Abstract
Minute virus of mice NS1 protein is a multifunctional phosphoprotein endowed with a variety of enzymatic and regulatory activities necessary for progeny virus particle production. To regulate all of its different functions in the course of a viral infection, NS1 has been proposed to be modulated by posttranslational modifications, in particular, phosphorylation. Indeed, it was shown that the NS1 phosphorylation pattern is altered during the infectious cycle and that the biochemical profile of the protein is dependent on the phosphorylation state of the polypeptide. Moreover, in vitro approaches have identified members of the protein kinase C (PKC) family, in particular, atypical PKC, as regulators of viral DNA replication through the phosphorylation of NS1 residues T435 and S473, thereby activating the protein for DNA unwinding activities. In order to substantiate these findings in vivo, we produced NS1 in the presence of a dominant-negative PKCλ mutant and characterized the purified protein in vitro. The NS1 protein produced under these conditions was found to be only partially phosphorylated and as a consequence to be deficient for viral DNA replication. However, it could be rescued for this viral function by treatment with recombinant activated PKCλ. Our data clearly demonstrate that NS1 is a target for PKCλ phosphorylation in vivo and that this modification is essential for the helicase activity of the viral polypeptide. In addition, the phosphorylation of NS1 at residues T435 and S473 appeared to occur mainly in the nucleus, providing further evidence for the involvement of PKCλ which, unlike PKCζ, accumulates in the nuclear compartment of infected cells.
Minute virus of mice (MVM), an autonomous parvovirus, is a small nonenveloped spherical particle containing single-stranded linear DNA. This 5.1-kb DNA encodes two structural and at least four nonstructural proteins, of which only NS1, the 83-kDa, mainly nuclear polypeptide, is essential for virus propagation in all cell types. This multifunctional protein is endowed with regulatory as well as enzymatic activities essential for various processes involved in progeny virus production. Besides regulating its own promoter, NS1 trans-activates the promoter for capsid protein production, initiates viral DNA amplification, and interferes with host cell physiology, thereby allowing efficient virus propagation to occur. The capacities of NS1 to modulate host cell transcription, translation, and posttranslational modifications and to interact physically with a variety of host proteins are thought to contribute to the overall cytostatic and cytotoxic effects imposed by the viral polypeptide on host cells (for a review, see reference 37).
In order to exert its various functions in a timely and coordinated manner, NS1 was proposed to be regulated by posttranslational modifications, in particular, phosphorylation. In fact, by monitoring NS1 phosphorylation through metabolic labeling, it was shown that the phosphopeptide pattern of the protein undergoes consecutive changes in the course of a viral infection (8). Furthermore, the biochemical functions of NS1 proved to be tightly dependent on the phosphorylation state of the polypeptide (27). These findings argue strongly for NS1 regulation by discrete phosphorylation and dephosphorylation events. By use of a kinase-free in vitro replication system, it was demonstrated that the initiation of viral DNA amplification indeed requires NS1 phosphorylation by members of the protein kinase C (PKC) family (29) and that distinct NS1 replicative functions are independently regulated by phosphorylation (7, 12, 26, 27, 29).
Unlike double-stranded DNA viruses, such as simian virus 40, parvoviruses use a rolling-circle mode of DNA replication (RCR) similar to that described for bacteriophages and single-stranded plasmids. Thus, replication intermediates are produced by unidirectional strand displacement synthesis starting from single-strand nicks produced by parvovirus initiator protein NS1, which becomes covalently attached to the 5′ end of the nicked strand. The free 3′-hydroxyl produced by this cleavage reaction then serves as a primer for DNA polymerase activity (for details, see references 5 and 10). Initiation and consecutive DNA amplification can be studied in vitro by using plasmids containing the minimal (left-end) origins of replication in the presence of cellular extracts, deoxynucleoside triphosphates (dNTPs), and purified NS1 (9). Recently, it was found that this in vitro replication reaction could be reconstituted by using recombinant polypeptides of NS1, polymerase δ, PCNA, RF-C, RPA, and nicking accessory protein PIF (5). In order to obtain a kinase-free in vitro replication system, we replaced the polymerase fraction with T4 bacteriophage DNA polymerase and used a subcellular fraction consecutively purified on phosphocellulose and threonine affinity columns to supply PIF, PCNA, and RPA. This modified in vitro system allowed us to study the regulation of NS1 replicative functions through phosphorylation (29) and, as a consequence, to show that recombinant PKCλ and an additional phorbol ester-stimulated component are sufficient to render dephosphorylated NS1 active for RCR (12).
Biochemical analyses of NS1 and site-directed mutants of NS1 have been performed to investigate the mechanisms of replication initiation at the protein level. NS1 is endowed with multiple enzymatic activities, such as ATP binding and hydrolysis (17), oligomerization (32), site-specific interaction with the origin (11, 21), site- and strand-specific endonuclease activity (6, 28), and helicase function (17, 26, 28), all of which proved to be essential for parvovirus DNA replication (26, 28). These developments allowed us to characterize the phosphorylation-dependent regulation of NS1 replicative functions on the biochemical level. Two NS1 target sites for atypical PKCλ or PKCζ phosphorylation, mapping at residues T435 and S473, were found to be essential for MVM DNA replication (7, 12). Phosphorylation of these residues appears to control two specific NS1-dependent events: (i) initiation of viral DNA replication through origin unwinding prior to nicking and trans-esterification (26) and (ii) viral DNA amplification through a processive helicase function in front of the replication fork (12, 26), ensuring polymerase δ-driven strand displacement synthesis.
All investigations performed so far regarding the involvement of PKCs in the regulation of MVM DNA replication have been based on either in vitro phosphorylation or the use of NS1 mutants to target PKC phosphorylation sites. The present investigation was performed to elucidate the role of PKC phosphorylation of NS1 under in vivo conditions and to analyze its impact on NS1 replicative functions. To obtain NS1 deprived of atypical PKC phosphorylation, we expressed the protein in mouse A9 cells in the presence of a dominant-negative PKCλ mutant. This strategy resulted in a partially phosphorylated NS1 protein which specifically lacked atypical PKCλ-induced modifications and which could be further purified and analyzed for its activities in vitro. In addition, we compared the pattern of phosphorylation of a purely cytoplasmic NS1 variant with that of the mainly nuclear wild-type protein, in relation to the subcellular distributions of atypical PKCs in mock- and MVM-infected A9 fibroblasts. The data presented here show that NS1 is a target for atypical PKCλ in vivo and that the resulting modifications control the replicative functions of NS1 at the level of its helicase activity. PKCλ phosphorylation of NS1 takes place predominantly in the nuclear compartment, in keeping with the accumulation of this kinase in the nucleus of the host cell. However, a significant fraction of PKCλ is still present in the cytoplasm, suggesting that additional factors mediate the compartmentalization of NS1 phosphorylation and activation.
MATERIALS AND METHODS
Viruses and cells.
Recombinant vaccinia viruses were generated in CV-1 cells, propagated in monolayer cultures of BSC-40 or HeLa cells, and purified over a sucrose cushion as previously described (25, 28). If no suitable antibodies were available for the detection of a recombinant protein (e.g., vλDN), recombinant vaccinia viruses harboring the appropriate gene were identified by PCR with a rightward primer located within the encephalomyocarditis virus leader sequence (5′-GATGCCCAGAAGGTACCCCATTG-3′) and a leftward primer specific for the coding sequence of the appropriate gene. Vaccinia virus vTF7-3 has been described elsewhere (15). The production of wild-type and mutant His6-tagged NS1 as well as PKCλ has been reported elsewhere (7, 12, 28). Wild-type MVM was propagated in A9 cells. A9 and BSC-40 cells were grown as monolayer cultures in Dulbecco's modified Eagle's medium containing 10% fetal calf serum. HeLa-S3 cells were grown in suspension in Joklik's medium containing 10% fetal calf serum in Spinner bottles.
NS1and PKC mutants. (i) NS1 variants.
The nucleotide binding site mutant K405R (25), the cytoplasmic deletion mutant dl158 (24), and the phosphorylation site mutants T435A and S473A (7, 12, 26) have been described and characterized elsewhere.
(ii) PKCλ dominant-negative mutant. The PKCλ-DN clone, encoding the regulatory domain of PKCλ (amino acids 27 to 236), was generated from the corresponding wild-type cDNA clone (12) by PCR with the primer pair 5′-ATACCATGGTAACACACTTTGAGCCTT-3′ and 5′-ATGCTCGAGCGACGCTTTACCACTCTCC-3′ and subcloned into pCR2.1 (Invitrogen, Groningen, The Netherlands) prior to transfer into pTMHis1.
Expression vectors and substrate plasmids.
Plasmids pTM-1 (20) and pTHis-1 (27), used to generate recombinant vaccinia viruses, have been described elsewhere. Plasmids containing the active and inactive left-end origins of MVM DNA replication (pL1-2TC and pL1-2GAA, respectively) have been described by Cotmore and Tattersall (9).
Production and purification of recombinant proteins.
PKCλ and NS1 polypeptides were produced from recombinant vaccinia viruses in suspension cultures of HeLa-S3 cells (27) by using vTF7-3 and the appropriate recombinant vaccinia virus containing the NS1 or the PKCλ gene under the control of the T7 promoter at 15 PFU/cell each. When needed, vaccinia virus expressing PKCλ-DN (or either of the control proteins NS2 and PKCλ) at 15 PFU/cell was added to achieve a dominant-negative effect directed against endogenous PKCλ. Infected cultures were harvested 18 h postinfection, whole (PKCλ) or nuclear (NS1) extracts were prepared, and His-tagged recombinant proteins were purified by using Ni2+-NTA-agarose (Qiagen) columns (28). Protein preparations were analyzed by discontinuous sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and Coomassie blue staining and tested for distinct biochemical properties.
NS1 metabolic labeling, purification, and phosphopeptide analyses.
Metabolic labeling of NS1 and tryptic phosphopeptide analyses were performed essentially as previously described (27). A9 cell cultures (107 cells) were infected with recombinant vaccinia viruses (15 PFU/cell each) and incubated for 3 h before the labeling medium (complete medium lacking phosphate and complemented with 10−10 Ci of [32P]orthophosphate [ICN]/cell) was added. After 4 h, the labeled cells were harvested directly in 1 ml of radioimmunoprecipitation assay buffer (20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 1% sodium deoxycholate, 1% Triton X-100) containing protease and phosphatase inhibitors. NS1 immunoprecipitation was carried out with 10 μl of anti-NSN antiserum (27) for 2 h at room temperature. Immune complexes were precipitated with protein A-Sepharose, washed with radioimmunoprecipitation assay buffer, and further purified by SDS-10% PAGE. 32P-labeled proteins were revealed by autoradiography after blotting on polyvinylidene fluoride membranes, and the band corresponding to NS1 was excised. Digestion of membrane-bound NS1 was performed with 50 U of trypsin for 18 h at 37°C. Tryptic peptides contained in the supernatant were recovered by lyophilization and analyzed on thin-layer cellulose plates in two dimensions, first by electrophoresis with pH 1.9 buffer and then by chromatography with phosphochromatography buffer.
Subcellular localization of PKCs.
For determination of the subcellular localization of endogenous PKC isoforms, A9 cells were grown on “spot slides.” Infections with MVM (30 PFU/cell) were performed for various periods of time. Cells were then fixed with paraformaldehyde, immunostained with various PKC-specific antibodies, and stained with either fluorescein isothiocyanate-, Cy2-, Texas red-, or Cy3-conjugated secondary immunoglobulin G (IgG) antibodies directed against the respective immunoglobulins (see legends to figures). Analyses were performed by standard immunofluorescence microscopy and by laser scanning confocal microscopy (Leica TCS SP laser fitted to a Leica IMRBE microscope).
Helicase assays.
Helicase assays were carried out as described previously (28). M13-VAR was used as a substrate and was prepared by annealing the M13rev primer (Amersham) to M13 single-stranded DNA followed by extension for 5 min at room temperature in the presence of T7 polymerase and dNTPs, including [α-32P]dATP. 32P-labeled fragments of various lengths were obtained by the addition of ddGTP and further incubation for 20 min. Purified NS1 was incubated with 20 ng of substrate for 40 min in the presence of 2 mM ATP. The reactions were stopped by the addition of SDS and EDTA, and the products were analyzed by 7% nondenaturing PAGE in the presence of 0.1% SDS.
Replication assays.
Replication assays were carried out as described previously (29) in the presence of subcellular fraction P1-Thr, 3 U of T4 DNA polymerase, and approximately 200 ng of His-tagged vaccinia virus-produced NS1 (as determined by Coomassie blue staining after SDS-PAGE). P1-Thr consists of the flowthrough fraction of 293 cell extracts purified on phosphocellulose columns and relieved of endogenous serine-threonine kinases by l-Thr affinity chromatography. This fraction contains the replication factors RPA, PCNA, and PIF. The assays were carried out with a 20-μl total volume containing 20 mM HEPES-KOH (pH 7.5), 5 mM MgCl2, 5 mM KCl, 1 mM dithiothreitol, 0.05 mM each dNTP, 2 mM ATP, 40 mM creatine phosphate, 1 μg of phosphocreatine kinase, 10 μCi of [α-32P]dATP (3,000 mCi/mmol), and 20 ng of the appropriate DNA template (pL1-2TC or pL1-2GAA) (12). After incubation at 37°C for 2 h, the reaction was stopped by the addition of 60 μl of 20 mM Tris (pH 7.5)-10 mM EDTA-0.2% SDS and incubation at 70°C for at least 30 min. The reaction products were analyzed by agarose gel electrophoresis after immunoprecipitation with anti-NSN antiserum and digestion with HindIII.
RESULTS
NS1 phosphorylation and regulation by PKCλ in vivo.
MVM NS1 is a multifunctional phosphoprotein that is thought to be primed for its various tasks by differential phosphorylation events in a timely coordinated manner exerted by distinct cellular protein kinases (8). In fact, a variety of in vitro analyses have shown that PKCs (29), particularly PKCλ (12, 26), are able to activate NS1 for its replicative functions, regulating the DNA unwinding activity of the viral product by phosphorylation at residues T435 and S473. However, the relevance of this kinase for NS1 regulation under in vivo conditions has remained elusive.
To rule out the possible involvement of alternative, as-yet-undefinded cellular protein kinases with the same substrate specificity as atypical PKCλ in NS1 phosphorylation in vivo, we designed a specific knockout strategy for living cells based on the genuine regulatory properties of PKCs. Atypical PKCs in the inactive state are thought to be in a closed conformation, in which the substrate binding site is blocked due to its interaction with the N-terminally located pseudosubstrate region. Upon interaction with acidic lipids, such as phosphatidylserine (PS), or the appropriate membranes in the cellular environment, a conformational switch disconnects the pseudosubstrate region from the catalytic center of the enzyme, thereby allowing binding and phosphorylation of the substrate to occur (22). In order to specifically inactivate endogenous PKCλ, we decided to supply the regulatory domain containing the pseudosubstrate region in excess to block the catalytic site of the enzyme. This complex of PKCλ with the artificially produced peptide, mimicking a closed conformation, is unable to respond to stimulation with the cofactor PS. The PKCλ regulatory region should thus behave in a dominant-negative manner, hence, its designation here as PKCλ-DN.
To produce the PKCλ-DN mutant, the cDNA fragment coding for the regulatory domain of this kinase was expressed by means of recombinant vaccinia viruses in mammalian cells. It should be noted that although PKCλ and PKCζ pseudosubstrate sites are identical, it is likely that differences within the sequences surrounding the regulatory domain, particularly around the zinc finger region, are sufficient to discriminate between the two atypical PKCs in vivo. Thus, the greater part of the PKCλ regulatory domain, corresponding to amino acids 27 to 236, was cloned into pTM-1, and recombinant vaccinia viruses expressing this PKCλ variant under the control of the bacteriophage T7 promoter and an encephalomyocarditis virus leader sequence were generated. For comparison, the NS1 protein was also coexpressed with wild-type PKCλ or the parvovirus NS2 protein in order to rule out side effects due to randomly expressed polypeptides.
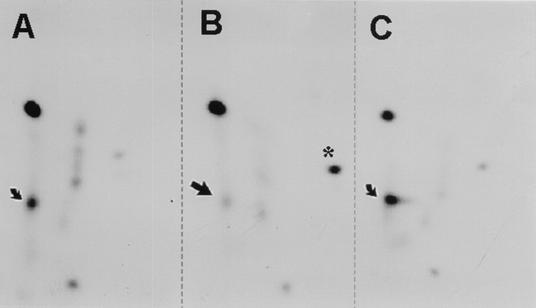
To investigate the involvement of PKCλ in NS1 phosphorylation in vivo, NS1 was expressed either alone or in the presence of PKCλ-DN or wild-type PKCλ, metabolically 32P labeled, and processed for tryptic phosphopeptide analyses. As shown in Fig. 1B versus Fig. 1A and C, the coexpression of PKCλ-DN correlated with the suppression of a major spot in the NS1 phosphopeptide map, while allowing the other phosphorylation events to occur. Since this spot was previously obtained upon in vitro phosphorylation of NS1 by recombinant PKCλ and was assigned to NS1 peptides harboring the phosphorylation sites T435 and S473 (7, 12), we were able to demonstrate that these two regulatory sites in NS1 are indeed targeted by PKCλ in vivo. Interestingly, an additional NS1 phosphopeptide became apparent upon inhibition of PKCλ (Fig. 1B), illustrating the interconnection of different phosphorylation pathways. It should also be noted that no increase in NS1 phosphorylation was observed upon overexpression of recombinant wild-type PKCλ (Fig. 1C), indicating that the amount of endogenous PKCλ present in the cells was not limiting for NS1 phosphorylation.
FIG. 1.
In vivo phosphorylation of NS1 in the presence and absence of active PKCλ. Wild-type NS1 was expressed either alone (A) or in the presence of PKCλ-DN (B) or recombinant PKCλ (C) by infecting A9 cells with recombinant vaccinia viruses. After metabolic 32P labeling, NS1 was purified by immunoprecipitation and SDS-PAGE. Phosphorylation of the individual NS1 polypeptides was determined by tryptic phosphopeptide analyses with two-dimensional thin layer electrophoresis and chromatography and was revealed by autoradiography. Arrows indicate spots which showed a reduced intensity after the coexpression of NS1 with PKCλ-DN, while the asterisk indicates a phosphopeptide which was induced under these conditions.
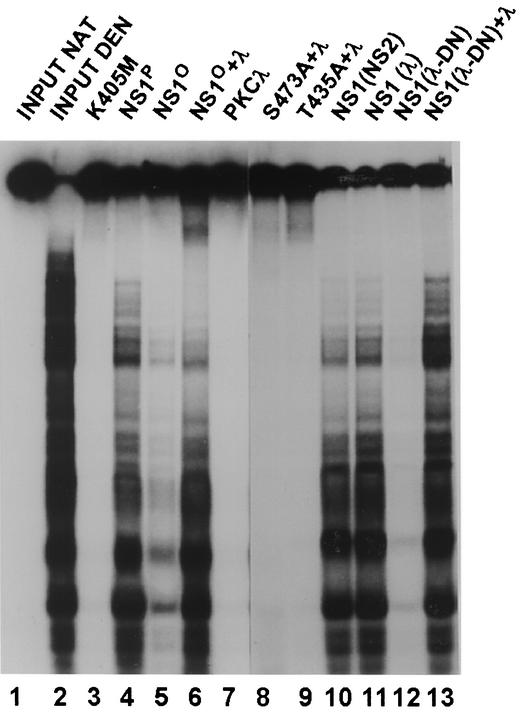
The inhibition of distinct NS1 phosphorylation events caused by the coexpression of PKCλ-DN prompted us to further characterize partially phosphorylated NS1 for its biochemical activities. Wild-type NS1 was expressed in the presence of PKCλ-DN in HeLa cells by means of recombinant vaccinia viruses. For the sake of comparison, the NS1 protein was also coexpressed with either wild-type PKCλ or the parvovirus NS2 protein, serving as a negative control for the specificity of the dominant-negative effect exerted by the N-terminal PKCλ polypeptide. His-tagged NS1 protein was purified by using Ni2+-NTA columns and was further characterized in various in vitro assays. As previously reported (12, 26) and shown in Fig. 2, lanes 1 to 7, PKCλ is able to regulate NS1 DNA unwinding activities in vitro. Therefore, we first tested purified NS1 polypeptides, which were generated in the presence of the above-mentioned regulatory proteins, in standard helicase assays. As shown in Fig. 2, lane 12, NS1 coexpressed with PKCλ-DN was devoid of detectable helicase activity. This defect could be assigned to the lack of in vivo phosphorylation of NS1 by PKCλ (as observed in Fig. 1), since the partially phosphorylated polypeptide recovered its helicase activity upon the addition of activated recombinant PKCλ (Fig. 2, lane 13). In contrast, the PKCλ phosphorylation site mutants T435A and S473A could not be rescued by recombinant PKCλ (Fig. 2, lanes 8 and 9). No inhibition of NS1 helicase activity was observed upon coexpression of NS1 with NS2 or wild-type PKCλ (Fig. 2, lanes 10 and 11), indicating that this inhibition was due to the dominant-negative effect of PKCλ-DN. Altogether, the data demonstrate the relevance of PKCλ phosphorylation of NS1 in vivo and its concomitant regulation of a major function of the viral product, namely, DNA unwinding activity.
FIG. 2.
The helicase activity of NS1 depends on its phosphorylation by PKCλ. The unwinding activity of purified NS1 polypeptides was determined in standard helicase assays in the presence of 2 mM ATP by using M13-VAR as a substrate. The reaction products were analyzed by 7% PAGE in the presence of 0.1% SDS. Lanes: 1 and 2, native and heat-denatured input substrates, respectively; 3, NS1 mutant K405M (nucleotide binding site mutant), serving as a negative control; 4, wild-type fully phosphorylated NS1; 5 and 6, wild-type NS1 dephosphorylated by treatment with alkaline phosphatase; 7, PKCλ alone; 8, NS1 mutant S473A; 9, NS1 mutant T435A; 10, NS1 produced in the presence of NS2; 11, NS1 produced in the presence of recombinant PKCλ; 12 and 13, NS1 produced in the presence of PKCλ-DN. Reactivation reactions shown in lanes 6 to 9 and 13 were performed in the presence of activated recombinant PKCλ.
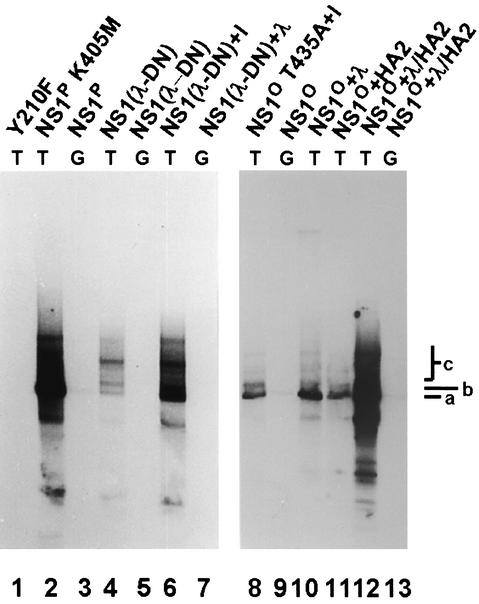
While PKCλ is sufficient to activate NS1 for helicase activity, at least one additional member of the PKC family appears to be required for NS1 to initiate RCR under in vitro conditions (12, 29). Accordingly, PKCλ and the subcellular fraction HA-2 (containing a tetradecanoyl phorbol acetate-responsive PKC) were both necessary to rescue fully dephosphorylated NS1 for RCR (Fig. 3, lanes 8 to 13). This result allowed us to test the specificity of the dominant-negative effect of PKCλ-DN by determining whether NS1 produced in the presence of this inhibitor was still phosphorylated by the HA-2 component and needed only PKCλ to become competent for RCR. The partially phosphorylated NS1 protein produced in the presence of PKCλ-DN [NS1(λ-DN)] was tested for its ability to initiate viral DNA amplification in a kinase-free in vitro replication system (29). As expected from its lack of helicase activity, NS1(λ-DN) proved to be deficient for RCR in the absence of any further treatment (Fig. 3, lanes 1 to 5). In contrast to alkaline phosphatase-treated NS1O, which required both HA-2 and PKCλ for reactivation (Fig. 3, lane 12), NS1(λ-DN) recovered its RCR function upon the addition of activated PKCλ only (lanes 6 and 7), demonstrating the specificity of our dominant-negative approach for PKCλ inhibition.
FIG. 3.
Reactivation of dephosphorylated NS1 in replication assays. NS1-dependent RCR of plasmids containing the left-end active (T) or inactive (G) origin was determined in a kinase-free in vitro system based on P1-Thr and T4 DNA polymerase (29). The reaction products were analyzed by 0.8% agarose gel electrophoresis after immunoprecipitation with anti-NSN antiserum, HindIII restriction digestion, and deproteination. The migration positions of the linearized plasmid (a), a replication intermediate produced by dephosphorylated NS1 (b), and higher-molecular-weight species that represent replication products with displaced single-stranded tails (c) are indicated on the right. Lanes: 1, mutant Y210F (linkage tyrosine mutant), serving as a negative control; 2 and 3, fully phosphorylated NS1; 4 to 7, NS1 produced in the presence of PKCλ-DN; 8 to 13, fully dephosphorylated NS1 (alkaline phosphatase treatment). Reactions in lanes 6, 7, 10, 12, and 13 were performed in the presence of activated recombinant PKCλ; reactions in lanes 11 to 13 were performed in the presence of activated subcellular fraction HA-2.
NS1 phosphorylation at residues T435 and S473 is achieved mainly in the nucleus.
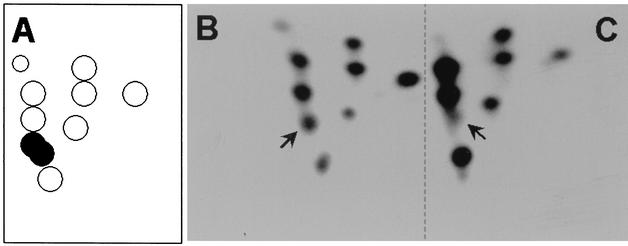
NS1 is a multifunctional phosphoprotein that is thought to be primed for its various tasks by differential phosphorylation events, as suggested by its modulation of biochemical activities or its modification by multiple protein kinases in vitro (27). This information raises the possibility that NS1 may be targeted by different kinases depending on its location within the cell. NS1 may be primed for distinct functions according to the cellular compartment in which it is located. To test this possibility, we determined whether differences in phosphorylation patterns could be detected between the mainly nuclear wild-type NS1 and a cytoplasmic variant lacking the nuclear localization signal. Previous investigations showed that the phosphorylation of NS1 expressed from recombinant vaccinia viruses closely mimics the pattern observed during the replicative phase of a normal MVM infection (8). This result allowed us to use the vaccinia virus expression system in order to compare the tryptic phosphopeptide maps of wild-type NS1 (the greater part of which is nuclear) and cytoplasmic mutant dl158 (lacking amino acids 98 to 256) (24) after metabolic 32P labeling in A9 cells. The patterns of phosphorylation of wild-type NS1 (Fig. 4B) and the cytoplasmic form, dl158 (Fig. 4C), could be distinguished only by the suppression of a few phosphopeptides in the cytoplasmic polypeptide. Interestingly, one of these suppressed spots was previously shown to contain two comigrating NS1 peptides harboring PKCλ phosphorylation sites T435 and S473 (7, 12). These results provided evidence to suggest that the majority of NS1 phosphorylation occurred in the cytoplasm of A9 cells, irrespective of nuclear translocation, while atypical PKCλ phosphorylation at T435 and S473 was achieved mainly in the nuclear compartment. These results are especially interesting because the latter phosphorylation events both target the NS1 helicase domain and regulate the NS1-dependent initiation of viral DNA replication (26), an activity confined to the nuclei of MVM-infected cells (1).
FIG. 4.
Tryptic phosphopeptide analyses of wild-type NS1 and cytoplasmic NS1 variant dl158. (A) Schematic pattern. (B and C) Peptide analyses. Wild-type NS1 (B) and mutant dl158 (C) were expressed by means of recombinant vaccinia viruses in A9 cells, metabolically 32P labeled, and purified by immunoprecipitation and SDS-PAGE. Tryptic phosphopeptide maps were obtained by two-dimensional thin-layer electrophoresis and chromatography and were revealed by autoradiography. Arrows indicate a spot that was significantly reduced in intensity in the dl158 map and that was previously assigned to NS1 peptides harboring the phosphorylation sites T435 and S473 (7, 12), as indicated by filled dots in the schematic pattern.
Subcellular localization of atypical PKCs in A9 cells.
In a variety of systems, PKCs are redistributed within cells upon activation, leading in many situations to the nuclear translocation of activated PKC isoforms (16). This compartmentalization of PKCs, together with our data concerning the nuclear location of NS1 phosphorylation by PKCλ in A9 cells, prompted us to investigate the subcellular distributions of endogenous atypical PKCs in the presence and absence of MVM. The above-mentioned results obtained with the dominant-negative kinase variant strongly argued for a role of PKCλ, rather than the related PKCζ isoform, in NS1 regulation under in vivo conditions. It was therefore interesting to test whether this conclusion was consistent with the respective intracellular distributions of the two atypical PKCs.
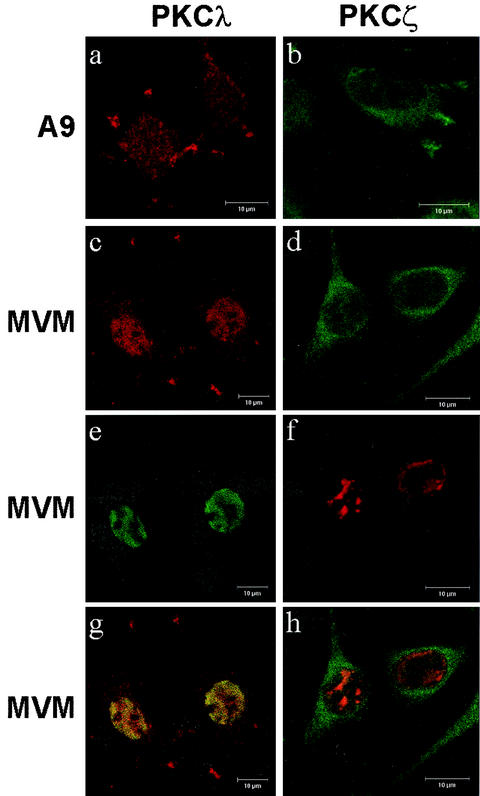
To this end, A9 cells were grown on spot slides, infected or not infected with MVM for 24 h, and fixed with paraformaldehyde. Individual PKCs were then identified with isoform-specific antibodies. NS1 was detected by polyclonal antiserum NS1C, raised against the C-terminal 16 amino acids of NS1 (24). The subcellular distributions of the PKCs and NS1 were then determined by using standard and confocal laser scanning microscopy. Figure 5 shows confocal images of representative cells (>80% of the cell population) obtained repeatedly after independent infections. PKCλ was found in both the cytoplasmic and the nuclear compartments of both mock- and MVM-infected cells. However, MVM-infected cells could be distinguished from controls by the nuclear localization of the PKCλ signal, which colocalized at least in part with the NS1 signal (Fig. 5, left column). Since no increase in the total protein content of PKCλ was observed upon MVM infection of A9 cells (8), this intensification of nuclear staining likely reflected the association of the kinase with membranes (or other nuclear structures) as a consequence of activation processes. Surprisingly, PKCλ did not appear to concentrate in distinct nuclear structures, such as the autonomous parvovirus-associated replication (APAR) bodies, which were recently shown to constitute parvovirus DNA replication centers (1). Therefore, the sites at which NS1 becomes phosphorylated and activated by atypical PKCs may be spatially dissociated from those at which NS1 initiates viral DNA replication, even though both events take place in the nuclear compartment.
FIG. 5.
Subcellular localization of endogenous atypical PKCλ and PKCζ in mock- and MVM-infected A9 cells. A9 cells were grown on spot slides, infected or not infected with MVM (30 PFU/cell), and fixed 24 h postinfection with paraformaldehyde. PKCλ was detected by using an isoform-specific mouse monoclonal antibody (Becton Dickinson P22520) and revealed with Cy3-conjugated anti-mouse IgG. PKCζ was detected by using a specific goat polyclonal antiserum (Santa Cruz) and revealed with fluorescein isothiocyanate-conjugated anti-goat IgG. For double-immunofluorescence staining, NS1 was detected with rabbit antiserum NS1C (7). Subcellular localization of PKCs and NS1 was analyzed by using laser scanning confocal microscopy. (a and b) Uninfected A9 cells. (c to h) MVM-infected A9 cells. (a and c) Anti-PKCλ (red). (e) Anti-NS1 (green). (g) Merge. (b and d) Anti-PKCζ (green). (f) Anti-NS1 (red). (h) Merge. Bars, 10 μm.
In contrast to PKCλ, endogenous PKCζ was detected predominantly in the cytoplasm of both mock- and MVM-infected A9 cells (>70% of a representative cell population) (Fig. 5, right column). Similarly, a recombinant constitutively active mutant form of PKCζ was found to be predominantly located in the cytoplasm of MVM-infected A9 cells (data not shown). These observations suggest that if PKCζ is active or becomes activated during MVM infection, its regulatory impact concerns cytoplasmic steps of the viral life cycle. Together with the PKCλ-DN data described above, the concordance of PKCλ but not PKCζ subcellular localization with NS1 phosphorylation at residues T435 and S473 strongly argues for the involvement of the PKCλ isoform in NS1 regulation through these modifications under in vivo conditions.
It is worth mentioning that the subcellular distributions of PKCλ and PKCζ were also characterized after the expression of these isoforms by means of recombinant vaccinia viruses. These analyses confirmed both the specificity of the antibodies for the respective PKC isoforms and the genuine intracellular distributions of these kinases (data not shown). Together with the similarity of the intracellular NS1 phosphorylation patterns in recombinant vaccinia virus- and MVM-infected cells (8), these observations support the use of the vaccinia virus expression system to investigate the interplay between NS1 and atypical PKCs.
DISCUSSION
Previous in vitro studies showed that phosphorylation of MVM major nonstructural protein NS1 is involved in the modulation of its biochemical activities (27). In consequence, distinct phosphorylation events are essential to endow the viral protein with properties enabling it to achieve distinct functions required for progeny virus production (8, 12, 29). In particular, atypical PKCλ was found to control NS1 DNA unwinding functions, i.e., enzymatic activities involved in viral DNA replication at both the initial nicking reaction and the subsequent strand displacement synthesis (12, 26, 27, 29). Although supported by investigations with NS1 variants obtained by site-directed mutagenesis at consensus PKC phosphorylation sites (7, 12, 26), the relevance of PKC-driven NS1 phosphorylation and regulation for MVM DNA replication remained to be demonstrated under in vivo conditions. This question was addressed in this work by using a dominant-negative construct to generate partially dephosphorylated NS1 in vivo and subsequently analyzing the purified protein in vitro. The data presented provide direct evidence for NS1 phosphorylation by PKCλ under in vivo conditions and, moreover, the regulation of NS1-dependent viral DNA replication by modulation of its helicase activity through these modifications. In addition, our results exemplify the interconnection between the activation of PKCs and their compartmentalization inside host cells.
PKCλ was shown to play an important role in the organization of the cell. Recently, PKCλ and its homologue in Caenorhabitis elegans, PAR3, were identified as signaling components regulating the shape and polarity of epithelial and neuronal cells (13, 23, 30, 35); they are involved in the regulation of the disassembly of actin stress fibers in ras-transformed cells (36). Furthermore, PKCλ proved able to trigger signaling cascades involving its own PDK-1/PKC pathway (31) or the signaling cascade of MEK/ERK protein kinases (2, 34). Hence, the experimental modulation of PKCλ may have significant consequences for cell physiology and thereby additional indirect influences on progeny virus production. Therefore, we used a PKCλ knockout approach in which the lack of in vivo phosphorylation of NS1 was further analyzed for its effect on the biochemical activities of the viral polypeptide outside the cellular environment. The recombinant vaccinia virus expression system makes this strategy possible, since it allows NS1 to be produced in mammalian cells both in sufficient amounts and with a phosphorylation pattern highly similar to that of the genuine NS1 polypeptide produced by MVM in mouse cells (8, 28). Thus, NS1 produced in the presence of a dominant-negative mutant, PKCλ-DN, was found to be only partially phosphorylated and to lack helicase and replication activities, defects that could be restored by supplementation of the in vitro assay mixtures with activated PKCλ. These experiments clearly demonstrated the direct involvement of this atypical PKC isoform in the phosphorylation of NS1 and the regulation of its replicative functions. This complementation in vitro excludes possible indirect effects of the PKCλ knockout through alterations of host cell physiology.
In vitro reactivation of NS1 helicase activity can be achieved with either one of the atypical PKCs, PKCλ or PKCζ (12). Although the involvement of PKCζ in general cannot be ruled out, it is not supported by the differences shown for the kinases in our experiments. (i) The differences in the primary structure between the regulatory domains of PKCλ and PKCζ imply specific inhibition of PKCλ by our dominant-negative mutant. (ii) The intracellular distributions of the two kinases are different. Nuclear phosphorylation of regulatory sites T435 and S473 likely is targeted by nuclear PKCλ, while mainly cytoplasmic PKCζ is a very doubtful candidate for these phosphorylation events.
During the course of an MVM infection, viral protein NS1 not only drives viral DNA replication but also is directly responsible for cell disturbances that eventually lead to cell lysis and release of progeny particles (4). To ensure the optimal production of viruses, NS1 functions are thought to become temporally regulated so that cytotoxic activities are delayed and do not interfere with virus replication. This regulation could be achieved by time-dependent changes in the pattern of phosphorylation of the viral polypeptide (8). However, some NS1 modifications, such as PKCλ phosphorylation at residues T435 and S473, activate the capacity of NS1 for both replication (12, 26) and alterations of host cell morphology (7). Therefore, additional means to regulate NS1 activities besides phosphorylation may be necessary to dissociate these conflicting activities.
Sequential functioning of NS1 could also be achieved through spatial regulation based on compartmentalization of the viral product and/or its regulators within infected cells. This possibility is consistent with results of the present investigation showing that PKCλ phosphorylation at residues T435 and S473 occurs predominantly in the nucleus. This evidence suggests that newly synthesized NS1 polypeptides remain underphosphorylated until they migrate to the nucleus, in which they are further modified and activated for their replicative functions. Despite the finding that the same modifications endow NS1 with the ability to induce alterations of host cell morphology (7), this toxic activity becomes apparent only after NS1 has been exported from the nucleus to interact with the cytoskeleton. In keeping with this proposal, an NS1 mutant with a C-terminal deletion was almost fully sequestered in the nucleus, proved to be competent for replication (24), but had less cytotoxic activity than the wild-type (30% cytoplasmic) polypeptide (18). Although this scenario is only hypothetical at this time, it raises the intriguing possibility that NS1 priming for distinct functions is controlled, at least in part, at the level of nuclear-cytoplasmic transport. Again, as already well described for DNA replication activities, the functionally related simian virus 40 large T antigen provides an interesting precedence for this type of regulation (33). Moreover, recent findings concerning the regulatory properties of small parvovirus protein NS2 also support NS1 regulation by compartmentalization. NS2 was recently shown to directly interact with nuclear export factor CRM1 (3), which proved to be essential for the nuclear egress of newly assembled capsids (14, 19). By analogy, NS2 could also be involved in the regulation of the export of other cellular and viral constituents, such as NS1 polypeptides. However, whether the nuclear export of NS1 is further subject to regulation by posttranslational modifications remains to be shown.
Acknowledgments
We are indebted to Bernard Moss (NIH) for making the T7-driven vaccinia virus expression system available to us. We are most grateful to Peter Tattersall and Susan Cotmore for plasmid constructs and helpful discussions.
This work was supported by the Commission of the European Communities.
Footnotes
This article is dedicated to Harald zur Hausen on the occasion of his retirement as head of the Deutsches Krebsforschungszentrum, with gratitude and appreciation for 20 years of leadership.
REFERENCES
- 1.Bashir, T., J. Rommelaere, and C. Cziepluch. 2001. In vivo accumulation of cyclin A and cellular replication factors in autonomous parvovirus minute virus of mice-associated replication bodies. J. Virol. 75:4394-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berra, E., M. M. Municio, L. Sanz, S. Frutos, M. T. Diaz-Meco, and J. Moscat. 1997. Positioning atypical protein kinase C isoforms in the UV-induced apoptotis signaling cascade. Mol. Cell. Biol. 17:4346-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bodendorf, U., C. Cziepluch, J.-C. Jauniaux, J. Rommelaere, and N. Salome. 1999. Nuclear export factor CRM1 interacts with nonstructural proteins NS2 from parvovirus minute virus of mice. J. Virol. 73:7769-7779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caillet-Fauquet, P., M. Perros, A. Brandenburger, P. Spegelaere, and J. Rommelaere. 1990. Programmed killing of human cells by means of an inducible clone of parvoviral genes encoding non-structural proteins. EMBO J. 9:2989-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christensen, J., and P. Tattersall. 2002. Parvovirus initiator protein NS1 and RPA coordinate replication fork progression in a reconstituted DNA replication system. J. Virol. 76:6518-6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen, J., S. F. Cotmore, and P. Tattersall. 2001. Minute virus of mice initiator protein NS1 and a host KDWK family transcription factor must form a precise ternary complex with origin DNA for nicking to occur. J. Virol. 75:7009-7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corbau, R., V. Duverger, J. Rommelaere, and J. P. F. Nüesch. 2000. Regulation of MVM NS1 by protein kinase C: impact of mutagenesis at consensus phosphorylation sites on replicative functions and cytopathic effects. Virology 278:151-167. [DOI] [PubMed] [Google Scholar]
- 8.Corbau, R., J. P. F. Nüesch, N. Salome, and J. Rommelaere. 1999. Phosphorylation of the viral non-structural protein NS1 during MVMp infection of A9 cells. Virology 259:402-415. [DOI] [PubMed] [Google Scholar]
- 9.Cotmore, S. F., and P. Tattersall. 1994. An asymmetric nucleotide in the parvoviral 3′hairpin directs segregation of a single active origin of DNA replication. EMBO J. 13:4145-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cotmore, S. F., and P. Tattersall. 1995. DNA replication in the autonomous parvoviruses. Semin. Virol. 6:271-281. [Google Scholar]
- 11.Cotmore, S. F., J. Christensen, J. P. F. Nüesch, and P. Tattersall. 1995. The NS1 polypeptide of the murine parvovirus minute virus of mice b inds to DNA sequences containing the motif [ACCA]2-3. J. Virol. 69:1652-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dettwiler, S., J. Rommelaere, and J. P. F. Nüesch. 1999. DNA unwinding functions of minute virus of mice NS1 protein are modulated by the lambda isoform of protein kinase C. J. Virol. 73:7410-7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ebnet, K., A. Suzuki, Y. Horikoshi, T. Hirose, M.-K. Meyer zu Brickwedde, S. Ohno, and D. Vestweber. 2001. The cell polarity protein ASIP/PAR-3 directly associates with junctional adhesion molecule (JAM). EMBO J. 20:3738-3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eichwald, V., L. Daeffler, M. Klein, J. Rommelaere, and N. Salomé. 2002. The NS2 proteins of minute virus of mice are required for efficient nuclear egress of progeny virions in mouse cells. J. Virol. 76:10356-10364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eucaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodnight, J.-A., H. Mischak, W. Kolch, and J. F. Mushinski. 1995. Immunocytochemical localization of eight protein kinase C isozymes overexpressed in NIH3T3 fibroblasts. J. Biol. Chem. 270:9991-10001. [DOI] [PubMed] [Google Scholar]
- 17.Jindal, H., C. B. Yong, G. M. Wilson, P. Tam, and C. R. Astell. 1994. Mutations in the NTP-binding motif of minute virus of mice (MVM) NS1 protein uncouple ATPase and DNA helicase functions. J. Biol. Chem. 269:3283-3289. [PubMed] [Google Scholar]
- 18.Legendre, D., and J. Rommelaere. 1992. Terminal regions of the NS1 protein of the parvovirus minute virus of mice are involved in cytotoxicity and promoter trans-inhibition. J. Virol. 66:5705-5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller, C. L., and D. J. Pintel. 2002. Interaction between parvovirus NS2 protein and nuclear export factor Crm1 is important for viral egress from the nucleus of murine cells. J. Virol. 76:3257-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moss, B., O. E. Stein, T. Mizukami, W. A. Alexander, and T. R. Fuerst. 1990. Product review. New mammalian expression vectors. Nature 348:91-92. [DOI] [PubMed] [Google Scholar]
- 21.Mouw, M., and D. J. Pintel. 1998. Amino acids 16-275 of minute virus of mice NS1 include a domain that specifically binds (ACCA)2-3-containing DNA. Virology 251:123-131. [DOI] [PubMed] [Google Scholar]
- 22.Newton, A. 1997. Regulation of protein kinase C. Curr. Opin. Cell Biol. 9:161-167. [DOI] [PubMed] [Google Scholar]
- 23.Noda, Y., R. Takeya, S. Ohno, S. Naito, T. Ito, and H. Sumimoto. 2001. Human homologues of the Caenorhabditis elegans cell polarity protein PAR6 as an adaptor that links the small GTPases Rac and Cdc42 to atypical protein kinase C. Genes Cells 6:107-119. [DOI] [PubMed] [Google Scholar]
- 24.Nüesch, J. P. F., and P. Tattersall. 1993. Nuclear targeting of the parvoviral replicator protein molecule NS1: evidence for self-association prior to nuclear transport. Virology 196:637-651. [DOI] [PubMed] [Google Scholar]
- 25.Nüesch, J. P. F., S. F. Cotmore, and P. Tattersall. 1992. Expression of functional parvoviral NS1 from recombinant vaccinia virus: effects of mutations in the nucleotide-binding motif. Virology 191:406-416. [DOI] [PubMed] [Google Scholar]
- 26.Nüesch, J. P. F., J. Christensen, and J. Rommelaere. 2001. Initiation of minute virus of mice DNA replication is regulated at the level of origin unwinding by atypical protein kinase C phosphorylation of NS1. J. Virol. 75:5730-5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nüesch, J. P. F., R. Corbau, P. Tattersall, and J. Rommelaere. 1998. Biochemical activities of minute virus of mice nonstructural protein NS1 are modulated by the phosphorylation state of the polypeptide. J. Virol. 72:8002-8012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nüesch, J. P. F., S. F. Cotmore, and P. Tattersall. 1995. Sequence motifs in the replicator protein of parvovirus MVM essential for nicking and covalent attachment to the viral origin: identification of the linking tyrosine. Virology 209:122-135. [DOI] [PubMed] [Google Scholar]
- 29.Nüesch, J. P. F., S. Dettwiler, R. Corbau, and J. Rommelaere. 1998. Replicative functions of minute virus of mice NS1 protein are regulated in vitro by phosphorylation through protein kinase C. J. Virol. 72:9966-9977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohno, S. 2001. Intercellular junctions and cellular polarity: the PAR-aPKC complex, a conserved core cassette playing fundamental roles in cell polarity. Curr. Opin. Cell Biol. 13:641-648. [DOI] [PubMed] [Google Scholar]
- 31.Parekh, D., W. Ziegler, and P. J. Parker. 2000. Multiple pathways control protein kinase C phosphorylation. EMBO J. 19:496-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pujol, A., L. Deleu, J. P. F. Nüesch, C. Cziepluch, J.-C. Jauniaux, and J. Rommelaere. 1997. Inhibition of parvovirus minute virus of mice replication by a peptide involved in the oligomerization of the nonstructural protein NS1. J. Virol. 71:7393-7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rihs, H., D. A. Jans, H. Fan, and R. Peters. 1991. The rate of nuclear cytoplasmic protein transport is determined by the casein kinase II site flanking the nuclear localization sequence of the SV40 T-antigen. EMBO J. 10:633-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schönwasser, D., R. M. Marais, C. J. Marschall, and P. J. Parker. 1998. Activation of the mitogen-activated protein kinase/extracellular signal-regulated kinase pathway by conventional, novel, and atypical protein kinase C isotypes. Mol. Cell. Biol. 18:790-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki, A., T. Yamanaka, T. Hirose, N. Manabe, K. Mizuno, M. Shimizu, K. Akimoto, Y. Izumi, T. Ohnishi, and S. Ohno. 2001. Atypical protein kinase C is involved in the evolutionarily conserved PAR protein complex and plays a critical role in establishing epthelia-specific junctional structures. J. Cell Biol. 152:1183-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ueberall, F., K. Hellbert, S. Kampfer, K. Maly, A. Villunger, M. Spitaler, J. Mwanjewe, G. Baier-Bitterlich, G. Baier, and H. H. Grunicke. 1999. Evidence that atypical protein kinase C-L and atypical protein kinase C-Z participate in ras-mediated reorganization of the F-actin cytoskeleton. J. Cell Biol. 144:413-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vanacker, J.-M., and J. Rommelaere. 1995. Non-structural proteins of autonomous parvoviruses: from cellular effects to molecular mechanisms. Semin. Virol. 6:291-297. [Google Scholar]