Abstract
Phosducin or phosducin-like protein (PhLP) is a positive regulator of Gβγ activity. The Gβ (SfaD) and Gγ (GpgA) subunits function in vegetative growth and developmental control in the model filamentous fungus Aspergillus nidulans. To better understand the nature of Gβγ-mediated signaling, phnA, encoding an A. nidulans PhLP, has been studied. Deletion of phnA resulted in phenotypes almost identical to those caused by deletion of sfaD, i.e., reduced biomass, asexual sporulation in liquid submerged culture, and defective fruiting body formation, suggesting that PhnA is necessary for Gβ function. The requirement for the RGS protein FlbA in asexual sporulation could be bypassed by the ΔphnA mutation, indicating that PhnA functions in FlbA-controlled vegetative growth signaling, primarily mediated by the heterotrimeric G protein composed of FadA (Gα), SfaD, and GpgA. However, whereas deletion of fadA restored both asexual sporulation and the production of sterigmatocystin (ST), deletion of sfaD, gpgA, or phnA failed to restore ST production in the ΔflbA mutant. Further studies revealed that SfaD, GpgA, and PhnA are necessary for the expression of aflR, encoding the transcriptional activator for the ST biosynthetic genes, and subsequent ST biosynthesis. Overexpression of aflR bypassed the need for SfaD in ST production, indicating that the results of SfaD-mediated signaling may include transcriptional activation of aflR. Potential differential roles of FadA, Gβγ, and FlbA in controlling ST biosynthesis are further discussed.
Vegetative growth signaling in the model filamentous fungus Aspergillus nidulans is primarily mediated by a heterotrimeric G protein composed of FadA (Gα), SfaD (Gβ), and GpgA (Gγ) (29, 33, 41). Activated FadA-GTP transduces vegetative growth signals in part through a cyclic AMP (cAMP)-dependent protein kinase (PkaA), which results in stimulation of hyphal proliferation and inhibition of asexual sporulation as well as production of the carcinogenic mycotoxin sterigmatocystin (ST), the penultimate precursor of the notorious aflatoxins (15, 34, 35, 41). FlbA is an RGS (regulator of G protein signaling) protein that negatively regulates FadA-mediated growth signaling, likely by enhancing the intrinsic GTPase activity of FadA (41). A loss of flbA function and FadA dominant activating mutations both result in the fluffy-autolytic phenotype, which is characterized by an accumulation of undifferentiated hyphal mass followed by colony disintegration (21, 41, 43). As if FadA is the primary target of FlbA activity, deletion or dominant interfering fadA mutations bypass the need for FlbA in asexual sporulation and ST production (21, 41, 43).
As shown for other eukaryotes (reviewed in reference 24), Gβγ subunits play an important role in A. nidulans. A. nidulans Gβγ (SfaD-GpgA) is required for normal vegetative growth, the formation of sexual fruiting bodies, and proper down-regulation of asexual development (29, 33). A recent study demonstrated that the heterotrimer comprised of GanB (another Gα subunit) and SfaD-GpgA is associated with spore germination and carbon source sensing in A. nidulans (20). Deletion of sfaD or gpgA resulted in restricted vegetative growth and rescued the asexual developmental defects caused by the absence of FlbA function, providing genetic evidence that SfaD and GpgA function in the FadA-mediated vegetative growth signaling pathway negatively controlled by FlbA. Moreover, deletion of sfaD and gpgA caused severely impaired formation of sexual fruiting bodies (cleistothecia), implying that SfaD-GpgA also plays an important role in sexual reproduction (29, 33).
Phosducin and phosducin-like proteins (PhLPs) are a group of evolutionarily conserved proteins that were initially thought to regulate Gβγ activity negatively by binding and sequestering the Gβγ heterodimer from its interaction with Gα or downstream effectors (2, 4, 11; reviewed in reference 30). However, recent genetic studies with the chestnut blight fungus Cryphonectria parasitica (17) and the social amoeba Dictyostelium discoideum (3) showed that PhLP is a positive regulator of Gβγ signaling. Furthermore, biochemical studies of PhLP in humans (23) and D. discoideum (18) clearly demonstrated that PhLP is essential for Gβγ dimer assembly and for normal (protein) levels of Gβ and Gγ. In particular, Lukov et al. (23) showed that PhLP bound nascent Gβ without Gγ, which stabilizes the Gβ subunit until Gγ can associate with the PhLP-Gβ complex. PhLP-Gβγ formation is transient, and PhLP is expected to be rapidly displaced from Gβγ. Free PhLP can then catalyze another round of Gβγ assembly, which is necessary for proper translation of the Gβ and Gγ mRNAs (23). Collectively, recent genetic and biochemical studies provide a new model for PhLP function in that PhLP is an essential positive regulator of Gβγ signaling via acting as a molecular chaperone for Gβγ assembly.
We have been dissecting the functions of heterotrimeric G protein components (Gαβγ) and their regulators (FlbA and RgsA) in coordinating vegetative growth, development, stress responses, and secondary metabolism in A. nidulans (14, 29, 33, 41, 43). Coupling the crucial roles of PhLP in G protein signaling and a partial understanding of the roles of the Gβγ heterodimer led us to identify potential PhLPs in A. nidulans and to further investigate the nature of SfaD-GpgA signaling. Three potential PhLPs (PhnA, PhnB, and PhnC) have been identified in the genome of A. nidulans, where PhnA is most similar to Bdm-1 in C. parasitica (17). Functional characterization of phnA and genetic and metabolic studies of sfaD, gpgA, and phnA deletion mutants revealed that PhnA is an essential positive regulator of SfaD-GpgA signaling required for vegetative growth and proper regulation of sexual/asexual development in A. nidulans. Moreover, we demonstrate that PhnA, SfaD, and GpgA are necessary for the production of ST and that SfaD-dependent ST production occurs through the expression of aflR, encoding a transcriptional activator of the ST biosynthetic genes.
MATERIALS AND METHODS
Aspergillus strains, culture conditions, and genetics.
The A. nidulans strains used for this study are listed in Table 1. Standard culture and genetic techniques were used (16, 27). Aspergillus strains were grown on solid or in liquid minimal medium with supplements (MM) at 37°C as previously described (16, 27). Measurements of growth rates, dry weights, and sporulation levels were done in triplicate in MM and YM (MM plus 0.1% yeast extract). Radial growth rates were determined by point inoculating individual strains at the center of solid MM and YM plates and incubating them at 37°C for 5 days. Numbers of conidia and Hülle cells per plate were counted as previously described (33). To measure dry weights, 5 × 107 conidia of each strain were inoculated in liquid YM and incubated at 37°C and 250 rpm for 24 h, followed by drying of the collected mycelium samples at room temperature for more than 5 days. To check submerged sporulation, 5 × 107 conidia of each strain were inoculated in 100 ml of liquid YM, incubated at 37°C at 250 rpm, and observed from 18 to 30 h under a microscope.
TABLE 1.
A. nidulans strains used for this study
| Strain | Genotypea | Source or reference |
|---|---|---|
| FGSC4 | veA+ | FGSCb |
| FGSC26 | biA1 | FGSC |
| PW1 | biA1 argB2 metG1 | P. Weglenski |
| JAS30 | biA1 metG1 | This study |
| RJA56.25 | pabaA1 yA2 | 32 |
| RKH51.117 | pabaA1 yA2 | 14 |
| RJY1.12 | pabaA1 yA2 flbA98 metG1 sfaA1 | 43 |
| RJY67.3 | pabaA1 yA2 flbA98 argB2 metG1 sfaC67 | 43 |
| RJY83.21 | pabaA1 yA2 flbA98 argB2 metG1 sfaE83 | 43 |
| TJAP3c | biA1 argB2 metG1 ΔphnA::argB+ | This study |
| TJAPF36d | ΔflbA::metG+biA1 argB2 metG1 ΔphnA::argB+ | This study |
| RJA5.9 | pyrG89 ΔflbA::argB+pyroA4 | 33 |
| RJYE07 | biA1 ΔflbA::argB+ ΔfadA::argB+trpC801 | 15 |
| RJA15.48e | biA1 ΔflbA::argB+ ΔfadA::argB+ | 33 |
| RJAG19.6f | pabaA1 yA2 ΔgpgA::argB+ | 33 |
| RJA55.4 | pabaA1 yA2 ΔgpgA::argB+ ΔsfaD::argB+ | 33 |
| RSRB1.15 | biA1 ΔsfaD::argB+ | S. Rosén and J.-H. Yu |
| RJA55.11 | pabaA1 yA2 ΔsfaD::argB+ | 33 |
| RSRAB.1g | pabaA1 yA2 metG1 ΔsfaD::argB+alcAp::aflR | S. Rosén and J.-H. Yu |
| RSRAB.4g | biA1 yA2 metG1 ΔsfaD::argB+alcAp::aflR | S. Rosén and J.-H. Yu |
All strains carry the veA1 mutation except for FGSC4.
FGSC, Fungal Genetics Stock Center.
TJAP1, -2, and -3 are isogenic and were examined for radial growth rates, dry weights, and sporulation levels. TJAP3 was used in Northern blot and ST analyses.
TJAPF8, -36, -38, and -73 are isogenic.
Other ΔfadA ΔflbA strains were also crossed with ΔphnA strains.
RJAG19.6, -19.8, and -19.9 are isogenic and were used in ST analysis.
Both strains were used in ST analysis.
Examinations of sexual development were carried out as previously described (32). Briefly, 106 conidia of multiple ΔphnA (TJAP1, -2, and -3), wild-type (RKH51.117), and ΔsfaD (RSRB1.15) strains were spread on solid MM in triplicate. The plates were sealed with plastic film and aluminum foil to prevent exposure to air and light and incubated at 37°C for 7 to 10 days. To enhance the sexual cross success rates of the ΔphnA mutant in the generation of the ΔphnA ΔflbA double mutant, spores of ΔphnA (TJAP1, -2, and -3) and ΔfadA ΔflbA (RJA15.48 and RJYE07) strains were mixed at various ratios (3:1, 2:1, 1:1, 1:2, and 1:3), and individual mycelial mats were torn and spread on sexual induction medium containing 20 mM glycine and 2% glucose (9). However, due to the lack of cleistothecium formation from the 20 crosses, the ΔphnA ΔflbA double mutant was generated by deleting flbA from the ΔphnA mutant (see below).
For Northern blot analyses, samples from liquid submerged and developmentally induced cultures were collected at designated time points as described previously (13, 31). Approximately 5 × 107 conidia of wild-type or relevant mutant strains were inoculated in 100 ml liquid YM in 250-ml flasks and incubated at 37°C and 250 rpm. Individual mycelial samples were collected, squeeze dried, and stored at −80°C until needed for total RNA isolation. For induction of sexual and asexual development, mycelia grown vegetatively for 16 h were transferred to solid MM, and the plates were exposed to air for asexual development induction or tightly sealed from air and light for sexual development induction. To check the mRNA levels of stcU and aflR, the hyphal mat was collected from a 2-ml stationary culture in a liquid complete medium (CM) enhancing ST production (31, 40) from 1 to 4 days postinoculation, and total RNA was isolated and subjected to Northern blot analyses.
The effects of aflR overexpression on ST production in the absence of SfaD function were examined by growing wild-type, ΔsfaD, and alcAp::aflR ΔsfaD strains (RSRAB.1 and RSRAB.4; S. Rosén and J.-H. Yu, unpublished data) in 2 ml liquid CM containing 2% glucose at 37°C for 24 h, replacing the medium with 2 ml liquid CM with 100 mM threonine (inducing conditions) or with 2% glucose (noninducing conditions), and further incubating the strains at 37°C for 1 to 4 days (or up to 7 days in glucose-CM). ST was extracted from individual samples as described below.
Generation of phnA deletion mutant.
The deletion of phnA was accomplished by multiple cloning steps due to the incomplete development of the double-joint PCR (DJ-PCR) technique (44) at that time. The 5′ (∼1.3 kb) and 3′ (∼1.2 kb) flanking regions of the phnA open reading frame (ORF) were amplified with the oligonucleotide pairs OJA6-OJA8 and OJA7-OJA9, respectively. The amplicons were digested with XhoI and self-ligated. The fused DNA fragment was cloned into the pGEM-T Easy vector (Promega), giving rise to an ∼2.5-kb joined fragment (ΔphnA/pGEM-T Easy plasmid). The XhoI-digested argB+ marker (∼1.8 kb) was generated by restriction enzyme digestion of the plasmid pJW88 (J. Wieser and T. H. Adams, unpublished). The argB+ marker was then ligated to the XhoI-cut ΔphnA/pGEM-T Easy plasmid. The final ΔphnA construct (pJAB2) consisted of the 5′ (∼1.3 kb) flanking region, argB+ (∼1.8 kb), and the 3′ flanking region (∼1.2 kb) in the pGEM-T Easy vector. The pJAB2 plasmid was introduced directly into A. nidulans PW1 to generate the ΔphnA mutant strains TJAP1, -2, and -3 (Table 1). The ΔphnA genotype was confirmed by PCR amplification of the phnA coding region with the primer pair OJA10 and OJA202 followed by restriction enzyme digestion of the amplicons and genomic DNA Southern blot analyses (44). Phenotypic changes caused by the deletion of phnA were linked with the ΔphnA genotype confirmed by PCR amplicon size and digestion patterns.
Generation of ΔphnA ΔflbA double mutant.
Due to the impairment of fruiting body formation caused by the ΔphnA mutation, even in outcrosses, the ΔphnA ΔflbA double mutant was generated by deleting the flbA gene from the ΔphnA mutant (TJAP2). The double-joint PCR method was used to generate the flbA deletion construct (44). An flbA deletion construct containing the metG+ marker (36) (amplified with the primer pair OJA232-233), with approximately 1 kb each of the 5′ and 3′ flanking regions of flbA (amplified with primer pairs OJA234-236 and OJA237-238; see Table 2), was constructed as described previously (44) and introduced into the recipient strain TJAP2 (biA1 argB2 metG1 ΔphnA::argB+). The resulting transformants were randomly screened for the deletion of flbA (using the primer pair OJA234-241) and phnA (primer pair OJA10-202) as described previously (44). All the ΔphnA ΔflbA double mutant strains isolated exhibited sporulation levels similar to those of the ΔphnA mutants.
TABLE 2.
Oligonucleotides used for this study
| Primer | Sequence | Position and/or purpose |
|---|---|---|
| OJA6 | cg G GAT CCg cat tgt acg ata atc ttg | Forward 5′ flanking region of phnA ORF for phnA deletion with BamHI tail |
| OJA7 | cg G GAT CCa gac gtc cat ttt gca att c | Reverse 3′ flanking region of phnA ORF for phnA deletion with BamHI tail |
| OJA8 | ccg CTC GAG agg agg cct gta gtt gg | Reverse 5′ flanking region of phnA ORF for phnA deletion with XhoI tail |
| OJA9 | ccg CTC GAG tga cat caa cat aac tct cc | Forward 3′ flanking region of phnA ORF for phnA deletion with XhoI tail |
| OJA10 | gcg cag act gat tca gca tgg | 5′ Forward region of phnA; confirmation of ΔphnA mutation |
| OJA202 | gaa gct cac aac atg ttg | Sequencing of phnA; confirmation of ΔphnA mutation |
| OJA232 | agc gca agt tcg atg ata tgg acc | 5′ Forward region of metG |
| OJA233 | aag atg tga tcg atg agg cat | 3′ Reverse region of metG |
| OJA234 | cga agc cat cca gaa ggt gat | 5′ Forward region of flbA; confirmation of ΔflbA mutation |
| OJA236 | ggt cca tat cat cga act tgc gct a gaa gag tgc agg tcg gag tgg | 5′ Flanking region of flbA with metG tail (underlined) |
| OJA237 | atg cct cat cga tca cat ctt a gct cac aac gtt cat gac agt aat | 3′ Flanking region of flbA with metG tail (underlined) |
| OJA238 | tcg tgt cag tca ctt gaa tga | 3′ Reverse region of flbA |
| OJA235 | cta gca tag cac cat acc cat | 5′ Nested primer for flbA deletion |
| OJA239 | gga tgg tag gta tgt gaa tca | 3′ Nested primer for flbA deletion |
| OJA240 | ttc gtc atc ccg tac tgt att | 5′ Forward region of flbA |
| OJA241 | gtg cct aga aag taa tat cga | 3′ Reverse region of flbA; confirmation of ΔflbA mutation |
| OJA144 | ttc cta tgt cat ggt agc | 5′ Forward region of stcU |
| OJA145 | gta cgg tgt gac taa ctg | 3′ Reverse region of stcU |
| OJA242 | gct gtc gat ctt tgt acc ctg | 5′ Forward region of laeA |
| OJA243 | cgt tcc tgg atg tgg tcg cct | 3′ Reverse region of laeA |
| OJA244 | cct gta tca tat gca aat atc | 5′ Forward region of aflR |
| OJA245 | atc att cca gat tat tca acc | 3′ Reverse region of aflR |
Nucleic acid manipulation.
Genomic DNA and total RNA isolation was carried out as previously described (31, 44). Probes for phnA, nsdD, aflR, stcU, and laeA were prepared by PCR amplification of individual ORFs from wild-type (FGSC4) genomic DNA by using the oligonucleotides listed in Table 2. Each amplicon was labeled with [32P]dCTP. A MagnaProbe membrane (Osmonics, MN) was used for blotting of the nucleic acids after the separation of ∼6 μg of total RNA in a 1.1% agarose gel containing 3% formaldehyde. 32P-labeled probes were used for hybridization, using modified Church buffer (15), at 63°C for 20 h.
ST analyses.
Conidia (∼106) of individual strains were inoculated into 2 ml liquid CM in 8-ml tubes, and the stationary cultures were incubated at 37°C for 7 days or 1 to 4 days. ST was extracted as previously described (31, 40). Approximately 5-μl aliquots of concentrated samples in 50 μl of CHCl3 were applied to a thin-layer chromatography (TLC) silica plate containing a fluorescence indicator (Kiesel gel 60 [20 cm × 20 cm × 0.25 mm]; E. Merck). An ST standard was purchased from Sigma, and 5 to 10 μg ST was applied to TLC plates. To identify the most optimum solvent system, TLC plates were developed in (i) toluene-ethyl acetate-90% formic acid (6:3:1 [vol/vol]), (ii) hexane-ethyl acetate (4:1 [vol/vol]), and (iii) toluene-ethyl acetate-acetic acid (6:3:1 [vol/vol]). We found that the hexane-ethyl acetate (4:1 [vol/vol]) solvent system clearly separated ST from various compounds, where the Rf value of ST was about 0.3. To enhance the visibility and detection limit of ST, aluminum chloride (20% AlCl3 · 7H2O in 95% ethanol [wt/vol]) was sprayed onto the TLC plates, and the plates were incubated at 90°C for 5 to 10 min. Bright light-green ST is clearly visible by this process (37). To correlate ST production with stcU and aflR mRNA levels, duplicate samples were prepared and collected from days 1 through 4, with one used for total RNA isolation and the other used for ST extraction as described above.
Microscopy and photographs.
Colony pictures on solid media were taken with a Sony digital camera (DSC-F828). Microscopic photographs were taken using an Olympus BH2 compound microscope with an Olympus DP-70 digital imaging system.
RESULTS
A. nidulans PhLPs.
TBLASTN and BLASTP searches of the A. nidulans genome (Broad Institute [http://www.broad.mit.edu/annotation/fungi/aspergillus/index.html/]), employing the C. parasitica Bdm-1 protein (17) and the three D. discoideum PhLPs (3) as queries, have identified three potential PhLPs, designated PhnA (AN0082.2), PhnB (AN4561.2), and PhnC (AN8847.2), for phosducin-like protein in A. nidulans. PhnA shows 42 to 47% similarity to Bdm-1 and PhLP1, whereas PhnB displays 43 to 44% similarity to both PhLP1 (class I) and PhLP2 (class II), but not Bdm-1. PhnC is similar (47%) to PhLP3 (class III) (3) only. Due to the high similarity of PhnA to Bdm-1, the phnA gene has been further studied.
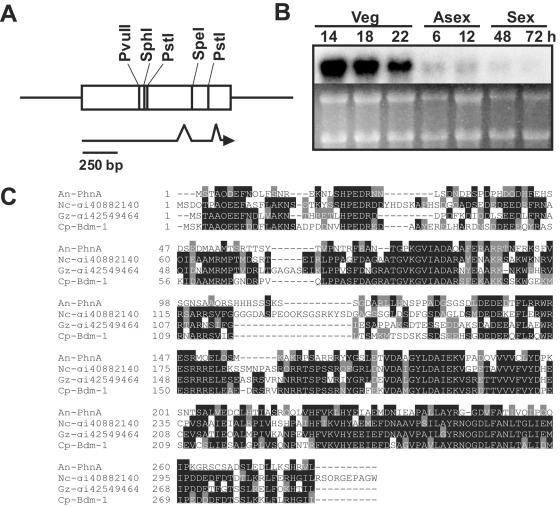
The phnA gene maps to chromosome VIII and is juxtaposed (1.4 kb apart) to sfaD (29). Sequencing of a reverse transcription-PCR amplicon of phnA revealed that it carries an ORF of 1,033 bp with two introns (72 and 113 bp; Fig. 1A), and the predicted PhnA protein consists of 281 amino acids, identical to AN0082.2 (XP_404219). The level of phnA mRNA is high in the vegetative growth stage and relatively low in the asexual and sexual development phases, indicating that phnA might be subject to transcriptional control and kept at low levels during asexual and sexual development (Fig. 1B). PhnA is highly similar (47 to 74% similarity) to Bdm-1 and class I PhLPs in Aspergillus fumigatus (gi:66851067), Neurospora crassa (gi:40882140), Magnaporthe grisea (gi:38106164), and Gibberella zeae (gi:42549464) (see Fig. 1C for an alignment). PhnA has the typical TGPKGVIADA (P instead of V) motif at the N terminus (amino acids 63 to 72), which is predicted to be necessary for interaction between PhLP and the Gα/Gβ subunit (38).
FIG. 1.
Summary of PhnA. (A) Partial restriction enzyme map of a phnA region with the PhnA ORF (open box) and introns (discontinuities in the arrow) shown. (B) phnA mRNA levels during various growth and developmental stages of the wild type (FGSC4). Numbers indicate times (hours) of incubation in liquid submerged culture (veg) and after asexual (asex) or sexual (sex) development induction. Equal loading of total RNAs was evaluated by measuring the amounts of RNA with a spectrophotometer and by ethidium bromide staining of rRNA. (C) Alignment of A. nidulans (An) PhnA with PhLPs of C. parasitica (Cp-bdm-1; gi6714950), N. crassa (Nc-gi40882140), and G. zeae (Gz-gi42549464). The alignment was done using ClustalW (8) with default settings and displayed by BoxShade (http://www.ch.embnet.org/software/BOX_form.html).
Role of PhnA in vegetative growth signaling.
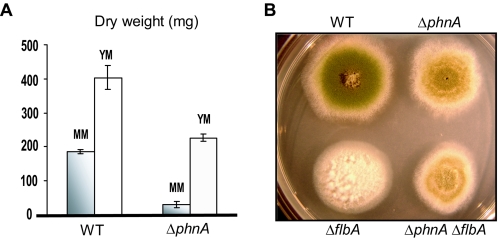
Our primary hypothesis was that PhnA plays a crucial role in the FadA-mediated growth signaling pathway by positively modulating SfaD-GpgA activity. The phnA gene was deleted, and the role of PhnA in vegetative growth signaling was examined by determining the dry weights of ΔphnA (TJAP1, TJAP2, and TJAP3) and wild-type (RKH51.117 and RJA56.25) cells in liquid MM and YM. As presented in Fig. 2A, the ΔphnA mutant strains exhibited significantly reduced vegetative growth (only 16% of the wild-type dry weight in liquid MM). However, radial growth rates of the ΔphnA mutant on solid medium were almost identical to those of the wild type (quantitative data not shown, but see Fig. 2B for reference). These growth phenotypes caused by the ΔphnA mutation were almost identical to those caused by the ΔsfaD mutation, indicating that PhnA may function in the FadA/SfaD/GpgA-mediated vegetative growth signaling pathway via modulating SfaD activity.
FIG. 2.
PhnA functions in FlbA-controlled vegetative growth signaling pathway. (A) Dry weights of ΔphnA mutant (TJAP3) and wild-type (WT; RJA56.25) strains grown in liquid MM and YM for 24 h (averages of triplicate cultures/measurements with standard error bars). (B) Deletion of phnA suppressed the fluffy-autolytic phenotype caused by the ΔflbA mutation. Relevant mutant and wild-type (WT; JAS30) strains were point inoculated onto solid MM and incubated at 37°C for 3 days. While the ΔflbA mutant (RJA5.9) exhibited the fluffy-autolytic phenotype, the ΔphnA ΔflbA mutant (TJAPF36) showed restored asexual sporulation and enhanced Hülle cell formation and was indistinguishable from the ΔphnA mutant (TJAP3).
To further test whether PhnA is involved in FadA-mediated growth signaling, we attempted to generate the ΔphnA ΔflbA double mutant via sexual crosses. However, we were unable to isolate cleistothecia from 20 independent crosses. Therefore, the ΔphnA ΔflbA double deletion mutants were generated by deleting the flbA gene from the ΔphnA mutant. A ΔflbA::metG+ cassette was constructed by the DJ-PCR method (44) and introduced into a ΔphnA strain (TJAP2). Of 80 transformants, 20 were randomly screened for the ΔphnA ΔflbA genotype. Four transformants were confirmed to be ΔphnA ΔflbA double mutants (TJAPF8, -36, -38, and -73; Table 1). As shown in Fig. 2B, the ΔphnA ΔflbA mutant and the ΔphnA mutant sporulated at the same level (quantitative data not shown), i.e., the absence of phnA is sufficient to bypass the need for FlbA in asexual development, suggesting that PhnA may function in the FlbA-controlled vegetative growth signaling pathway.
PhnA defined as the seventh flbA suppressor.
Five flbA loss-of-function suppressors were previously isolated (43). While sfaB (fadAG205R) (43) and sfaD (29) were identified, the rest of the suppressors (sfaA, sfaC, and sfaE) have yet to be defined. To test whether phnA can identify sfaA1, sfaC67, or sfaE83, the phnA ORF and promoter regions were amplified from genomic DNAs of sfaA1, sfaC67, and sfaE83 mutants (RJY1.12, RJY67.3, and RJY83.21), and amplicons were directly sequenced. Whereas no mutations were found in the phnA ORFs and the promoters of the sfaA1 and sfaE83 mutants, one phnA silent mutation (a C-to-T transition resulting in an unaltered proline at the 33rd amino acid position) was identified in the sfaC67 mutant. To test whether this silent mutation defined the sfaC67 mutant allele, sexual crosses were carried out between the ΔphnA mutant strains (TJAP1, -2, and -3) and the sfaC67 mutant. Of >20 crosses, three fragile fruiting bodies (cleistothecia) were formed. Approximately 12.5% of progeny exhibited the fluffy-autolytic phenotype resulting from the flbA98 phnA+ sfaC+ genotype, suggesting that phnA and sfaC are unlinked and that the phnA silent mutation (P33P) found in the sfaC67 mutant does not delineate sfaC67. Taken together, these data define phnA as the seventh flbA suppressor locus, in addition to fadA, sfaD, gpgA, sfaA, sfaC, and sfaE.
PhnA is associated with negative regulation of asexual sporulation.
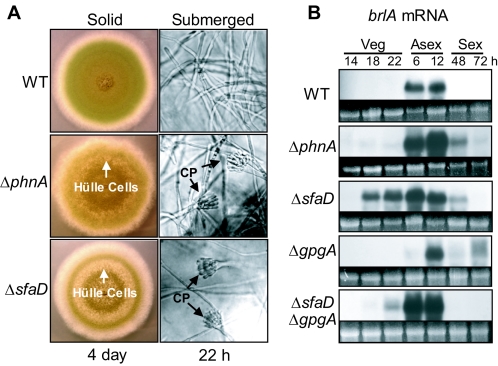
The ΔsfaD mutant elaborated complete conidiophore structures within 22 h postinoculation in liquid submerged culture (29). To examine the role of PhnA in controlling asexual sporulation, the ΔphnA mutant was cultured in liquid YM. As with the ΔsfaD mutant, deletion of phnA caused the formation of conidiophores in liquid submerged cultures as early as 18 h postinoculation in YM, whereas wild-type strains did not sporulate (Fig. 3A). We then examined the mRNA levels of brlA, encoding a key C2H2-type transcription factor required for conidiophore development (1). As shown in Fig. 3B, Northern blot analyses revealed that the brlA mRNA accumulated at various levels in the ΔphnA, ΔsfaD, and ΔsfaD ΔgpgA mutants (TJAP3, RSRB1.15, and RJA55.4), but not in the wild type or the ΔgpgA mutant (RJAG19.9), during vegetative growth stages. Furthermore, compared to the wild type, much higher levels of brlA mRNA accumulated in the ΔphnA, ΔsfaD, and ΔsfaD ΔgpgA mutants when they were induced for asexual development. In addition, brlA mRNA was detected in the ΔphnA and ΔsfaD mutants even when they were grown under conditions that preferentially induce sexual development (Fig. 3B, sex, 48 h). However, the accumulation of brlA in the ΔgpgA mutant was delayed about 6 h when it was induced for asexual development. This is consistent with the previous observation that the deletion of gpgA resulted in delayed asexual sporulation (33). Collectively, these data show that PhnA plays an important role in the negative regulation of asexual sporulation, likely via modulating SfaD activity.
FIG. 3.
Developmental changes caused by ΔphnA mutation. (A) Enhanced Hülle cell formation on solid MM and development of conidiophores (CP) in liquid submerged culture caused by ΔphnA mutation. Whereas the wild type (WT; RJA56.25) did not produce conidiophores even at 30 h, the ΔphnA (TJAP3) and ΔsfaD (RSRB1.15) mutant strains began to elaborate conidiophores as early as 18 h in liquid YM. The colony and microscopic photographs shown were taken after 4 days on solid MM and after 22 h in liquid YM, respectively. (B) Northern blot analysis of brlA in various mutants. While no brlA mRNA was detected during the vegetative growth stage of the WT (RJA56.25) or ΔgpgA (RJAG19.9) strain, a precocious and elevated accumulation of brlA mRNA was evident for the ΔphnA, ΔsfaD, and ΔsfaD ΔgpgA mutant strains (TJAP3, RSRB1.15, and RJA55.4). Equal loading of total RNAs was evaluated by ethidium bromide staining of rRNA.
Deletion of phnA causes impairment in sexual fruiting body formation.
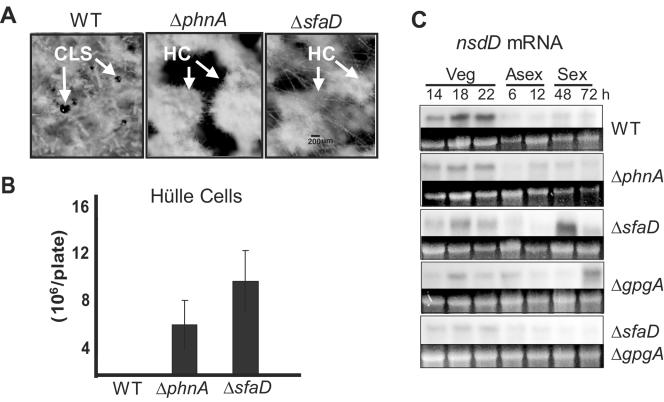
Our previous studies demonstrated that SfaD and GpgA are required for the formation of cleistothecia under self-fertilized and outcrossed conditions (29, 33). To test a potential role for PhnA in sexual reproduction, we examined the development of Hülle cells and cleistothecia in the ΔphnA mutant and found that, despite enhanced Hülle cell formation, the ΔphnA mutant was unable to produce cleistothecia under self-fertilized (homothallic) conditions (Fig. 4A and B) or in outcrosses with the wild type or other mutants (not shown). These sexually defective phenotypes are identical to those caused by ΔsfaD (29) and ΔgpgA (33) mutations. These results indicate that PhnA, SfaD, and GpgA are crucial for sexual fruiting body formation and balanced Hülle cell production.
FIG. 4.
Deletion of phnA resulted in impairment in sexual reproduction. (A) Close-up views (magnification, ∼×40) of wild-type (WT; RKH51.117) and ΔphnA (TJAP3) and ΔsfaD (RSRB1.15) mutant strains grown under sexually induced conditions for 10 days. While the WT abundantly produced both cleistothecia (CLS) and Hülle cells under self-fertilized conditions, the ΔphnA and ΔsfaD mutant strains did not form any cleistothecia yet exhibited an enhancement of aggregated Hülle cells (HC). (B) Numbers of Hülle cells were counted in 5-day-old cultures on solid MM under air-exposed conditions (averages of triplicate cultures/measurements with standard error bars). While the WT did not produce any Hülle cells under these conditions, the ΔphnA and ΔsfaD mutant strains (TJAP3 and RSRB1.15) formed large numbers of Hülle cells. (C) Northern blot analysis of nsdD in various mutants. No clear differences were observed in nsdD mRNA levels between the WT (RKH51.117) and the ΔphnA, ΔsfaD, ΔgpgA, and ΔsfaD ΔgpgA mutants. Equal loading of total RNAs was evaluated by ethidium bromide staining of rRNA.
To test whether the requirement for PhnA, SfaD, and GpgA in sexual reproduction is associated with proper expression of nsdD, encoding a GATA-type transcriptional activator for sexual development (12), levels of nsdD mRNA were examined in ΔphnA (TJAP3), ΔsfaD (RSRB1.15), ΔgpgA (RJAG19.9), and ΔsfaD ΔgpgA (RJA55.4) strains. As presented in Fig. 4C, the nsdD mRNA accumulated at similar levels in the relevant mutant and wild-type strains throughout the life cycle, indicating that the lack of sexual fruiting in the mutants was not due to altered expression of nsdD.
PhnA, SfaD, and GpgA are required for ST production.
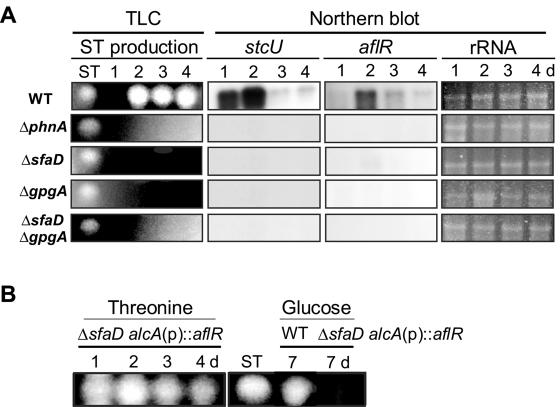
A. nidulans produces a carcinogenic mycotoxin, ST, as one of its secondary metabolites. Previous studies revealed that a key prerequisite for the biosynthesis of ST is inhibition of the FadA/PkaA pathway, which requires the RGS protein FlbA (15, 34, 35). Loss-of-function flbA mutants are unable to produce ST, and deletion of fadA bypasses the need for FlbA in asexual sporulation and ST production (15). The fact that the deletion of phnA, sfaD, or gpgA could restore asexual sporulation in the ΔflbA mutant (29, 33; this study) led us to test whether individual double mutants regained the ability to produce ST. Somewhat surprisingly, we found that the absence of phnA, sfaD, or gpgA function did not restore the production of ST in the ΔflbA mutant (data not shown). These findings further led us to examine the potential direct roles of PhnA, SfaD, and GpgA in ST production. This was done by inoculating individual deletion mutants and the wild type in 2 ml liquid CM for 1 to 4 days and analyzing ST production in each stationary culture as described previously (40). As if PhLP and Gβγ are necessary for the biosynthesis of ST, the ΔphnA (TJAP3), ΔsfaD (RSRB1.15), ΔgpgA (RJAG19.9), and ΔsfaD ΔgpgA (RJA55.4) mutants were unable to produce ST (Fig. 5A).
FIG. 5.
Requirement of PhnA, SfaD, and GpgA in ST biosynthesis. (A) Results of TLC and Northern blot analyses of the wild type (WT; FGSC26) and the mutants for ST biosynthesis. Conidia were inoculated in 2 ml of liquid CM and incubated for 1 to 4 days. Numbers indicate the times (days) of incubation. While the WT began to produce ST at 2 days, the ΔphnA (TJAP3), ΔsfaD (RSRB1.15), ΔgpgA (RJAG19.9), and ΔsfaD ΔgpgA (RJA55.4) mutants did not produce detectable levels of ST until 4 days. Moreover, no aflR or stcU mRNA accumulation was clearly detectable in the mutants. The ST standard is indicated. Equal loading of total RNAs was evaluated by ethidium bromide staining of rRNA. (B) The WT (FGSC26) and ΔsfaD alcAp::aflR (RSRAB.4 and RSRAB.1) and ΔsfaD (RSRB1.15; not shown but did not produce ST under any conditions) mutant strains were grown in 2 ml of liquid CM with 2% glucose for 24 h, and then the medium was replaced with liquid CM with threonine or glucose for 1 to 4 days (up to 7 days for glucose-CM). Under noninducing conditions (glucose), only the WT produced ST (only results for 7 days are shown). Note that aflR overexpression in threonine-CM restored ST production in the ΔsfaD mutant.
The A. nidulans genome contains a cluster of 25 genes (stc) required for ST biosynthesis, the expression of which is dependent on the activity of AflR, a Zn(II)2Cys6 transcription factor (6, 10, 42). To test whether the lack of ST production in the three mutants was associated with alterations in the expression of aflR and stc genes, we examined the steady-state mRNA levels of aflR and stcU, encoding an ST biosynthetic enzyme (6), under the above-mentioned conditions. As presented in Fig. 5A, no aflR or stcU mRNA accumulation occurred in the four mutants, implying that PhnA, SfaD, and GpgA are required for (proper) expression of the aflR and stc genes. We also examined mRNA levels of laeA, encoding a regulator of secondary metabolism (5), and found that they were not altered in the four mutants (data not shown).
To further dissect the regulatory role of SfaD in ST production, we tested the effect of overexpression of aflR in the absence of sfaD. Two sets of wild-type and ΔsfaD alcAp::aflR strains (RSRAB.1 and RSRAB.4; Table 1) were cultured in 2 ml liquid CM (stationary) with glucose (noninducing conditions) for 1 day, the medium was replaced by liquid CM with threonine (inducing) or glucose (noninducing), and strains were incubated at 37°C for an additional 1 to 4 days (or up to 7 days in glucose-CM). As shown in Fig. 5B, ST production in ΔsfaD alcAp::aflR strains (only data for RSRAB.1 are shown) grown under inducing conditions (threonine) was fully restored to the wild-type level, whereas no ST was detected in the same strains grown under noninducing (glucose) conditions, even 7 days after replacement of the medium. These data indicate that SfaD-dependent ST biosynthesis occurs through the transcriptional activation of aflR, not through modification of the AflR protein (see Discussion).
DISCUSSION
Phosducin is a cytosolic phosphoprotein first identified in photoreceptor cells of the retina (22) and the developmentally related pineal gland (28). The further identification of PhLPs in vertebrates and lower eukaryotes underscores the existence of a family of proteins characterized as cytosolic regulators of G protein signaling (reviewed in reference 30).
While several functions of phosducin and PhLPs have been suggested, their most critical role was thought to be high-affinity sequestration with the Gβγ heterodimer of heterotrimeric G proteins, thereby negatively controlling G protein-mediated intracellular signaling (reviewed in reference 30). However, a series of recent biochemical studies demonstrated that PhLP is required for normal levels of Gβ and Gγ subunits as well as assembly of the Gβγ dimer and thus is an essential positive regulator of G protein signaling (17, 18, 23). Genetic data from the filamentous fungus C. parasitica and the social amoeba D. discoideum were in accordance with the newly proposed positive role of PhLP (3, 17). Our present study shows that the A. nidulans phnA gene, encoding a PhLP, is required for SfaD-GpgA-mediated signaling for vegetative growth, regulation of development, and production of ST, further supporting the essential positive role of PhLP in Gβγ signaling.
The fact that the deletion of phnA reduced the biomass and suppressed the asexual developmental defects caused by the absence of FlbA function provides genetic evidence that PhnA is an active participant in FadA (Gα)- and SfaD-GpgA (Gβγ)-mediated vegetative growth signaling. Moreover, the phnA deletion mutant exhibited phenotypes almost identical to those of the ΔsfaD mutant but different from those of the ΔgpgA mutant (33), suggesting that PhnA is essential for SfaD functionality and supporting the idea that, in addition to functioning as a heterodimer, SfaD and GpgA may have distinct signaling roles (33). Our previous studies showed that both SfaD and GpgA are required for sexual fruiting body (cleistothecium) formation in a somewhat dominant manner (29, 33). As PhnA is needed for Gβγ-mediated signaling for cleistothecium development, the ΔphnA mutant was found to be severely impaired in sexual reproduction. It was shown that the two putative G protein-coupled receptors GprA and GprB were required for self-fertilization and that enhanced expression of nsdD could not bypass the need for GprA/B in ascospore formation under homothallic conditions (32). In this study, we demonstrated that the requirement for PhnA in sexual fruiting is not due to altered expression of nsdD (12). This result is consistent with our previous proposal that the G protein-coupled receptors → G protein (yet to be identified) and NsdD might function in separate regulatory branches (32).
Probably the most remarkable finding in this study is that while deletion of sfaD, gpgA, or phnA overcame developmental defects caused by the ΔflbA mutation, it did not rescue ST production in the ΔflbA mutant. Further investigation revealed that SfaD, GpgA, and PhnA themselves are necessary for the biosynthesis of ST and expression of aflR. These results suggest potential differential (or opposite) roles of individual G protein components in controlling ST production. Previous studies showed that activated (GTP-bound) FadA primarily mediates vegetative proliferation signaling and that constitutively active (FadAd+) FadA mutations (G42R, Q204L, and R178C) (41, 43) and a loss of flbA function both result in the fluffy-autolytic phenotype as well as a lack of development and ST production (15, 41). However, it is important to note that FadA itself is not a direct negative regulator of ST biosynthesis because, while deletion of fadA bypassed FlbA function in both asexual sporulation and ST production, it did not affect ST biosynthesis (15). In summary, whereas FadA-mediated signaling results in an inhibition of ST biosynthesis, SfaD and GpgA are essential for aflR expression and subsequent ST biosynthesis.
The absence of ST production in FadAd+ or flbA mutants was partially clarified by the characterization of the pkaA gene, encoding a protein kinase A (PKA) catalytic subunit (34) which functions downstream of FadA/FlbA. Deletion of pkaA suppressed both developmental and ST biosynthetic defects caused by the ΔflbA mutation, whereas overexpression of pkaA resulted in enhanced vegetative growth, reduced asexual sporulation, and inhibition of aflR expression (34). These results imply that signaling mediated by the FadA→PkaA branch might be primarily responsible for the negative regulation of ST production and that the absence of FadA or PkaA function is sufficient to bypass the need for FlbA in ST biosynthesis (15; reviewed in reference 45). The role of PkaA in negatively controlling ST production was further supported by a recent study which showed that AflR could be phosphorylated by PkaA in vitro (35). Furthermore, the potential posttranscriptional negative regulation of AflR activity by PkaA-dependent phosphorylation in vivo was demonstrated by mutation of the putative phosphorylation target Ser to Ala in AflR (35). Such targeted amino acid replacements abolished the inhibitory effects of pkaA overexpression on AflR activity. Collectively, it has been proposed that the FadA→PkaA branch negatively regulates the expression of aflR mRNA and the activity of the AflR protein (Fig. 6).
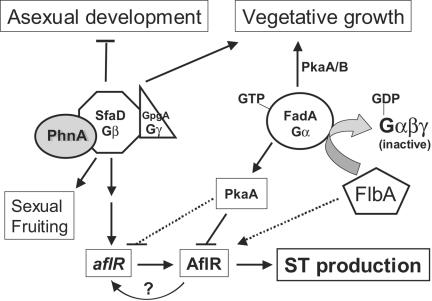
FIG. 6.
Proposed differential roles of FadA, FlbA, PhnA, and Gβγ in controlling ST production. We propose that the A. nidulans PhLP PhnA is an essential component of SfaD-GpgA-mediated signaling, which is required for normal vegetative growth, sexual fruiting body formation, ST production, and proper down-regulation of asexual development. FadA-mediated vegetative growth signaling is transduced in part by the primary PKA PkaA (34). PkaB is the secondary (backup) PKA catalytic subunit, playing a role in hyphal growth and germination (25). The FadA→PkaA signaling pathway is thought to be primarily responsible for the negative control of ST biosynthesis via modulating the activity of the AflR protein (35). The potential inhibitory role of the FadA→PkaA pathway on the transcription of aflR is indicated by a dotted line (34, 35). It is proposed that the results of SfaD-GpgA signaling include transcriptional activation of aflR and subsequent ST production. A dotted arrow presents the predicted posttranscriptional/posttranslational activation of AflR by FlbA. Potential autoactivation of aflR transcription by the AflR protein (10, 35) is also indicated.
However, there is a complicating factor in interpreting the precise role of FlbA in controlling ST production. Shimizu et al. (35) showed that the requirement for FlbA in AflR-mediated ST production was PkaA independent, suggesting a potential direct posttranslational regulation of AflR by FlbA. Moreover, overexpression of aflR could not restore ST production in the ΔflbA mutant (35; J. K. Hicks and N. P. Keller, unpublished data). This is consistent with the hypothesis that, while the primary role of FlbA in development and ST production is inactivating the FadA→PkaA pathway, FlbA has additional roles in asexual development and ST biosynthesis (15, 41). It has been speculated that FlbA may be necessary for the activity of the AflR protein via unknown posttranslational mechanisms (35) (Fig. 6).
Without information on the potential effector proteins of SfaD-GpgA, it is premature to devise the mechanisms underlying the requirement for SfaD-GpgA in ST biosynthesis. Previously, we speculated on the potential involvement of a mitogen-activated protein kinase(s) in transducing SfaD-GpgA-mediated signals for sexual reproduction (32, 33). In addition, because SfaD and GpgA are the only Gβ and Gγ subunits found in A. nidulans, it is possible that the absence of PhnA, SfaD, or GpgA may affect signaling mediated by the three Gα subunits, FadA, GanB, and GanA (7, 41). In fact, deletion of the Gβ or Gγ subunit has been shown to cause a reduction of Gα proteins in other filamentous fungi (19, 26, 39). However, deletion of fadA, ganB, or ganA did not cause the lack of ST production (14, 15; K.-Y. Jahng, personal communication), indicating that no single Gα protein is essential for ST biosynthesis. The effects of the absence of all three Gα proteins on growth, morphogenesis, and secondary metabolism in A. nidulans remain to be studied, though. Nonetheless, the fact that the overexpression of aflR was sufficient to bypass the requirement for SfaD in ST production indicates that cellular responses to SfaD-mediated signaling may include transcriptional activation of aflR (Fig. 6).
Bok and Keller (5) identified LaeA, which is an upstream positive regulator of aflR expression and ST production. To begin to dissect the mechanisms underlying the necessity of SfaD in ST biosynthesis, we examined the mRNA levels of laeA in wild-type and phnA, sfaD, and gpgA deletion mutant strains and found no differences (data not shown), suggesting that SfaD signaling and LaeA may function in separate ways. The identification of downstream components transducing signals mediated by SfaD-GpgA is crucial for further understanding the differential roles of individual G protein components in controlling morphogenesis coupled with secondary metabolism.
Acknowledgments
We thank the Broad Institute for the A. nidulans genome database and Ellin Doyle for critically reviewing the manuscript.
This work was supported by a National Science Foundation grant (MCB-0421863) to J.H.Y.
REFERENCES
- 1.Adams, T. H., M. T. Boylan, and W. E. Timberlake. 1988. brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell 54:353-362. [DOI] [PubMed] [Google Scholar]
- 2.Bauer, P., S. Müller, M. Puzicha, S. Pippig, B. Obermaier, E. J. M. Helmreich, and M. J. Lohse. 1992. Phosducin is a protein kinase A-regulated G-protein regulator. Nature 358:73-76. [DOI] [PubMed] [Google Scholar]
- 3.Blaauw, M., J. C. Knol, A. Kortholt, J. Roelofs, Ruchira, M. Postma, A. J. W. G. Visser, and P. J. M. Van Haastert. 2003. Phosducin-like proteins in Dictyostelium discoideum: implications for the phosducin family of proteins. EMBO J. 22:5047-5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blüml, K., W. Schnepp, S. Schröder, M. Beyermann, M. Macias, H. Oschkinat, and M. J. Lohse. 1997. A small region in phosducin inhibits G-protein βγ-subunit function. EMBO J. 16:4908-4915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bok, J. W., and N. P. Keller. 2004. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell 3:527-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, D. W., J.-H. Yu, H. S. Kelkar, M. Fernandes, T. C. Nesbitt, N. P. Keller, T. H. Adams, and T. J. Leonard. 1996. Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 93:1418-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, M.-H., K.-S. Chae, D.-M. Han, and K.-Y. Jahng. 2004. The GanB Gα-protein negatively regulates asexual sporulation and plays a positive role in conidial germination in Aspergillus nidulans. Genetic 167:1305-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D'Souza, C. A., B. N. Lee, and T. H. Adams. 2001. Characterization of the role of the FluG protein in asexual development of Aspergillus nidulans. Genetics 158:1027-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandes, M., N. P. Keller, and T. H. Adams. 1998. Sequence-specific binding by Aspergillus nidulans AflR, a C6 zinc cluster protein regulating mycotoxin biosynthesis. Mol. Microbiol. 28:1355-1365. [DOI] [PubMed] [Google Scholar]
- 11.Flanary, P. L., P. R. DiBello, P. Estrada, and H. G. Dohlman. 2000. Functional analysis of Plp1 and Plp2, two homologues of phosducin in yeast. J. Biol. Chem. 275:18462-18469. [DOI] [PubMed] [Google Scholar]
- 12.Han, K.-H., K.-Y. Han, J.-H. Yu, K.-S. Chae, K.-Y. Jahng, and D.-M. Han. 2001. The nsdD gene encodes a putative GATA-type transcription factor necessary for sexual development of Aspergillus nidulans. Mol. Microbiol. 41:299-309. [DOI] [PubMed] [Google Scholar]
- 13.Han, K.-H., J.-A. Seo, and J.-H. Yu. 2004. A putative G protein-coupled receptor negatively controls sexual development in Aspergillus nidulans. Mol. Microbiol. 51:1333-1345. [DOI] [PubMed] [Google Scholar]
- 14.Han, K.-H., J.-A. Seo, and J.-H. Yu. 2004. Regulators of G-protein signaling in Aspergillus nidulans: RgsA downregulates stress response and stimulates asexual sporulation through attenuation of GanB (Gα) signaling. Mol. Microbiol. 53:529-540. [DOI] [PubMed] [Google Scholar]
- 15.Hicks, J. K., J.-H. Yu, N. P. Keller, and T. H. Adams. 1997. Aspergillus sporulation and mycotoxin production both require inactivation of the FadA Gα protein-dependent signaling pathway. EMBO J. 16:4916-4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Käfer, E. 1977. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv. Genet. 19:33-131. [DOI] [PubMed] [Google Scholar]
- 17.Kasahara, S., P. Wang, and D. L. Nuss. 2000. Identification of bdm-1, a gene involved in G protein β-subunit function and α-subunit accumulation. Proc. Natl. Acad. Sci. USA 97:412-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knol, J. C., R. Engel, M. Blaauw, A. J. Visser, and P. J. van Haastert. 2005. The phosducin-like protein PhLP1 is essential for Gβγ dimer formation in Dictyostelium discoideum. Mol. Cell. Biol. 25:8393-8400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krystofova, S., and K. A. Borkovich. 2005. The heterotrimeric G-protein subunits GNG-1 and GNB-1 form a Gβγ dimer required for normal female fertility, asexual development, and Gα protein levels in Neurospora crassa. Eukaryot. Cell 4:365-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lafon, A., J.-A. Seo, K.-H. Han, J.-H. Yu, and C. d'Enfert. 2005. The heterotrimeric G-protein GanB(α)-SfaD(β)-GpgA(γ) is a carbon source sensor involved in early cAMP-dependent germination in Aspergillus nidulans. Genetics 171:71-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, B. N., and T. H. Adams. 1994. Overexpression of flbA, an early regulator of Aspergillus asexual sporulation leads to activation of brlA and premature initiation of development. Mol. Microbiol. 14:323-334. [DOI] [PubMed] [Google Scholar]
- 22.Lee, R. H., B. S. Lieberman, and R. N. Lolley. 1990. Retinal accumulation of the phosducin/T beta gamma and transducin complexes in developing normal mice and in mice and dogs with inherited retinal degeneration. Exp. Eye Res. 51:325-333. [DOI] [PubMed] [Google Scholar]
- 23.Lukov, G. L., T. Hu, J. N. Mclaughlin, H. E. Hamm, and B. M. Willardson. 2005. Phosducin-like protein acts as a molecular chaperone for G protein βγ dimer assembly. EMBO J. 24:1965-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCudden, C. R., M. D. Hains, R. J. Kimple, D. P. Siderovski, and F. S. Willard. 2005. G-protein signaling: back to the future. Cell. Mol. Life Sci. 62:551-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ni, M., S. Rierson, J.-A. Seo, and J.-H. Yu. 2005. The pkaB gene encoding the secondary PKA catalytic subunit has a synthetic lethal interaction with pkaA and plays overlapping and opposite roles in Aspergillus nidulans. Eukaryot. Cell 4:1465-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parsley, T. B., G. C. Segers, D. L. Nuss, and A. L. Dawe. 2003. Analysis of altered G-protein subunit accumulation in Cryphonectria parasitica reveals a third Galpha homologue. Curr. Genet. 43:24-33. [DOI] [PubMed] [Google Scholar]
- 27.Pontecorvo, G., J. A. Roper, L. M. Hemmons, K. D. Macdonald, and A. W. J. Bufton. 1953. The genetics of Aspergillus nidulans. Adv. Genet. 5:141-238. [DOI] [PubMed] [Google Scholar]
- 28.Reig, J. A., L. Yu, and D. C. Klein. 1990. Pineal transduction. Adrenergic-cyclic AMP-dependent phosphorylation of cytoplasmic 33-kDa protein (MEKA) which binds beta gamma-complex of transducin. J. Biol. Chem. 265:5816-5824. [PubMed] [Google Scholar]
- 29.Rosén, S., J.-H. Yu, and T. H. Adams. 1999. The Aspergillus nidulans sfaD gene encodes a G protein β subunit that is required for normal growth and repression of sporulation. EMBO J. 18:5592-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schulz, R. 2001. The pharmacology of phosducin. Pharmacol. Res. 43:1-10. [DOI] [PubMed] [Google Scholar]
- 31.Seo, J.-A., Y. Guan, and J.-H. Yu. 2003. Suppressor mutations bypass the requirement of fluG for asexual sporulation and sterigmatocystin production in Aspergillus nidulans. Genetics 165:1083-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seo, J.-A., K.-H. Han, and J.-H. Yu. 2004. The gprA and gprB genes encode putative G protein-coupled receptors required for self-fertilization in Aspergillus nidulans. Mol. Microbiol. 53:1611-1623. [DOI] [PubMed] [Google Scholar]
- 33.Seo, J.-A., K.-H. Han, and J.-H. Yu. 2005. Multiple roles of a heterotrimeric G protein γ subunit in governing growth and development of Aspergillus nidulans. Genetics 171:81-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimizu, K., and N. P. Keller. 2001. Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics 157:591-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimizu, K., J. K. Hicks, T. P. Huang, and N. P. Keller. 2003. Pka, Ras and RGS protein interactions regulate activity of AflR, a Zn(II)2Cys6 transcription factor in Aspergillus nidulans. Genetics 165:1095-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sienko, M., and A. Paszewski. 1999. The metG gene of Aspergillus nidulans encoding cystathionine beta-lyase: cloning and analysis. Curr. Genet. 35:638-646. [DOI] [PubMed] [Google Scholar]
- 37.Stack, M., and J. V. Rodricks. 1971. Method for analysis and chemical confirmation of sterigmatocystin. J. Assoc. Off. Anal. Chem. 54:86-90. [PubMed] [Google Scholar]
- 38.Xu, J., D. Wu, D. V. Z. Slepak, and M. I. Simon. 1995. The N terminus of phosducin is involved in binding of βγ subunits of G protein. Proc. Natl. Acad. Sci. USA 92:2086-2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang, Q., S. I. Poole, and K. A. Borkovich. 2002. A G-protein β subunit required for sexual and vegetative development and maintenance of normal Gα protein levels in Neurospora crassa. Eukaryot. Cell 1:378-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu, J.-H., and T. J. Leonard. 1995. Sterigmatocystin biosynthesis in Aspergillus nidulans requires a novel type I polyketide synthase. J. Bacteriol. 177:4792-4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu, J.-H., J. Wieser, and T. H. Adams. 1996. The Aspergillus FlbA RGS domain protein antagonizes G-protein signaling to block proliferation and allow development. EMBO J. 15:5184-5190. [PMC free article] [PubMed] [Google Scholar]
- 42.Yu, J.-H., R. A. E. Butchko, M. Fernandes, N. P. Keller, T. J. Leonard, and T. H. Adams. 1996. Conservation of structure and function of the aflatoxin regulatory gene aflR from Aspergillus nidulans and A. flavus. Curr. Genet. 29:549-555. [DOI] [PubMed] [Google Scholar]
- 43.Yu, J.-H., S. Rosén, and T. H. Adams. 1999. Extragenic suppressors of loss-of-function mutations in the Aspergillus FlbA RGS domain protein. Genetics 151:97-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu, J.-H., Z. Hamari, K.-H. Han, J.-A. Seo, Y. Reyes-Domínguez, and C. Scazzocchio. 2004. Double-joint PCR (DJ-PCR): a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41:973-981. [DOI] [PubMed] [Google Scholar]
- 45.Yu, J.-H., and N. P. Keller. 2005. Regulation of secondary metabolism in filamentous fungi. Annu. Rev. Phytopathol. 43:437-458. [DOI] [PubMed] [Google Scholar]