepressed GCN4 mRNA translation eightfold in the S288C mutant but only threefold in W303 relative to that in cells carrying the empty vector. Expression of the soluble α1(TM4/TM5) protein induced GCN4 mRNA translation fivefold, but only in the S288C background. We therefore decided to characterize the response in this background.
FIG. 4.
GCN4 transcription or translation in yeast cells expressing α1β1 Na,K-ATPase, α1(TM4/TM5), or an empty expression vector. The S288C mutant (A) or W303 (B) strain carrying the translational GCN4-lacZ reporter fusion, the S288C mutant strain carrying the GCN4Δuorf-lacZ fusion (C) and a plasmid for galactose-inducible expression of α1β1 Na,K-ATPase or α1(TM4/TM5) or an empty vector, and the W303 strain carrying the translational GCN4-lacZ reporter fusion and a plasmid for tetracycline-inducible expression (D) of Na,K-ATPase or the empty vector were grown in amino acid-supplemented minimal medium at 30°C as described in Materials and Methods. At an OD450 of 1, each culture was induced with 2% galactose (A, B, C) or various concentrations of doxycycline (D) dissolved in growth medium lacking glucose but containing amino acids. For panels A, B, and C, specific β-galactosidase activities 0, 4, 24, 48, and 72 h after induction were determined in purified cytosolic fractions. For panel D, β-galactosidase activities were determined 24 h after induction with doxycycline. Data are given as mean values ± standard deviations from three independent growth experiments with double determinations of each data point (A, B, D) or as mean values from two independent growth experiments with double determinations of each data point (C).
To rule out that heterologous expression increased the GCN4 mRNA level, we determined the time-dependent accumulation of β-galactosidase activity in the S288C mutant strain carrying the GCN4Δuorf-lacZ reporter. It can be seen from Fig. 4C that heterologous expression did not influence GCN4 transcription relative to that in cells carrying the empty vector control but increased the density of Gcn4p by derepressing GCN4 mRNA translation.
The high level of β-galactosidase activity prior to galactose induction seen with the GCN4-lacZ reporter strain expressing α1β1 or α1(TM4/TM5) is in agreement with the leaky transcription from the galactose-inducible promoters that is evident in Fig. 2A and B, as ouabain binding sites and α1(TM4/TM5) protein were present before the addition of galactose. The GCN4 response seen with this expression system is therefore not an acute response but may, to some extent, reflect adaptation to heterologous expression. To study the acute impact of heterologous expression on GCN4 mRNA translation, a more tightly regulated system was required. We therefore used a dual expression system (2) to produce the Na,K-ATPase α1 and β1 subunits from tetracycline-inducible promoters (Fig. 1) in the W303 strain carrying the GCN4-lacZ reporter. The data in Fig. 4D demonstrate that the induction of Na,K-ATPase expression by doxycycline derepressed GCN4 mRNA translation up to 70-fold relative to that with the empty vector. The observed translational derepression is qualitatively in agreement with what was seen for the galactose-inducible system but is quantitatively much stronger. Importantly, it can be seen that the GCN4-lacZ activity prior to induction was indistinguishable from that of the empty vector control when the tetracycline-inducible expression system was used. Furthermore, by comparing Fig. 4B and D, it can be seen that GCN4-lacZ activity obtained after the induction of heterologous expression was three- to fourfold higher for the tetracycline-inducible expression system than for the galactose-inducible expression system.
Derepression of GCN4 mRNA translation by heterologous expression requires Gcn2p-mediated eIF-2α phosphorylation.
Translational derepression of GCN4 mRNA translation generally requires phosphorylation of eIF-2α at serine 51 by the Gcn2p protein kinase. To determine whether derepression of GCN4 mRNA translation during heterologous protein production of α1β1 or α1(TM4/TM5) Na,K-ATPase relies on GCN2, expression analysis of these proteins was performed with a gcn2Δ mutant strain carrying the GCN4-lacZ or GCN4Δuorf-lacZ reporter. The results in Fig. 5A show that deletion of GCN2 prevented the accumulation of β-galactosidase from the translational lacZ fusion but not from the transcriptional fusion. Derepression of GCN4 mRNA translation in response to heterologous expression therefore requires the eIF-2α kinase Gcn2p.
FIG. 5.
GCN4 transcription and translation in a gcn2Δ mutant strain (A) and GCN4 translation in a sui2S51A mutant strain (B) expressing either Na,K-ATPase, α1(TM4/TM5), or no recombinant protein. gcn2Δ mutant cells carrying either the translational reporter GCN4-lacZ or the transcriptional reporter GCN4Δuorf-lacZ or the sui2S51A mutant strain carrying the translational reporter GCN4-lacZ was grown and induced with galactose at time zero, as described in Materials and Methods. β-Galactosidase activities were determined at the indicated time points in purified cytosolic fractions. Columns marked with asterisks refer to β-galactosidase activities in gcn2Δ mutant cells carrying the transcriptional reporter. The isogenic wild-type strain (SUI2) expressing the α1β1 Na,K-ATPase was included as a positive control in panel B. The results in panel B are from at least three independent growth experiments with triple determinations of each data point.
To determine whether phosphorylation of serine 51 in eIF-2α is essential for derepression of GCN4 mRNA translation in response to heterologous expression, we generated a yeast strain that only produces eIF-2α with an alanine at position 51. The data in Fig. 5B show that serine 51 is essential for derepression of GCN4 mRNA translation in response to heterologous expression, strongly indicating that phosphorylation of eIF-2α at this position is required to increase the density of Gcn4p.
Heterologous expression increases the density of phosphorylated eIF-2α.
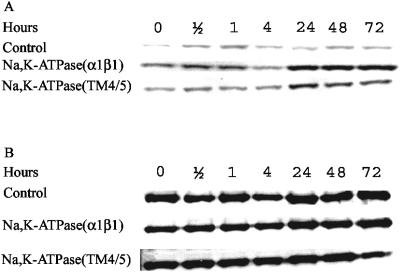
To determine whether heterologous expression affects the density of phosphorylated eIF-2α, Western blots were made by using a polyclonal antibody that specifically recognizes eIF-2α phosphorylated at serine 51 or a polyclonal antibody recognizing phosphorylated as well as nonphosphorylated eIF-2α. Quantification of the data in Fig. 6 demonstrated that the ratios between phosphorylated eIF-2α and total eIF-2α were 18 and 12 times higher for cells expressing α1β1 Na,K-ATPase or the soluble α1(TM4/TM5) loop, respectively, than for cells containing only the empty expression vector. Expression in the isogenic gcn2Δ mutant strain prevented the accumulation of phosphorylated eIF-2α (data not shown). These data provide strong evidence that the Gcn2p protein kinase mediates phosphorylation of the α subunit of eIF-2α at serine 51 in response to heterologous expression of the α1β1 Na,K-ATPase.
FIG. 6.
Time-dependent phosphorylation of eIF-2α by Gcn2p kinase during heterologous expression of α1β1 Na,K-ATPase or empty vector. Yeast cells were grown as described in Materials and Methods and harvested 0, 4, 24, 48, and 72 h after galactose induction. Cytosolic fractions were isolated, and equal amounts of total cytoplasmic protein (40 μg) were analyzed by Western blotting using a polyclonal antibody that specifically recognizes eIF-2α phosphorylated at serine 51 (A) or a polyclonal antibody that recognizes phosphorylated and nonphosphorylated forms of eIF-2α (Β).
The tRNA binding domain of Gcn2p is required for derepression of GCN4 mRNA translation.
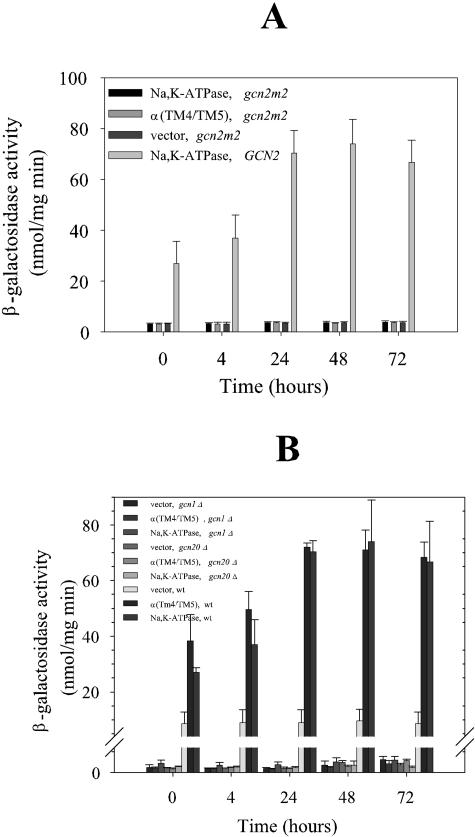
Gcn2p is activated by amino acid starvation through binding of uncharged tRNA to a regulatory domain showing homology to histidyl-tRNA synthetase (40). We expressed our model proteins in a yeast GCN4-lacZ reporter strain expressing the gcn2m2 allele. This allele produces a protein that is unable to bind uncharged tRNA and therefore unable to sense the presence of empty tRNA molecules. The data in Fig. 7A show that the tRNA binding domain is also required for the induction of GCN4 translation in response to heterologous expression, suggesting that heterologous expression increases the density of uncharged tRNA.
FIG. 7.
GCN4 mRNA translation in a gcn2m2 mutant strain (A), a gcn1Δ mutant strain (B), a gcn20Δ mutant strain (B), or the isogenic wild-type strain expressing either the α1β1 Na,K-ATPase, α1(TM4/TM5), or no recombinant protein. Yeast cells carrying the translational reporter GCN4-lacZ were grown and induced with galactose for heterologous expression at time zero, as described in Materials and Methods. β-Galactosidase activities were determined at the indicated time points in purified cytosolic fractions. The isogenic wild-type strain expressing α1β1 Na,K-ATPase was included as a control. The results are from at least three independent growth experiments with triple determinations of each data point.
The Gcn1p-Gcn20p complex is required for derepression of GCN4 mRNA translation.
The activation of Gcn2p kinase activity in response to amino acid starvation requires interactions between the N terminus of Gcn2p and the Gcn1p-Gcn20p protein complex (25). To determine if heterologous expression also derepresses GCN4 mRNA translation in a Gcn1p- and Gcn20p-dependent manner, we analyzed GCN4-lacZ activity in gcn1Δ and gcn20Δ knockout strains in response to heterologous expression. The results shown in Fig. 7B demonstrate that both proteins are essential for derepression of GCN4 translation after heterologous expression.
GCN4 mRNA translation is derepressed at a very low density of heterologous protein.
To determine how the level of recombinant membrane protein production influences GCN4 mRNA translation during heterologous expression, the α1β1 Na,K-ATPase expression plasmid or the empty vector was integrated into the yeast chromosome or selected for a low (20 to 30 copies per cell) or high (200 to 300 copies per cell) plasmid copy number (6). Figure 8A shows that derepression of GCN4 translation is independent of the copy number, as there were no differences in β-galactosidase activity, regardless of whether the expression plasmid was present at a low or high copy number or integrated into the yeast chromosome. To determine how the observed Gcn4p-β-galactosidase activity depends on the produced amount of recombinant protein, we determined the density of functional Na,K-ATPase molecules in crude yeast membranes by [3H]ouabain binding. The data in Fig. 8B show that the highest density of [3H]ouabain binding sites was seen in yeast cells with a high copy number of the expression plasmid and that integration of the plasmid into the yeast chromosome resulted in very low ouabain binding (<0.05 pmol/mg, or ∼1‰ of total membrane protein). Regardless of the density of ouabain binding sites, similar Gcn4p-β-galactosidase activities were detected, indicating that a very low density of Na,K-ATPase molecules is sufficient to activate a strong GCN4 response.
FIG. 8.
Influence of expression level on GCN4 mRNA translation. S288C mutant cells carrying the translational GCN4-lacZ reporter fusion and the expression plasmid for α1β1 Na,K-ATPase or the empty vector were grown in amino acid-supplemented minimal medium, 0.5% glucose, and 3% glycerol at 30°C in the absence of leucine, in the presence of leucine, or with the expression plasmid integrated into the yeast chromosome. At an OD450 of 1.0, cultures were induced with 2% galactose dissolved in growth medium lacking glucose but containing amino acids (as described in Materials and Methods). After 0, 4, 24, 48, and 72 h, specific β-galactosidase activities were determined in cytoplasmic fractions, and [3H]ouabain binding was determined in crude membranes with a [3H]ouabain concentration of 14.7 nM. Specific ouabain binding was corrected for nonspecific binding by subtracting the values obtained for the empty vector.
Misfolding of a heterologous membrane protein or general interference with ER folding does not affect translational expression of GCN4.
We have previously shown that amino acid alterations in the phylogenetically conserved soluble 708-TGDGVNDSPALKK-720 segment of Na,K-ATPase result in temperature-sensitive folding mutants (21). These mutants accumulate at 15°C but not at 35°C due to misfolding in the endoplasmic reticulum (ER), as monitored by induction of the unfolded protein response (UPR). To assess whether protein folding in the ER affects GCN4 translation, the wild type and two temperature-sensitive mutants, α1(N713A)β1 and α1(S715A)β1, were produced at temperatures ranging from 15°C to 35°C in a GCN4-lacZ reporter strain (PAP2861, Table 1) or a UPR-lacZ reporter strain (PAP2571, Table 1). The data in Fig. 9A show that the induction of GCN4 translation is independent of the temperature and the Na,K-ATPase allele being expressed. The data in Fig. 9B show how the expression temperature affected induction of the unfolded protein response for wild-type Na,K-ATPase, the α1(N713A)β1 mutant, or the α1(S715A)β1 mutant. It can be seen that folding of wild-type Na,K-ATPase, in contrast to that of the mutants, was not compromised at temperatures between 15°C and 35°C. In addition, it can be seen from Fig. 9C that the accumulation of mutant proteins, in contrast to that of the wild type, was compromised when the temperature was increased above 20°C. These results show that the observed increases in eIF-2α phosphorylation and GCN4 mRNA translation in response to heterologous membrane protein production are not due to misfolding of a single protein in the ER and that misfolding does not increase the derepression of GCN4 mRNA translation further.
FIG. 9.
Effects of protein misfolding on GCN4 translation. S288C mutant yeast cells carrying either the translational GCN4-lacZ reporter fusion (A) or the UPR-lacZ fusion (B) and an expression plasmid for α1β1 Na,K-ATPase, α1(N713A)β1-Na,K-ATPase, or α1(S715A)β1-Na,K-ATPase or the empty vector were grown as described in Materials and Methods to an OD450 of 1.0. One-fifth of each culture was transferred to 15, 20, 25, 30, or 35°C and induced after 30 min with 2% galactose dissolved in growth medium lacking glucose but containing amino acids. After 48 h, specific β-galactosidase activities were determined in purified cytoplasmic fractions, and ouabain binding capacities were determined in purified membrane fractions (C). The ouabain binding capacity was defined as 100% for membranes isolated from cells grown at 15°C. S288C mutant yeast cells carrying the GCN4-lacZ fusion or the UPR-lacZ reporter were separated into three portions (D). Each culture was supplemented with 1 μg/ml tunicamycin, 15 mM β-mercaptoethanol, or 50 mM 3-aminotriazole. β-Galactosidase activities were measured in cytosolic fractions from cells harvested after 18 h. The data shown are from two or three independent growth experiments with triple determinations of each data point.
The substances tunicamycin, β-mercaptoethanol, and 3-aminotriazole were added individually to exponentially growing yeast strains carrying either a UPR-lacZ fusion or a GCN4-lacZ fusion to see whether interference with overall protein folding in the ER induces GCN4 mRNA translation and if derepression of GCN4 mRNA translation affects the unfolded protein response. The data in Fig. 9D show that the addition of either tunicamycin or β-mercaptoethanol induced the unfolded protein response but did not derepress GCN4 mRNA translation. It can also be seen that derepression of GCN4 translation did not activate the unfolded protein response.
The Gcn2p kinase therefore does not respond to the presence of a single class of misfolded proteins in the ER or to conditions where protein folding in this compartment is compromised.
Induction of GCN4 translation is independent of Na,K-ATPase activity.
Aspartate 369 in the Na,K-ATPase α1 subunit is transiently phosphorylated during the reaction cycle and is absolutely required for enzyme activity but not for protein folding or targeting in yeast (30). We expressed a protein with the α1(D369A)β1 mutation in the GCN4-lacZ reporter strain to exclude the possibility that GCN4 mRNA translation was induced by a reduced ATP/ADP ratio and/or an unfavorable Na+ or K+ distribution across the plasma membrane due to in situ pump activity. The data in Fig. 10A exclude these possibilities, as expression of the mutant protein induced GCN4 translation to the same extent as expression of the wild type. The biological activity of the recombinant protein is therefore not required for translational induction of GCN4.
FIG. 10.
Induction of GCN4 translation in yeast cells expressing an inactive Na,K-ATPase (A) or the rat PACR1 receptor (B) or overexpressing the endogenous PMA1 gene (B). Yeast cells carrying the translational GCN4-lacZ reporter fusion and an expression plasmid for α1(D369A)β1-Na,K-ATPase, rat PACR1, or yeast PMA1 were grown at 30°C as described in Materials and Methods. Protein production was induced at an OD450 of 1.0 with 2% galactose dissolved in medium lacking glucose. β-Galactosidase activities were determined in purified cytosolic fractions. The results shown are from two or three independent growth experiments with triple determinations of each data point.
Expression of the 7TM receptor PACR1 or overexpression of the endogenous H-ATPase, Pma1p, also induces GCN4 mRNA translation.
To determine if the induction of GCN4 mRNA translation is specific for overexpression of the intact Na,K-ATPase or the soluble α1(TM4/TM5) part, we measured GCN4 mRNA translation during heterologous expression of the 7TM receptor PACR1 (pituitary adenylate cyclase-activating polypeptide receptor). This receptor can be expressed in a functional form in yeast (S. Rojko and P. A. Pedersen, unpublished results). The data in Fig. 10B show that expression of this 7TM receptor induces GCN4 mRNA translation to the same extent as the other tested proteins. The high GCN4-β-galactosidase activity before galactose induction was probably due to the previously observed leaky expression from the CYC-GAL promoters used.
To address whether the observed induction of GCN4 translation was specific for heterologous proteins, we overexpressed the endogenous plasma membrane H-ATPase from the expression plasmid used for the heterologous proteins. Western blotting showed that Pma1p was overexpressed two- to threefold (data not shown), and the data in Fig. 10B show that the excess amount of Pma1p induced GCN4 translation, but to a lesser extent than that induced by heterologous expression.
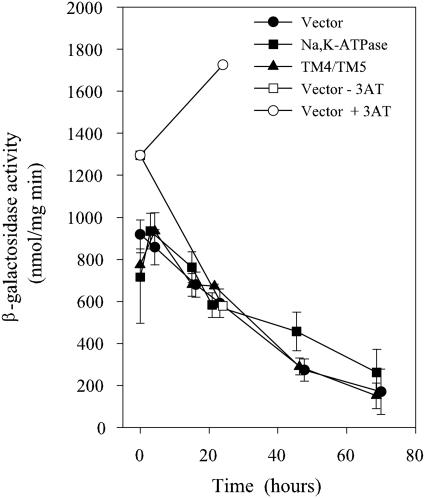
Gcn4p induced by heterologous protein production does not induce HIS4 transcription.
One of the target genes for the Gcn4p transcription factor under conditions of amino acid starvation is the HIS4 gene. To determine whether Gcn4p induced by heterologous gene expression is able to transactivate HIS4 transcription, we determined the time-dependent accumulation of β-galactosidase in a reporter yeast strain carrying a transcriptional HIS4-lacZ fusion. The data in Fig. 11 show that HIS4-lacZ activity actually decreased under conditions where GCN4-lacZ activity induced by heterologous expression increased. In contrast, the addition of 3-AT (an inhibitor of histidine biosynthesis) increased HIS4-lacZ activity. This indicates that Gcn4p produced in response to heterologous protein production does not target the exact same genes as Gcn4p produced in response to amino acid starvation.
FIG. 11.
Transactivation of HIS4 gene in response to heterologous expression. Yeast cells carrying the transcriptional HIS4-lacZ fusion and an expression plasmid for α1β1 Na,K-ATPase or α1(TM4/TM5) or the empty expression vector were grown and induced as described in Materials and Methods. Specific β-galactosidase activities were determined in isolated cytoplasmic fractions of cells harvested 0, 4, 24, 48, or 72 h after induction. A culture containing the empty vector was supplemented or not with 3-AT and harvested after 24 h. Specific β-galactosidase activities were determined in purified cytosolic fractions from three independent experiments with triple determinations of each data point.
Increased GCN4 mRNA translation induced by recombinant protein production is not affected by the glucose or amino acid concentration in the growth medium.
It is well established that starvation for glucose induces translational expression of GCN4 (41). It was therefore important to rule out that heterologous expression induced glucose starvation. We therefore induced Na,K-ATPase biosynthesis after growth of cells in either 0.5% or 2% glucose and included 0.5% or 2% glucose in the induction medium. The data in Fig. 12 show that translational induction of GCN4 was unaffected by the glucose concentration in the growth medium and that the glucose concentration did not affect the density of Na,K-ATPases in the membranes. To further rule out any influence of glucose, we induced heterologous protein production after growth with 3% raffinose as the sole carbon source. The data in Fig. 13 show, in agreement with those in Fig. 12, that the observed induction of GCN4 mRNA translation due to heterologous expression is unrelated to the glucose concentration in the growth medium. The data in Fig. 12 also exclude the possibility that the observed induction of GCN4 translation was due to faster consumption of amino acids in the growth medium of the strain expressing the Na,K-ATPase than in that of the control strain, as the experiments to collect the data in Fig. 13 were carried out in the absence of amino acids.
FIG. 12.
Effect of glucose and amino acids on translationally induced GCN4 expression and accumulation of functional Na,K-ATPases in yeast membranes. Cells were grown at 30°C in minimal medium with either 0.5% glucose or 2% glucose and 3% glycerol until the OD450 reached 1.0 and then induced with 2% galactose dissolved in growth medium. β-Galactosidase activities and capacities for [3H]ouabain binding were determined with purified cytoplasmic and crude membrane fractions, respectively, as described in Materials and Methods. A concentration of 14.5 nM [3H]ouabain was used. Specific ouabain binding was corrected for nonspecific binding by subtracting the values obtained for the empty vector.
FIG. 13.
Effect of carbon source on translationally induced GCN4 expression. Cells were grown at 30°C in amino acid-supplemented minimal medium with 2% raffinose until the OD450 reached 1.0 and then induced with 2% galactose dissolved in growth medium. Specific β-galactosidase activities were determined in purified cytoplasmic fractions.
The cytoplasmic and vacuolar amino acid pools are not affected by heterologous protein production.
It was important to rule out the possibility that differences in the cytoplasmic or vacuolar pools of amino acids in the Na,K-ATPase-expressing strain and the control strain caused the observed induction of GCN4 mRNA translation. We therefore determined these pool sizes prior to and after the induction of recombinant Na,K-ATPase. The data in Fig. 14 show that the cytoplasmic and vacuolar amino acid pools were very similar for the two strains and cannot explain the induced GCN4 mRNA translation in the Na,K-ATPase-expressing strain.
FIG. 14.
Measurement of cytoplasmic (A) and vacuolar (B) amino acid pools in the Na,K-ATPase-expressing strain and the control strain carrying the empty vector. Results from the Na,K-ATPase-expressing cells and the corresponding control cells are grouped. Cells were grown at 30°C in amino acid-supplemented minimal medium with 0.5% glucose and 3% glycerol until the OD450 reached 1.0 and then induced with 2% galactose dissolved in growth medium. The amino acid pools were determined by the ninhydrin method, as described in Materials and Methods. The results shown are from two independent experiments.
DISCUSSION
In the present study, we report the first response to heterologous protein production at the translational level. We demonstrated that heterologous expression of soluble or integral membrane proteins or overexpression of an endogenous membrane protein derepresses translation of GCN4 mRNA. Heterologous expression of the Na,K-ATPase integral membrane protein induced GCN4 translation between 4- and 70-fold compared to the control, depending on the strain background and the expression system. The soluble part of the Na,K-ATPase, α1(TM4/TM5), only induced GCN4 translation in the S288C mutant background. This is in accordance with results obtained for amino acid starvation, which caused a strain-dependent increase in GCN4 translation (15). The presented observations widen the spectrum of stress responses involving the Gcn4p transcription factor to include a response of considerable importance for basic and applied sciences.
A reduction in the rate of translation initiation is a well-characterized stress response in mammalian cells which is directed against viral infections or an unfavorable folding environment in the ER. These conditions reduce translation initiation in general through phosphorylation of eIF-2α. In yeast, however, eIF-2α phosphorylation caused by amino acid starvation does not substantially inhibit general protein synthesis but, rather, increases the translation of a subset of mRNA species (16). In accordance with this, heterologous expression also does not affect translation initiation within a time span where other stress situations such as glucose depletion and osmotic shock decrease translation initiation, as the polysome distribution was unaffected by heterologous expression. Our data also indicate that transcription in general is unaffected by heterologous expression, as the density of the short-lived PAB1 mRNA was grossly unaltered for 72 h after the induction of recombinant protein production. Our results therefore exclude overall reduced macromolecular synthesis as the cause of low heterologous membrane protein production.
Besides amino acid starvation, Gcn4p biosynthesis is induced by starvation for purines (31), glucose limitation (41), growth on ethanol (41), high salinity in the growth medium (8), treatment with methyl methanesulfonate (31), or treatment with the Tor1p and Tor2p inhibitor rapamycin (36). In these cases, translational induction of GCN4 is part of the cellular response to changes in the environment. However, our results demonstrate that internal stress factors like heterologous expression or endogenous membrane protein overexpression also induce translational induction of GCN4. This underscores the point made by Natarajan et al. (27) that Gcn4p is a master regulator of gene expression able to induce the transcription of a number of genes in response to a panoply of stress situations.
The induction of GCN4 translation usually requires the Gcn2p kinase (17). Heterologous expression also induces GCN4 translation in a Gcn2p-dependent manner, as translational induction was not observed in a gcn2Δ mutant strain.
The only known substrate for Gcn2p is eIF-2α, and all Gcn2p-dependent stress responses, except for those in response to UV irradiation (7), involve eIF-2α phosphorylation. This is also the case for heterologous expression, as derepression of GCN4 mRNA translation required serine 51 in eIF-2α and was accompanied by an increase in eIF-2α phosphorylation that was sustained for at least 72 h after the induction of recombinant protein production.
The only known way to activate Gcn2p is through binding of uncharged tRNA to its histidyl-tRNA-like domain. This domain is also essential for the derepression of GCN4 mRNA translation in response to heterologous expression, as the gcn2m2 allele prevented Gcn4p from accumulating to a high level.
The Gcn1p-Gcn20p complex that is essential for the activation of Gcn2p through interactions with its amino terminus was also found to be required for the stimulation of Gcn2p in the present study. It has been suggested that the ribosome-associated Gcn1p-Gcn20p complex may either facilitate binding of uncharged tRNA to the ribosomal A site or facilitate transfer from the A site to Gcn2p (40). Since translational derepression of GCN4 due to heterologous expression was shown to rely on the tRNA binding domain in Gcn2p, it could be speculated that heterologous expression of cDNA containing rare codons would increase the ratio between the relevant uncharged tRNA and charged tRNA and consequently stimulate Gcn2p activity. However, the expression of the Na,K-ATPase from a chromosomal copy of the expression plasmid derepressed GCN4 translation to the same extent as expression from a very-high-copy-number plasmid, although the density of Na,K-ATPase molecules in yeast membranes only reached 1‰. It would, however, be hard to imagine that such a low expression level would drain the cell of charged tRNA molecules specific for rare codons. At least in Escherichia coli, rare codons only seem to be prohibitive for very high expression levels (22).
The endogenous H-ATPase Pma1p is the most abundant plasma membrane protein in yeast, accounting for up to 25% of the total plasma membrane protein content (32). Therefore, yeast does have the capacity to deposit at least one membrane protein in the plasma membrane at a high density. This density of Pma1p is very high compared to the density of about 1% for recombinant Na,K-ATPase in the plasma membrane of yeast (29). The fact that a density of as little as 1‰ for Na,K-ATPase induced GCN4 translation as efficiently as 1%, while an expression level of 25% for Pma1p did not trigger the GCN4 stress response, illustrates the sensitivity of the system and its ability to discriminate between endogenous and heterologous protein production. The GCN4 system does, however, respond to a two- to threefold overexpression of the endogenous Pma1p protein. In view of the initially high Pma1p content, this represents a huge density of Pma1p in yeast membranes, demonstrating that yeast has the capacity to express huge amounts of membrane protein. These data point to a new role of GCN4 in responding to unbalanced expression of endogenous proteins.
To determine what step in the biosynthesis of membrane proteins was monitored by the GCN4 system, we took advantage of previously characterized temperature-sensitive Na,K-ATPase mutants. In contrast to wild-type Na,K-ATPase, these mutant proteins only accumulated to a wild-type level at 15°C, and not at 35°C, due to misfolding in the ER lumen, as determined by induction of the unfolded protein response. The mutant proteins were synthesized at rates comparable to that of the wild type at both temperatures (21). The observation that the expression of wild-type and mutant Na,K-ATPases induced GCN4 translation to the same extent irrespective of the temperature excludes the possibility that Gcn4p is synthesized in response to the presence of a single class of misfolded protein in the ER lumen. This scenario would also contradict the observation that expression of the cytoplasmic α1(TM4/TM5) loop protein induces GCN4 translation without postulating the existence of a cytoplasmic sensor. This was confirmed by the fact that interference with general protein folding in the ER lumen only induced the unfolded protein response and not GCN4 mRNA translation. Thus, the induction of GCN4 translation by heterologous expression is unrelated to the ability of the protein to fold in the ER lumen. Consequently, the experimental data indicate that a step in heterologous protein production or endogenous protein overexpression preceding entry into the ER lumen must activate Gcn2p and cause eIF-2α phosphorylation.
The Na,K-ATPase is responsible for maintaining Na+ and K+ gradients across the plasma membrane in all higher eukaryotes except for plants. Even though the Na+ concentration in yeast minimal medium is very low, in situ pump activity might interfere with ion homeostasis and cause an induction of GCN4 translation. This can be ruled out, however, as expression of an enzymatically inactive Na,K-ATPase (30) induced GCN4 translation to the same extent as the wild type. Imbalanced Na+ and/or K+ gradients are therefore not the cause of GCN4 induction.
Translational induction of GCN4 seems to be a general response to at least heterologous membrane protein production, as the 7TM receptor PACR1 induced translation to the same extent as the Na,K-ATPase.
What are the promoter targets for Gcn4p produced in response to heterologous expression? A classical Gcn4p target is the HIS4 promoter, which only relies on this transcription factor for activity (24). It was therefore surprising, to some extent, that the increased Gcn4p concentration was not followed by increased HIS4 transcription. On the contrary, HIS4 transcription was reduced after the induction of heterologous membrane protein production. This parallels the observations made under glucose starvation, which was shown to increase GCN4 translation but reduce HIS4 mRNA levels to near the detection limit (41). This indicates that the amount of Gcn4p protein cannot be taken as an indicator of Gcn4p transcriptional activity and that Gcn4p may be modified in response to the stimulus that triggered its biosynthesis. This modification might prevent localization to the nucleus or interactions with the basal transcriptional apparatus. Mbf1p is required for the induction of classical promoters targeted by Gcn4p (34). The observation that the classical HIS4 promoter was not activated in response to heterologous expression despite increased production of Gcn4p may indicate that interactions with Mbf1p at this promoter are prevented during heterologous expression. Posttranslational modifications could be a way to destroy Gcn4p-Mbf1p interactions and allow interactions with other, presently unknown, promoters.
The classical way to induce GCN4 translation is starvation for amino acids, but glucose starvation also increases GCN4 translation. Induction by heterologous expression might therefore be due to a higher rate of amino acid and/or glucose consumption in yeast expressing the heterologous protein than in the control. If this were the case, one would not expect a constant low level of GCN4 translation in the control strain over the entire 72 h of the experiment but rather a later onset of GCN4 translation than that in the Na,K-ATPase-expressing strain. Also, experiments performed in the presence of either 0.5% or 2% glucose or in the presence or absence of amino acids in the growth medium ruled out the possibility that starvation for amino acids or glucose is responsible for the observed induction of GCN4 translation. A further argument for this comes from the fact that growth in raffinose as the sole carbon source showed the same induction of GCN4 translation as growth in 0.5 or 2.0% glucose.
Since uncharged tRNA is the compound activating Gcn2p kinase, the induction of GCN4 translation by heterologous expression could be caused by reduced cytoplasmic levels of amino acids, as observed in the case of glucose limitation (41). Reductions in cytoplasmic levels of amino acids cannot be due to auxotrophy, as all yeast strains carrying expression plasmids or empty control vector used in the present study are prototrophs. Determinations of the cytoplasmic and vacuolar amino acid pools excluded the possibility that the observed induction of GCN4 translation was related to pool size, as these were similar for the heterologously expressing strain and the control. Thus, conditions previously described to induce GCN4 translation seem unable to explain the presently observed increase in GCN4 translation, although it cannot be ruled out by our experiments that the concentration of a single amino acid could have been limited. This is not likely, however, as these experiments were performed in the presence of amino acids and even the exclusion of amino acids from the growth medium did not evoke translational induction of GCN4 mRNA.
Acknowledgments
We thank David Sørensen and Safa Elizadeh for excellent technical assistance. A. Hinnebusch and R. Wek are thanked for their generous gifts of anti-yeast eIF-2α antibodies, GCN4 reporter plasmids, and a plasmid carrying the gcn2m2 allele, and we thank G. Fink for the HIS4-lacZ reporter plasmid.
The Danish Science Foundation and the Novo-Nordic Foundation supported the present work.
REFERENCES
- 1.Ashe, M. P., S. K. De Long, and A. B. Sachs. 2000. Glucose depletion rapidly inhibits translation initiation in yeast. Mol. Biol. Cell 11:833-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belli, G., E. Gari, L. Piedrafita, M. Aldea, and E. Herrero. 1998. An activator/repressor dual system allows tight tetracycline-regulated gene expression in budding yeast. Nucleic Acids Res. 15:942-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berben, G., J. Dumont, V. Gilliquet, P. A. Bolle, and F. Hilger. 1991. The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast 7:475-477. [DOI] [PubMed] [Google Scholar]
- 4.Campbell, I. D. 2002. The march of structural biology. Nat. Rev. Mol. Cell Biol. 3:377-381. [DOI] [PubMed] [Google Scholar]
- 5.Cereghino, G. P., J. L. Cereghino, C. Ilgen, and J. M. Cregg. 2002. Production of recombinant proteins in fermenter cultures of the yeast Pichia pastoris. Curr. Opin. Biotechnol. 13:329-332. [DOI] [PubMed] [Google Scholar]
- 6.Cesareni, G., and J. A. H. Murray. 1987. Plasmid vectors carrying the replication origin of filamentous single stranded phages, p. 135-149. In J. K. Setlow (ed.), Genetic engineering, principles and methods, vol. 9. Plenum Press, New York, N.Y. [Google Scholar]
- 7.Engelberg, D., C. Klein, H. Martinetto, K. Struhl, and M. Karin. 1994. The UV response involving the ras signaling pathway and AP-1 transcription factors is conserved between yeast and mammals. Cell 77:381-390. [DOI] [PubMed] [Google Scholar]
- 8.Goossens, A., T. E. Dever, A. Pascual-Ahuir, and R. Serrano. 2001. The protein kinase Gcn2p mediates sodium toxicity in yeast. J. Biol. Chem. 276:30753-30760. [DOI] [PubMed] [Google Scholar]
- 9.Grisshammer, R., and C. G. Tate. 1995. Overexpression of integral membrane proteins for structural studies. Q. Rev. Biophys. 28:315-422. [DOI] [PubMed] [Google Scholar]
- 10.Grundmann, O., H. U. Mösch, and G. H. Braus. 2001. Repression of GCN4 mRNA translation by nitrogen starvation in Saccharomyces cerevisiae. J. Biol. Chem. 276:25661-25671. [DOI] [PubMed] [Google Scholar]
- 11.Herrick, D., R. Parker, and A. Jacobsen. 1990. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:2269-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hill, J. E., A. M. Myers, T. J. Koerner, and A. Tzagoloff. 1986. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast 2:163-167. [DOI] [PubMed] [Google Scholar]
- 13.Hinnebusch, A. G. 1985. A hierarchy of trans-acting factors modulates translation of an activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 5:2349-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hinnebusch, A. G. 1986. The general control of amino acid biosynthetic genes in the yeast Saccharomyces cerevisiae. Crit. Rev. Biochem. 21:277-317. [DOI] [PubMed] [Google Scholar]
- 15.Hinnebusch, A. G. 1992. General and pathway-specific regulatory mechanisms controlling the synthesis of amino acid biosynthetic enzymes in Saccharomyces cerevisiae, p. 319-414. In E. W. Jones, J. R. Pringle, and J. R. Broach (ed.), The molecular and cellular biology of the yeast Saccharomyces, vol. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- 16.Hinnebusch, A. G. 2000. Mechanism and regulation of initiator methionyl-tRNA binding to ribosomes, p. 185-243. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Hinnebusch, A. G., and K. Natarajan. 2002. Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryot. Cell 1:22-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jia, M. H., R. A. Larosa, J. M. Lee, A. Rafalski, E. Derose, G. Gonye, and Z. Xue. 2000. Global expression profiling of yeast treated with an inhibitor of amino acid biosynthesis, sulfometuron methyl. Physiol. Genomics 3:83-92. [DOI] [PubMed] [Google Scholar]
- 19.Jidenko, M., R. C. Nielsen, T. L. Sorensen, J. V. Moller, M. le Maire, P. Nissen, and C. Jaxel. 2005. Crystallization of a mammalian membrane protein overexpressed in Saccharomyces cerevisiae Proc. Natl. Acad. Sci. USA 102:11687-11691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones, E. W. 1991. Tackling the protease problem in Saccharomyces cerevisiae. Methods Enzymol. 194:428-453. [DOI] [PubMed] [Google Scholar]
- 21.Jorgensen, J. R., and P. A. Pedersen. 2001. Role of phylogenetically conserved amino acids in folding of Na,K-ATPase. Biochemistry 40:7301-7308. [DOI] [PubMed] [Google Scholar]
- 22.Kane, J. F. 1995. Effect of rare codon clusters on high-level expression of heterologous proteins in Escherichia coli. Curr. Opin. Biotechnol. 6:494-500. [DOI] [PubMed] [Google Scholar]
- 23.Lenoir, G., T. Menguy, F. Corre, C. Montigny, P. A. Pedersen, D. Thinès, M. le Maire, and P. Falson. 2002. Overproduction in yeast and rapid and efficient purification of the rabbit SERCA1a Ca2+-ATPase. Biochim. Biophys. Acta 1560:67-83. [DOI] [PubMed] [Google Scholar]
- 24.Lucchini, G., A. G. Hinnebusch, C. Chen, and G. R. Fink. 1984. Positive regulatory interactions of the HIS4 gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 4:1326-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marton, M. J., C. R. Vazquez de Aldana, H. Qui, K. Chakraburtty, and A. Hinnebusch. 1997. Evidence that GCN1 and GCN20, translational regulators of GCN4, function on elongating ribosomes in activation of eIF-2α kinase GCN2. Mol. Cell. Biol. 17:4474-4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muller, G. 2000. Towards 3D structures of G protein-coupled receptors: a multidisciplinary approach. Curr. Med. Chem. 7:861-888. [DOI] [PubMed] [Google Scholar]
- 27.Natarajan, K., M. R. Meyer, B. M. Jackson, D. Slade, C. Roberts, A. G. Hinnebusch, and M. J. Marton. 2001. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 21:4347-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohsumi, Y., K. Kitamoto, and Y. Anraku. 1988. Changes induced in the permeability barrier of the yeast plasma membrane by cupric ion. J. Bacteriol. 170:2676-2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pedersen, P. A., J. H. Rasmussen, and P. L. Jorgensen. 1996. Expression in high yield of pig α1β1 Na,K-ATPase and inactive mutants D369N and D807N in Saccharomyces cerevisiae. J. Biol. Chem. 271:2514-2522. [DOI] [PubMed] [Google Scholar]
- 30.Pedersen, P. A., J. H. Rasmussen, and P. L. Jorgensen. 1996. Consequences of mutations to the phosphorylation site of the α-subunit of Na,K-ATPase for ATP binding and E1-E2 conformational equilibrium. Biochemistry 35:16085-16093. [DOI] [PubMed] [Google Scholar]
- 31.Rolfes, R. J., and A. G. Hinnebusch. 1993. Translation of the yeast transcriptional activator GCN4 is stimulated by purine limitation: implications for activation of the protein kinase GCN2. Mol. Cell. Biol. 13:5099-5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Serrano, R. 1991. Transport across yeast vacuolar and plasma membranes, p. 532-585. In J. R. Broach, J. R. Pringle, and E. W. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces, vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- 33.Swartz, J. R. 2001. Advances in Escherichia coli production of therapeutic proteins. Curr. Opin. Biotechnol. 12:195-201. [DOI] [PubMed] [Google Scholar]
- 34.Takemura, K., S. Harashima, H. Ueda, and S. Hirose. 1998. Yeast coactivator MBF1 mediates GCN4-dependent transcriptional activation. Mol. Cell. Biol. 18:4971-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uesono, Y., and A. Toh-E. 2002. Transient inhibition of translation initiation by osmotic stress. J. Biol. Chem. 277:13848-13855. [DOI] [PubMed] [Google Scholar]
- 36.Valenzuela, L., C. Aranda, and A. Gonzales. 2001. TOR modulates GCN4-dependent expression of genes turned on by nitrogen limitation. J. Bacteriol. 183:2331-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vlak, J. M., C. D. de Gooijer, J. Tramper, and H. G. Miltenburger. 1996. Insect cell cultures: fundamental and applied aspects. Kluwer Academic, Dordrecht, The Netherlands.
- 38.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 39.Wallin, E., and G. von Heijne. 1998. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 7:1029-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wek, S. A., S. Zhu, and R. C. Wek. 1995. The histidyl-tRNA synthetase-related sequence in the eIF-2 alpha protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol. Cell. Biol. 15:4497-4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang, R., S. A. Wek, and R. C. Wek. 2000. Glucose limitation induces GCN4 translation by activation of Gcn2 protein kinase. Mol. Cell. Biol. 20:2706-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]