Abstract
Mutations in EBS1 were identified in Saccharomyces cerevisiae that cosuppress missense, frameshift, and nonsense mutations. Evidence from studies of loss of function and overexpression of EBS1 suggests that Ebs1p affects gene expression by inhibiting translation and that a loss of EBS1 function causes suppression by increasing the rate of translation. Changes in EBS1 expression levels alter the expression of wild-type genes, but, in general, no changes in mRNA abundance were associated with a loss of function or overexpression of EBS1. Translation of a lacZ reporter was increased in strains carrying an ebs1-Δ mutant gene, whereas translation was decreased when EBS1 was overexpressed. The cap binding protein eIF-4E copurifies with Ebs1p in the absence of RNA, suggesting that the two proteins interact in vivo. Although physical and genetic interactions were detected between Ebs1p and Dcp1p, copurification was RNase sensitive, and changes in the expression of Ebs1p had little to no effect on decapping of the MFA2 transcript. The combined results suggest that Ebs1p inhibits translation, most likely through effects on eIF-4E rather than on decapping. Finally, EBS1 transcript levels are under the control of nonsense-mediated mRNA decay (NMD), providing the first example of an NMD-sensitive transcript whose protein product influences a step in gene expression required for NMD.
All eukaryotes have a nonsense-mediated mRNA decay (NMD) pathway that monitors the fidelity of gene expression at the level of RNA surveillance (6). One purpose served by NMD is to identify and degrade transcripts containing errors that cause premature termination of translation, which limits the accumulation of potentially deleterious truncated proteins (40). The UPF genes, which were first identified in Saccharomyces cerevisiae (8, 26, 27), are represented by orthologs in widely divergent evolutionary branches of the eukaryotes (6) but are absent in prokaryotes and archaea. In addition to RNA surveillance, the Upf proteins control the abundance of hundreds of transcripts in S. cerevisiae (18, 28).
The mechanism of nonsense transcript decay is intimately related to translation and decapping (31, 49). The Upf proteins and the translation termination factors eRF1 and eRF3 are assembled in a surveillance complex that is recruited to nonsense transcripts to catalyze termination at a premature stop codon, followed by decapping and decay (for a recent summary, see reference 7). Upf1p interacts with the decapping enzyme Dcp2p to promote Xrn1p-mediated decay from the 5′ end (17, 36) and with Ski7p to promote exosome-mediated decay from the 3′ end (32, 45). Although the broad outlines of the circuitry leading to NMD are known, the manner in which the surveillance complex affects the interplay between mRNA decay and translation is still unclear.
To better understand the relationship between NMD and translation, we devised a selection method to uncover rare hypomorphic mutations in genes coding for either translation factors or additional Upf proteins. The possibility of unidentified UPF genes is suggested by the fact that seven genes are required for NMD in Caenorhabditis elegans (1, 4, 15, 39), whereas only three have been found in S. cerevisiae (27). The UPF genes were originally found by selecting for allosuppressors of frameshift mutations in HIS4 and LEU2 that bring out-of-frame premature stop codons into register (8). The most common suppressors corresponded to mutations in genes coding for mRNA decay factors, i.e., UPF1, UPF2, and UPF3.
To reduce the frequency of recovery of known UPF genes, duplicate copies of UPF1, UPF2, and UPF3 were introduced on a plasmid to mask recessive mutations in these genes. Cosuppressors of the frameshift mutation his4-38 and the missense mutation ctf13-30 were selected. The latter mutation was included based on a previous study showing that most of the extragenic suppressors of the ctf13-30 mutation were mutations in UPF1, UPF2, and UPF3 (9). The introduction of UPF gene duplications dramatically altered the types of suppressors recovered, reducing the frequency of mutations in known UPF genes from nearly 100% to 18%.
To define an example of what might be represented among the suppressors obtained with the merodiploid selection scheme, two suppressors were characterized that correspond to mutations in the EBS1 gene. The evidence reported here suggests that EBS1 codes for a global inhibitor of translation. Interestingly, although Ebs1p is not required for NMD, its function is connected with NMD in two ways: NMD requires translation, and EBS1 mRNA abundance is controlled by the Upf proteins.
MATERIALS AND METHODS
Strains, plasmids, and genetic methods.
The strains used for this study are listed in Table 1, and the plasmids used are listed in Table 2. To reduce variation due to differences in genetic background to permit meaningful comparisons of growth rates or transcript levels, sets of congenic strains differing only at known loci were constructed by serial transformation with plasmids carrying appropriate wild-type genes. A typical congenic set consisted of a strain carrying the ebs1-Δ mutation and transformants of that strain carrying EBS1 on a single-copy centromeric (CEN) or multicopy 2μm plasmid.
TABLE 1.
Strains used for this study
| Strain | Relevant genotype | Reference |
|---|---|---|
| RGy13565a | ebs1-Δ | |
| ASFy1 | upf1-Δ::URA3 his4-38 SUF1-1 | |
| ASFy2 | upf1-Δ::his4-38 SUF1-1 | |
| ASFy5 | ebs1-1 leu2-2 tyr7-1 | |
| ASFy6 | ebs1-2 ura3 leu2-2 tyr7-1 | |
| ASFy22 | ebs1-Δ upf3-Δ leu2-1 tyr7-1 can1-100 | |
| ASFy26 | dcp1-Δ ebs1-Δ | |
| ASFy29 | ebs1-Δ upf1-Δ upf2-Δ upf3-Δ can1-100 | |
| ASFy30 | ebs1-Δ his4-38 SUF1-1 ctf13-30 | |
| ASFy31 | ebs1-Δ can1-100 | |
| ASFy32 | ebs1-Δ | |
| CSy4 | msc2::KAN leu2-2 tyr7-1 his4-38 | |
| MJL12 | his4-38 SUF1-1 ctf13-30 | |
| MJL13 | his4-38 SUF1-1 ctf13-30 | |
| MNY8 | CRM1::KAN(pCRM1T539C) | 37 |
| mutA | ebs1-1 his4-38 SUF1-1 ctf13-30 | |
| mutB | ebs1-2 his4-38 SUF1-1 ctf13-30 | |
| PJ6-94A | gal4-Δ gal80-Δ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ | 21 |
| QGy37 | upf1-Δ ebs1-Δ | |
| QGy40 | ebs1-Δ | |
| QGy50 | ebs1-Δ cup1::LEU2/PGK1pG/MFA2pG | |
| QGy51 | ebs1-Δ cup1::LEU2/PGK1pG/MFA2pG | |
| QGy52 | ebs1-Δ dcp1-2::TRP1 ski8-Δ | |
| QGy53 | ebs1-Δ dcp2-7::URA3 ski3::TRP1 | |
| QGy54 | ebs1-Δ dcp1-2::TRP1 cup1::LEU2/ PGK1pG/MFA2pG | |
| QGy55 | ebs1-Δ dcp2-7 cup1::LEU2/PGK1pG/ MFA2pG | |
| yRP840 | cup1::LEU2/PGK1pG/MFA2pG | 25, 46 |
| yRP841 | cup1::LEU2/PGK1pG/MFA2pG | 25, 46 |
| yRP1345 | dcp1-2 ski8-Δ | 25, 46 |
| yRP1502 | dcp2-7 ski3-Δ | 25, 46 |
| yRP1515 | dcp1-2 cup1::LEU2/PGK1pG/MFA2pG | 25, 46 |
| yRP1516 | dcp2-7 cup1::LEU2/PGK1pG/MFA2pG | 25, 46 |
From Research Genetics Inc.
TABLE 2.
Plasmids used for this study
| Plasmid | Relevant features and genes | Reference |
|---|---|---|
| pACTAA | lacZ-UAA-luc | 3 |
| pACTGA | lacZ-UGA-luc | 3 |
| pACTMV | lacZ-UAG-luc | 3 |
| pACTQ | lacZ-CAA-luc | 3 |
| pAA79 | CEN4 LEU2 UPF1 | |
| pAF10 | UPF3-BD (two-hybrid) | |
| pAF15 | CEN4 URA3 EBS1 | |
| pAF16 | 2 μm URA3 EBS1 | |
| pAF17 | CEN4 LEU2 EBS1 | |
| pAF18 | 2 μm LEU2 EBS1 | |
| pAF19 | EBS1-AD (two-hybrid) | |
| pAF52 | DCP1-AD (two-hybrid) | |
| pAF53 | DCP1-BD (two-hybrid) | |
| pAF54 | CEN4 EBS1-GFP | |
| pAF77 | CEN4 EBS1 TRP1 | |
| pAF78 | 2 μm TRP1 EBS1 | |
| pLS74 | CEN4 UPF3 | 42 |
| pLS80 | CEN4 UPF1 UPF2 | |
| pNB30 | TIF1-BD (eIF-4A-BD) (two-hybrid) | |
| pNB41 | CDC33-BD (eIF-4E-BD) (two-hybrid) | |
| pNB32 | TIF4631-BD (eIF-4G-BD) (two-hybrid) | |
| pNB42 | STO1-BD (Cbc1p-BD) (two-hybrid) | |
| pRP783 | CEN4 DCP1 | 46 |
| pRP877 | CEN4 dcp1-7 | 46 |
| pRP893 | CEN4 dcp1-31 | 46 |
| pML1 | CEN4 UPF1 UPF2 UPF3 | 28 |
| pML2 | CEN4 UPF2 UPF3 | |
| pNE53 | CEN4 CUP1 GFP1 3′UTR UPF2 | |
| pNE164a | 2 μm HA URA3 | |
| pNE179b | CEN HA URA3 | |
| pNE183b | CEN4 cMyc TRP1 | |
| pNE224 | 2 μm CUP1p-UPF1(1-241)-cMyc TRP1 | |
| pNE225 | 2 μm CUP1p-DCP1-HA URA3 | |
| pNE227 | CEN4 TRP1 eIF-4E-cMyc | |
| pNE238 | CEN4 URA3 HA-lacZ | |
| pQG63 | CEN4 ebs1-T-3A TRP1 | |
| pQG65 | CEN4 EBS1-HA URA3 | |
| pQG66 | CEN4 DCP1-cMyc TRP1 | |
| pRSL100 | CEN4 UPF1 UPF2 UPF3 |
Parent vector for HA-tagged fusions driven by the CUP1 promoter.
Parent vectors for c-Myc- and HA-tagged fusions driven by the ADH1 promoter.
Selection of suppressors in merodiploids.
Mutations in UPF1, UPF2, or UPF3 that cause a recessive loss of function suppress the frameshift mutation his4-38 in the presence of the temperature-sensitive frameshift suppressor SUF1-1 tRNA mutation, allowing growth in the absence of histidine at 30°C and 37°C (8, 27). Mutations in UPF1, UPF2, or UPF3 also suppress the temperature-sensitive growth of strains carrying the missense mutation ctf13-30, which codes for a defective kinetochore subunit (9). A twofold increase in ctf13 mRNA abundance resulting in the inactivation of NMD permits growth at 37°C.
Spontaneous suppressors of the his4-38 (SUF1-1) and ctf13-30 mutations that conferred growth on synthetic 2% dextrose (SD) medium lacking histidine at 37°C were isolated in strains MJL12 and MJL13 carrying plasmid pRLS100, which expresses UPF1, UPF2, and UPF3. Since chromosomal UPF genes were also present, duplicate UPF genes masked recessive mutations, thereby reducing the recovery of strains with mutations in known UPF genes. Merodiploid cells were grown in SD−Ura to select for the presence of pRLS100. Cells (1.5 × 109) were plated on SD−Ura medium and incubated at 37°C for 10 days. Colonies were patched on SD−Ura, incubated for 5 days at 37°C, replica plated on SD−Ura−His, and incubated at 37°C for 10 days. Corevertants were screened for suppression of the nonsense mutations leu2-2 (UGA) and tyr7-1 (UAA).
EBS1 cloning.
EBS1 was identified in a yeast genomic library containing DNA inserts in the CEN vector Ycp50 (obtained from ATCC). Plasmid 73-79-1, carrying EBS1, was digested with KpnI and BamHI to release a 3,553-bp fragment, which was subcloned into the KpnI and BamHI sites of pRS316 and pRS426, creating pAF15 and pAF16, respectively. The BamHI-SacI fragment from pAF15 was subcloned into pRS315 and pRS425 to create plasmids pAF17 and pAF18, respectively.
RNA methods.
Methods for RNA isolation and Northern blotting were described previously (42). The abundance and half-life of EBS1 mRNA were determined using RNAs isolated from the congenic strains W303a (UPF1) and AAY320 (upf1) (23). Transcription was inhibited with 25 μg/ml thiolutin (Pfizer). Riboprobes were prepared by in vitro transcription in the presence of [α-32P]CTP and [α-32P]UTP (3,000 Ci/mmol) (Perkin-Elmer). Signals were quantitated with a Typhon 9200 imager. EBS1 mRNA was detected on Northern blots by using a riboprobe complementary to nucleotides 182 to 2323. NMD was monitored by the accumulation of CYH2 pre-mRNA relative to CYH2 mRNA on Northern blots, using a riboprobe complementary to nucleotides 572 to 959. CYH2 pre-mRNA (encoding ribosomal protein L29) contains an intron that is inefficiently spliced (22). An in-frame stop codon in the intron triggers rapid decay of unspliced pre-mRNA, which is exported to the cytoplasm, translated, and degraded by NMD (19). CYH2 pre-mRNA accumulates to a higher level in Nmd− strains due to an increase in the pre-mRNA half-life.
IP, cytological, and two-hybrid methods.
Immunoprecipitation (IP) and Western blotting were performed as described previously (48), using protein extracts from strain W303a transformed with the following plasmids expressing epitope-tagged proteins: pQG66, expressing Dcp1-c-Myc; pQG65, expressing Ebs1-hemagglutinin (Ebs1-HA); pNE224, expressing Upf1 (amino acids 1 to 241)-c-Myc; and pNE225, expressing Dcp1-HA. When indicated, protein extracts were supplemented with 10 μg/ml RNase A. A translational fusion between Ebs1p and green fluorescent protein (GFP) was detected in live cells as described previously (41). Leptomycin B was provided by M. Yoshida. To test for two-hybrid interactions, strain PJ6-94A was cotransformed with pGBDU-C1 and pGAD, expressing full-length coding sequences for Ebs1p and other proteins to be tested fused to the Gal4 binding domain and Gal4 activation domain, respectively (21) (see Table 2 for a list of two-hybrid plasmids). PJ6-94A carries a Gal4-HIS3 reporter that is transcriptionally activated when a protein-protein interaction brings the DNA binding and activation domains together, leading to growth on SD−His.
Gap repair mapping.
To map the ebs1-1 mutation, pAF15 was digested with restriction enzymes, removing staggered 300- to 500-base-pair fragments covering the EBS1 open reading frame. Gapped linear plasmids were purified and transformed into strain ASFy5. When the ebs1-1 mutation lies within a gap, DNA repair synthesis using the chromosome as the template repairs the plasmid with DNA that carries the mutation. Mutant plasmids were identified by suppression of the leu2-2 and tyr7-1 mutations and transferred from yeast to Escherichia coli, and the DNA sequences were compared to that of the wild type to locate mutations. The entire gene was sequenced, and only one mutation was found.
Translation termination readthrough.
Stop codon readthrough was measured by using a dual reporter designed for this purpose (3, 38, 44). The readthrough frequency was calculated from the ratio of the luciferase/β-galactosidase activities. To determine the frequency of readthrough, the ratio for the nonsense reporters (UAA, UAG, and UGA) was divided by the ratio for a control reporter (CAA). The dual reporter automatically corrects for differences in mRNA levels or rates of translation initiation.
RESULTS
Selection for suppressors that improve gene expression.
In order to identify mutations that improve gene expression through transcript stabilization or by other mechanisms such as increased translation, cosuppressors of the his4-38 (SUF1-1) and ctf13-30 mutations were selected on SD−His medium at 37°C. Fifty independent corevertants were isolated from strain MJL12 or MJL13 transformed with plasmid pRLS100, which carries UPF1, UPF2, and UPF3. The presence of duplicate UPF genes was intended to mask recessive mutations in UPF1, UPF2, and UPF3 that normally comprise the majority of corevertants, thereby increasing the likelihood of obtaining mutations in other genes that arise at a lower frequency.
To distinguish between suppressors that affect NMD and those that improve gene expression by other mechanisms, the corevertants were analyzed by Northern blotting using a probe that detects CYH2 pre-mRNA and CYH2 mRNA. Changes in the CYH2 pre-mRNA/mRNA ratio serve as a diagnostic indicator of the activity of the NMD pathway (see Materials and Methods). Increased ratios were observed for 14 corevertants, making them candidates for harboring mutations that affect NMD. However, the change in ratio for one (a mutA mutant, described below) was small and of questionable significance, and it was later reclassified as unaltered.
pRLS100 (URA3) present in the parent strain was counterselected in the 14 corevertants by using 5-fluoroorotic acid to remove duplicate copies of UPF1, UPF2, and UPF3. Complementation tests were performed on the 5-fluoroorotic acid-resistant variants by transforming each with single-copy centromeric (CEN) and multicopy 2μm plasmids carrying UPF1, UPF2, or UPF3. Seven corevertants lost the suppression phenotype when transformed with a plasmid carrying UPF1, whereas two lost the suppression phenotype when transformed with a plasmid carrying UPF2. These corevertants carried mutations in UPF1 and UPF2, despite the presence of duplicate UPF genes in the parent strain. The mutations were recessive, suggesting that they were recovered because of homozygosis due to gene conversion.
In the remaining five corevertants, called mutA, mutB, mutF, mutH, and mutI, suppression of his4-38 (SUF1-1) and ctf13-30 was phenotypically unchanged after transformation with pRLS100, indicating the presence of suppressor mutations in genes other than UPF1, UPF2, and UPF3. Each corevertant was mated with a strain carrying the leu2-2 and tyr7-1 nonsense alleles. Null mutations in UPF1, UPF2, and UPF3 that abolish NMD suppress both alleles (42). In spore progeny from matings of mutF, mutH, and mutI mutants with a strain carrying the leu2-2 and tyr7-1 alleles, no suppression was detected, as indicated by a lack of growth on SD−Leu−Tyr medium. Analyses of these corevertants, which suppress only the his4-38 (SUF1-1) and ctf13-30 mutations, are being pursued separately. The experiments in this report focus on the remaining two corevertants, mutA and mutB. Spore progeny with the mutA leu2-2 tyr7-1 genotype grew on SD−Leu−Tyr medium due to suppression of the leu2-2 and tyr7-1 mutations. Spore progeny with the mutB leu2-2 tyr7-1 genotype grew on SD−Leu medium but failed to grow on SD−Leu−Tyr medium, indicating that mutB suppressed the leu2-2 but not tyr7-1 mutation.
mutA and mutB corevertants contain mutations in EBS1.
To identify the gene carrying the mutA suppressor mutation, a mutA leu2-2 tyr7-1 strain was transformed to a Ura+ phenotype with a CEN URA3 plasmid library containing yeast genomic DNA fragments. The growth rates of the library transformants were compared on SD−Ura and SD−Ura−Leu−Tyr medium. Three identical but independently isolated library plasmids complemented the mutA suppressor mutation, as indicated by reduced growth in the absence of uracil, leucine, and tyrosine, indicating a loss of leu2-2 and tyr7-1 suppression. DNA sequence analysis revealed coding sequences for the COQ4, MSC2, and EBS1 genes in the plasmid inserts. Subclones that carried each gene separately were tested for the ability to complement suppression. Only the plasmids carrying EBS1 complemented suppression of his4-38 (SUF1-1), ctf13-30, leu2-2, and tyr7-1 (Fig. 1A and B). The mutA mutation is referred to as ebs1-1.
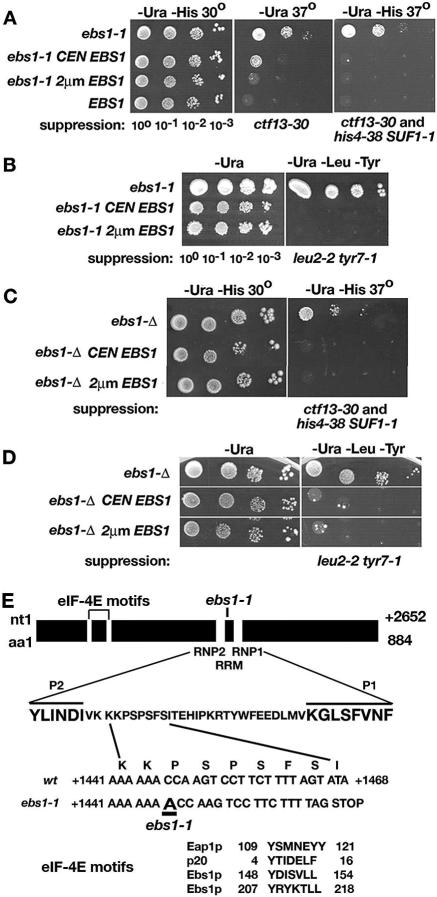
FIG. 1.
Phenotypes caused by ebs1-1 and ebs1-Δ mutations. Strains were transformed with single-copy centromeric (CEN) or multicopy 2μm plasmids carrying EBS1 or empty vectors as follows: A, mutA transformant (ebs1-1 his4-38 SUF1-1 ctf13-30); B, ASFy5 (ebs1-1 leu2-2 tyr7-1); C, ASFy30 (ebs1-Δ his4-38 SUF1-1 ctf13-30); and D, ASFy32 (ebs1-Δ). Growth was assayed using serial dilutions of overnight cultures as indicated. (E) Map of the EBS1 gene showing the location of the ebs1-1 mutation and eIF-4E and RRM motifs.
To establish that ebs1-1-mediated suppression is caused by a loss of EBS1 function, we examined the null allele ebs1-Δ containing an insertion of the KAN gene conferring G148 resistance. The ebs1-Δ mutation suppressed the his4-38 (SUF1-1), ctf13-30, leu2-2, and tyr7-1 alleles to the same extent as the ebs1-1 mutation (compare Fig. 1A with 1C and 1B with 1D). Suppression was abolished when wild-type EBS1 was expressed from a CEN or 2μm plasmid. The Leu and Tyr phenotypes segregated 4+:0− in tetrads from a cross between strains carrying the ebs1-1 leu2-2 tyr7-1 and ebs1-Δ leu2-2 tyr7-1 alleles, indicating that ebs1-1 is genetically linked to ebs1-Δ. Based on the relative strength of suppression in ebs1-Δ and ebs1-1 mutant strains, the ebs1-1 mutation most likely causes a complete loss of function.
The approximate location of the ebs1-1 mutation was determined by gap repair (seeMaterials and Methods). DNA sequence analysis of the local region predicted to contain the mutation revealed a +1 A/T insertion causing an extra A residue in the mRNA at position 1447 within codon 483 (Fig. 1E). The shift in reading frame brings an out-of-frame UAG codon into register six codons downstream of the insertion. The entire gene was sequenced to show that no other mutations were present. The site of the ebs1-1 mutation is flanked by conserved elements RNP2 and RNP1, characteristic of an RNA recognition motif (RRM). Ebs1p also contains two matches to consensus eIF-4E binding domains.
Analysis of a cross between ASFy30 (ebs1-Δ his4-38 SUF1-1 ctf13-30) and the mutB mutant (his4-38 SUF1-1 ctf13-30) indicated that the mutB corevertant contained two recessive suppressor mutations, namely, a hypomorphic allele of EBS1, designated ebs1-2, and a second mutation in an unlinked gene. In segregants carrying ebs1-2 without the second mutation, suppression of the his4-38 (SUF1-1) and ctf13-30 alleles was complemented by a plasmid expressing EBS1. To establish genetic linkage between ebs1-2 and EBS1, the KAN gene was inserted in the nonessential MSC2 gene immediately upstream of EBS1 (strain CSy4). The insertion did not affect EBS1 expression or function, as indicated by the strength of suppression. A cross between this strain and an ebs1-2 leu2-2 tyr7-1 strain revealed that the Kanr and ebs1-2 (scored as a suppressor of leu2-2) alleles were genetically linked, as expected for an allele of EBS1. Strains carrying ebs1-2 had unaltered CYH2 pre-mRNA/mRNA ratios, suggesting that the altered ratio in the original mutB corevertant was due to the unlinked suppressor or possibly another mutation. Overall, the results indicate that 2 of the 50 corevertants contained mutations in EBS1.
Mutations in EBS1 have no effect on nonsense mRNA abundance.
The original mutA strain carrying ebs1-1 was classified as one of the 14 corevertants showing increased CYH2 pre-mRNA/mRNA ratios compared to that of the MJL12 parent strain. However, the results were statistically marginal and not considered reliable. To more rigorously assess the effects of ebs1 mutations on RNA accumulation, the mutA mutant and ASFy5 strains, both of which carry the ebs1-1 mutation, were transformed separately with an empty vector, a single-copy CEN plasmid carrying EBS1, or a multicopy 2μm plasmid expressing EBS1. RNAs were isolated from transformants and analyzed on Northern blots using probes specific for CYH2, his4-38, and leu2-2 mRNA.
Strain ASFy5 (ebs1-1) transformed with an empty vector had a CYH2 pre-mRNA/mRNA ratio of 1.4 ± 0.3 compared with that of the transformant expressing EBS1 from the CEN plasmid (ratio of 1) (Fig. 2A). The transformant expressing EBS1 from the 2μm plasmid had a ratio of 1.1 ± 0.2. Similar results were obtained using transformants of the mutA mutant (ebs1-1) and ASFy31 (ebs1-Δ) strains (data not shown). Using PGK1 mRNA as the normalization signal, the relative abundance of his4-38 mRNA in the mutA (ebs1-1) mutant strain was 1.1 ± 0.1 compared to the transformant expressing EBS1 from the CEN plasmid (Fig. 2B). A similar relative abundance of 1.1 ± 0.5 was observed when EBS1 was expressed from the 2μm plasmid. The relative abundance of leu2-2 mRNA in ASFy5 (ebs1-1) was 1.1 ± 0.1 (Fig. 2C). A similar relative abundance of 1.1 ± 0.2 was observed when EBS1 was expressed from the 2μm plasmid. For comparison, a complete loss of UPF function causes 5- to 6-, 4- to 5-, and 2.2-fold increases in the CYH2 pre-mRNA/mRNA ratio and the accumulation of his4-38 mRNA and leu2-2 mRNA, respectively (27). Statistical analyses using t tests showed that at a P value of 0.05, a loss of EBS1 function fails to alter the accumulation of three NMD-sensitive RNAs known to be stabilized in upf− strains.
FIG. 2.

Effect of ebs1-1 mutation on accumulation of NMD-sensitive transcripts. (A) Northern blotting was used to determine the CYH2 pre-mRNA/mRNA ratio in an ebs1-1 strain transformed with an empty vector or a CEN or 2μm plasmid carrying EBS1. The amount of change (fold) in the CYH2 pre-mRNA/mRNA ratio is indicated, along with the standard error from three experiments (in parentheses). The relative accumulations of his4-38 mRNA (B) and leu2-2 mRNA (C) were determined in an ebs1-1 mutant strain transformed with the plasmids indicated above. The signals were normalized to the PGK1 and HIS4 mRNAs, respectively.
The results described above could be misinterpreted if the PGK1 and HIS4 transcripts used for normalization depend on EBS1 expression. This would invalidate them as adequate controls, as using them could mask changes in transcript levels. To test the adequacy of the normalization controls, RNA levels were monitored by Northern blotting as described above, but SCR1 RNA, an untranslated noncoding RNA component of the signal recognition particle (16), was used for normalization. The results confirmed that the his4-38 and leu2-2 transcript levels were unchanged when EBS1 was deleted or overexpressed (Table 3) and also validated the use of PGK1 and HIS4 as normalization signals.
TABLE 3.
Effects of changes in EBS1 expression on transcript levels
| mRNA | Genotype | Fold change (SD; P value)a |
|---|---|---|
| EBS1 | Chromosomal | 1 |
| CEN plasmid | 2.9 (0.06) | |
| 2μm plasmid | 30.8 (5.4) | |
| PGK1 | ebs1-1 | 1.2 (0.3; 0.33) |
| 2μm EBS1 | 1.1 (0.1; 0.17) | |
| his4-38 | ebs1-1 | 1.1 (0.4; 0.60) |
| 2μm EBS1 | 0.7 (0.2; 0.11) | |
| PET18 | ebs1-1 | 2.2 (0.6; 0.08) |
| 2μm EBS1 | 0.9 (0.3; 0.80) | |
| leu2-2 | ebs1-1 | 1.2 (0.5; 0.65) |
| 2μm EBS1 | 0.9 (0.2; 0.38) | |
| HIS4 | ebs1-1 | 1.3 (0.3; 0.26) |
| 2μm EBS1 | 1.2 (0.4; 0.48) | |
| CUP1 | ebs1-1 | 0.7 (0.3; 0.24) |
| 2μm EBS1 | 1.0 (0.1; 0.84) | |
| HIS3 | ebs1-1 | 0.6 (0.1; 0.02) |
| 2μm EBS1 | 1.2 (0.1; 0.04) | |
| CAN1 | ebs1-1 | 1.2 (0.3; 0.51) |
| 2μm EBS1 | 1.4 (0.6; 0.45) | |
| can1-100 | Nmd−ebs1-Δ | 1.1 (0.4; 0.70) |
| Nmd− 2μm EBS1 | 1.1 (0.2; 0.64) | |
| CYH2 | Nmd−ebs1-Δ | 1.1 (0.1; 0.23) |
| Nmd− 2μm EBS1 | 1.1 (0.1; 0.18) |
Transcript levels were normalized to noncoding SCR1 RNA detected with a riboprobe complementary to nucleotides 60 to 424. Nmd− strains carried upf1-Δ, upf2-Δ, and upf3-Δ. Values were calculated from three independent trials and normalized to values obtained from strains expressing EBS1 from a CEN plasmid.
Changes in EBS1 expression levels alter resistance to 3-aminotriazole, copper, and canavanine.
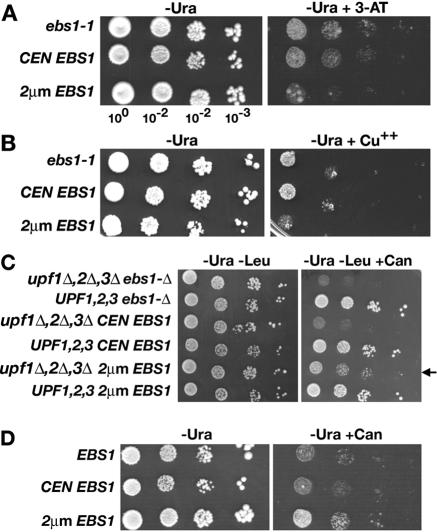
Given the results described above, growth in the presence of 3-aminotriazole (3-AT) or copper was used to qualitatively assess whether Ebs1p affects the expression of wild-type HIS3 and CUP1. 3-AT inhibits His3p, whereas Cup1p chelates excess copper. The degrees of resistance to 3-AT and copper correlate with the levels of His3p and Cup1p, respectively (29, 34). Resistance to 75 mM 3-AT or 0.5 mM copper was examined by comparing transformants of strain ASFy5 (ebs1-1 HIS3 CUP1) carrying an empty vector, a CEN plasmid carrying EBS1, or a 2μm plasmid expressing EBS1 (Fig. 3A and B). In both tests, relative growth rates were slower when EBS1 was expressed from a 2μm plasmid. Since the levels of expression of HIS3 and CUP1 are proportional to the degrees of 3-AT and copper sensitivity, respectively, the increased sensitivity resulting from EBS1 overexpression suggests that excess EBS1 decreases the expression of HIS3 and CUP1.
FIG. 3.
Effect of Ebs1p on gene expression. To assay the expression of HIS3 and CUP1, an Nmd+ ebs1-1 HIS3 CUP1 mutant strain was transformed with an empty vector, a CEN plasmid carrying EBS1, or a 2μm plasmid expressing EBS1. (A) Growth was assayed on SD−Ura−His containing 75 mM 3-AT. (B) Growth was assayed on SD−Ura containing 0.5 mM CuCl2. (C) EBS1 overexpression affects canavanine resistance in Nmd+ strains carrying can1-100. CEN and 2μm plasmids carrying EBS1 were transformed into an Nmd− strain, with or without plasmid pRSL100, expressing UPF1, UPF2, and UPF3. Growth was assayed on SD−Ura−Leu containing 100 μg/ml canavanine. The arrow (right) draws attention to the increased resistance of the upf− transformant expressing EBS1 from the 2μm plasmid. (D) EBS1 overexpression affects canavanine resistance in Nmd+ CAN1-expressing strains. A strain with the genotype UPF EBS1 CAN1 was transformed with an empty vector and a CEN plasmid or 2μm plasmid carrying EBS1. Growth was assayed on SD−Ura containing 20 μg/ml arginine and 100 μg/ml canavanine.
CAN1 codes for an arginine permease that promotes the lethal uptake of canavanine, whereas the can1-100 allele, which contains a UAA nonsense mutation, confers resistance by preventing uptake due to a nonfunctional arginine permease. The extent to which strains carrying CAN1 and can1-100 are resistant to canavanine was used to assess whether changes in EBS1 expression affect the expression of CAN1 and can1-100. A CEN plasmid and a 2μm plasmid carrying EBS1 were transformed into strain ASFy29, which carries the ebs1-Δ, can1-100, upf1-Δ, upf2Δ, and upf3-Δ alleles. The transformants either contained or lacked a plasmid expressing UPF1, UPF2, and UPF3. The ebs1-Δ transformant expressing wild-type UPF genes was canavanine resistant, indicating that a loss of EBS1 function alone does not visibly suppress the Canr phenotype of a can1-100 mutant (Fig. 3C). The loss of EBS1 function also failed to suppress another UAA nonsense mutation, leu2-1 (data not shown). The upf mutant transformants lacking EBS1 or expressing EBS1 from the CEN plasmid were canavanine sensitive, indicating that suppression due to the inactivation of NMD was unaffected by the absence of EBS1 or the presence of Ebs1p at normal levels. However, a upf− transformant overexpressing EBS1 from the 2μm plasmid displayed increased resistance (indicated by the arrow in Fig. 3C). This result suggests that overexpression of EBS1 partially inhibits the suppression of can1-100 caused by the inactivation of NMD.
To test whether overexpression of EBS1 inhibits the expression of the wild-type CAN1 gene, strain MJL12 (UPF EBS1 CAN1) was transformed with an empty vector, a CEN plasmid carrying EBS1, or a 2μm plasmid expressing EBS1. Growth was assayed on SD medium containing 20 μg/ml arginine and 100 μg/ml canavanine (Fig. 3D). The addition of arginine permits some growth in the presence of canavanine because arginine and canavanine compete for import. The transformant expressing EBS1 from the 2μm plasmid was more resistant to canavanine than transformants containing the empty vector or expressing EBS1 from the CEN plasmid. These results suggest that overexpressing EBS1 causes increased resistance to canavanine because the expression of CAN1 is reduced. This could explain the synthetic phenotype that results when EBS1 is overexpressed in a upf− background.
When transcript levels were examined using SCR1 RNA as a normalization control, no changes related to EBS1 expression levels were observed for HIS3, CUP1, CAN1, or can1-100 transcripts, even though expression from a 2μm plasmid was about 31-fold higher than expression from a single chromosomal EBS1 gene (Table 3). Taken together, the results suggest that changes in EBS1 expression levels have little to no effect on mRNA abundance. Therefore, changes in mRNA levels are not likely to underlie the observed changes in drug resistance.
Ebs1p inhibits translation without affecting the frequency of premature termination.
Reporters were used to monitor the relative rates of translation in strains that carry the ebs1-1 allele or that overexpress Ebs1p. β-Galactosidase activities produced by expressing lacZ driven by the ADH1 promoter from a CEN plasmid were measured in strain Asy5 (ebs1-1) transformed with an empty vector or a CEN or 2μm plasmid carrying EBS1. Enzyme activities were adjusted for differences in reporter mRNA levels after normalization to the SCR1 RNA level to compare the relative activities per equivalent numbers of transcripts. Compared to that of the ebs1-1 mutant strain, the β-galactosidase activity was reduced to 62% ± 5% or 46% ± 1% when Ebs1p was expressed from the CEN or 2μm plasmid, respectively (Fig. 4A). The differences were significant (P = 0.01). These results suggest that the rate of translation is sensitive to the EBS1 expression level and that Ebs1p negatively regulates translation.
FIG. 4.

Effects of EBS1 expression levels on translation. (A) Relative levels normalized to SCR1 RNA were determined for transformants carrying an empty vector or a CEN or 2μm plasmid carrying EBS1. Enzyme activities were adjusted relative to lacZ mRNA levels to compare relative rates of translation per transcript. (B) β-Galactosidase-to-luciferase enzyme activity ratios were calculated for transformants carrying dual reporters that contain an internal stop codon and either an empty vector or a CEN or 2 μm plasmid carrying EBS1. (C) GFP fluorescence in cells expressing Ebs1-GFP compared to DAPI (4′,6′-diamidino-2-phenylindole) staining of DNA. Bar = 5 μm.
To assess whether the ebs1-Δ mutation increases readthrough at premature translation termination codons similar to that observed for upf mutations (31, 49), a lacZ-luc dual-gene reporter that contains lacZ, an internal segment, and luc (encoding firefly luciferase) was used to measure readthrough frequencies for all three stop codons (3). Expression from a reporter lacking a stop codon in the internal segment produces a β-galactosidase-luciferase fusion protein that retains both enzymatic activities. Introduction of a termination codon into the internal segment causes the production of a protein with β-galactosidase activity but no luciferase activity when translation termination is 100% efficient. The rate of stop codon readthrough was measured by comparing the ratios of luciferase to β-galactosidase activities produced from reporters containing and lacking a stop codon in the internal segment.
Readthrough frequencies were measured in strain RGy13565 transformed with dual reporter plasmids and a CEN or 2μm plasmid carrying EBS1 or an empty vector. Although the frequencies varied for each of the three stop codons, no statistically significant changes were observed when EBS1 was deleted or overexpressed (Fig. 4B). These results suggest that Ebs1p function and the phenotypes of ebs1− mutants are not related to the efficiency of premature translation termination.
Ebs1p localizes throughout the cytoplasm.
A cytoplasmic distribution would be expected if Ebs1p plays a role in translation. To determine the cellular distribution of Ebs1p, an Ebs1-GFP translational fusion expressed in strain MNY8 was visualized in live cells. GFP fluorescence was distributed throughout the cytoplasm (Fig. 4C). Ebs1p contains a putative leucine-rich nuclear export sequence (residues 229 to 239). We examined the distribution of Ebs1-GFP in the presence of leptomycin B in strain MNY8, which carries a leptomycin-sensitive mutation in CRM1 (13, 20, 43). If leptomycin B inhibited the export of nucleus-localized Ebs1-GFP, then nuclear accumulation should occur in the presence of the drug. Leptomycin B had no detectable effect on the distribution of Ebs1-GFP (data not shown). The results suggest that Ebs1p is a cytoplasmic protein.
Interactions with Cap binding proteins and translation factors.
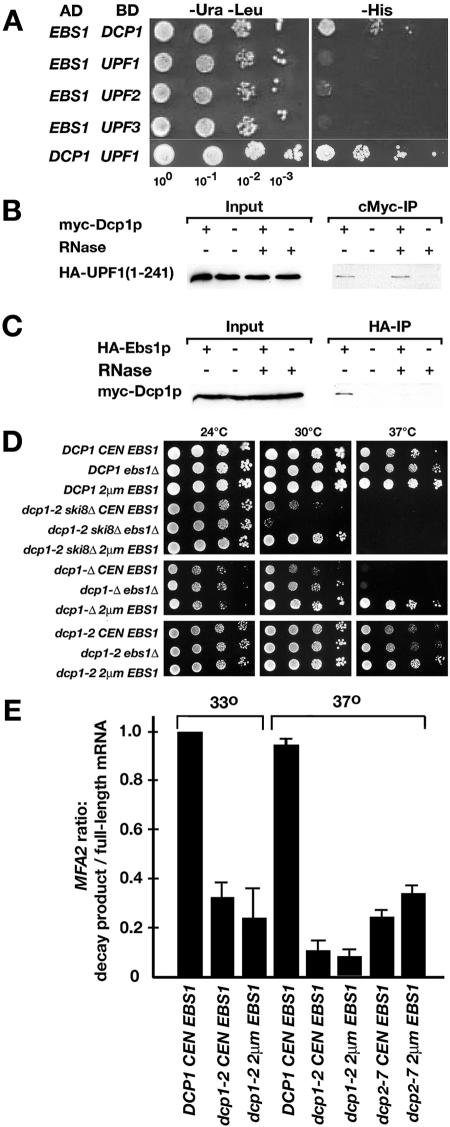
Since the results described above suggested a role for Ebs1p in translation and since Ebs1p contains two consensus motifs for binding to the cytoplasmic cap binding protein eIF-4E (Fig. 1E), we examined interactions with cap binding proteins and several translation initiation factors. Two-hybrid tests involving eIF-4A and eIF-4E were complicated by autoactivation of transcription of the HIS3 two-hybrid reporter. Little to no growth beyond the autoactivation level was detected on SD−His medium when strain PJ69-4A was cotransformed with two-hybrid plasmids expressing Ebs1-AD and eIF-4E-BD (Fig. 5A). Similarly, no two-hybrid interaction was detected between Ebs1p and the initiation factor eIF-4A or eIF-4G or the nuclear cap binding protein Cbc1p. However, when HA-Ebs1p was immunoprecipitated, Myc-eIF-4E was detected in the post-IP lysate by Western blotting, with or without pretreatment with RNase A prior to IP (Fig. 5B), indicating that the two proteins copurify in an RNA-independent manner. The nuclear cap binding proteins Myc-Cbc1p and Myc-Cbc2p were not detected in post-IP lysates (not shown).
FIG. 5.
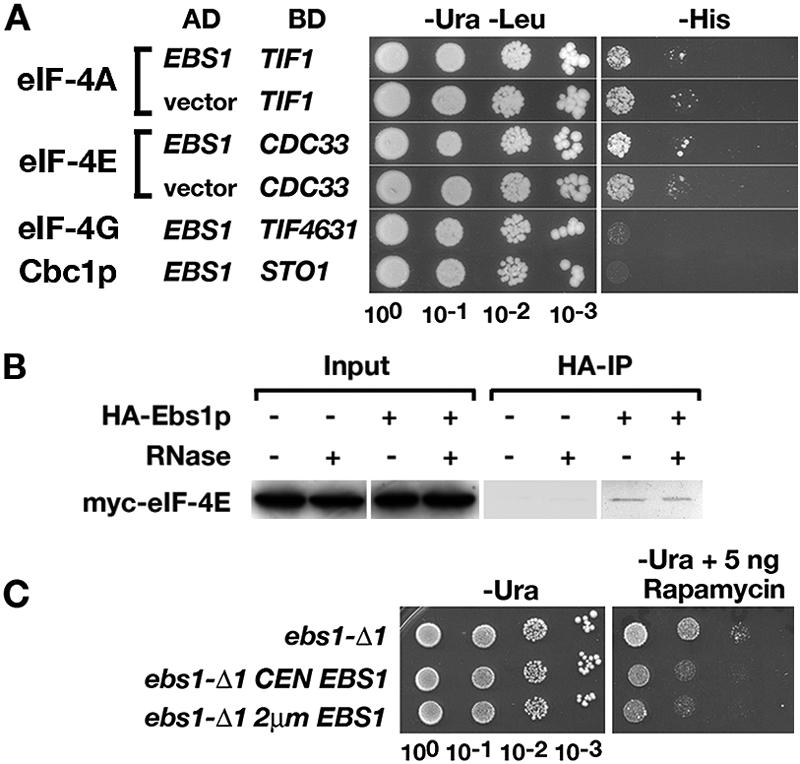
Relationship of Ebs1p to translation factors. (A) Two-hybrid interactions. Strain PJ69-4A was cotransformed with plasmids expressing an Ebs1p-AD fusion containing all of Ebs1p and one of the following: eIF-4A-BD (TIF1), eIF-4E-BD (CDC33), eIF-4G-BD (TIF4631), or Cbc1p-BD (STO1). The growth of vector control strains was due to autoactivation of transcription. (B) Co-IP of Myc-eIF-4E and HA-Ebs1p. Lysates contained 10 μg/ml RNase A where indicated. (C) Rapamycin resistance. Strain RGy13565 (ebs1-Δ) was transformed with the empty vector pRS316, the CEN plasmid pAF15 carrying EBS1, or the 2μm plasmid pAF16 carrying EBS1. Growth was monitored on medium lacking uracil and containing 5 ng/ml of rapamycin.
The eIF-4E binding protein Eap1p confers resistance to the immunosuppressant rapamycin because it functions in the TOR signaling pathway that controls translation initiation (5). We tested the sensitivity of ebs1 mutants to rapamycin (Fig. 5C). Strain RGy13565 (ebs1-Δ) was transformed with an empty vector, a CEN plasmid carrying EBS1, or a 2μm plasmid expressing EBS1. The transformants were plated on SD−Ura medium containing 1 or 5 ng/ml rapamycin. At 1 ng/ml, all of the transformants grew at similar rates. However, at 5 ng/ml, the ebs1-Δ mutant was more resistant than transformants carrying the CEN-EBS1 and 2μm EBS1 plasmids. The growth rates were the same regardless of whether Ebs1p was expressed from the CEN or the 2μm plasmid. Similar results were obtained with strains carrying ebs1-1.
Interactions with the decapping complex and the Upf proteins.
It was reported previously that Ebs1p interacts in the two-hybrid system with the decapping component Dcp1p (47). Experiments were performed to further assess the possibility of interactions with the decapping complex. The Upf proteins required for NMD were included in the experiments because Upf1p was shown previously to interact with Dcp2p (17). To test for two-hybrid interactions, strain PJ69-4A was cotransformed with plasmids expressing two-hybrid fusions encoding Ebs1-AD (Gal4 activation domain) along with Upf1-BD, Upf2-BD, Upf3-BD, or Dcp1-BD (Gal4 binding domain). Potential interactions were assessed on SD−His medium, where growth due to an interaction would activate transcription of the HIS3 two-hybrid reporter (Fig. 6A). No interactions were detected when PJ69-4A was cotransformed with plasmids expressing Ebs1-AD and Upf1-BD, Upf2-BD, or Upf3-BD. However, two-hybrid interactions were detected in transformants coexpressing Upf1-BD and Dcp1-AD, indicating that Upf1p interacts with Dcp1p, and in transformants coexpressing Ebs1-AD and Dcp1-BD, indicating that Ebs1p interacts with Dcp1p (Fig. 6A).
FIG. 6.
Relationship between Ebs1p, mRNA decapping, and mRNA decay. (A) Two-hybrid interactions. Strain PJ69-4A was cotransformed with plasmids expressing an Ebs1p-AD fusion containing all of Ebs1p and one of the following: Dcp1p-BD, Upf1p-BD, Upf2p-BD, or Upf3p-BD. The same strain was cotransformed with Dcp1p-AD and Upf1p-BD. Growth was monitored on SD−His to assay activation of the HIS3 two-hybrid reporter. (B) RNA-dependent co-IP of Myc-Dcp1p and HA-Ebs1p. (C) RNA-independent co-IP of Myc-Upf11-241 and HA-Dcp1p. Lysates contained 10 μg/ml RNase A where indicated. (D) Effects on growth when EBS1 was overexpressed in dcp1-2, ski8-Δ, and dcp1-Δ mutants. (E) Relative decapping rates were determined by Northern blotting using a probe that detects MFA2pG mRNA containing a G loop near the 3′ end (2, 12). Since decapping leads to 5′ decay up to the G loop, the ratio of the decay product to the full-length mRNA provides a measure of relative decapping rates. Cells were shifted from a permissive (25°C) to a restrictive (33°C or 37°C) temperature as indicated.
When HA-Dcp1p was immunoprecipitated, either full-length Myc-Upf1p (not shown) or a truncated version of Myc-Upf1p containing amino acids 1 to 241 (Fig. 6B) was detected by Western blotting with the post-IP lysate. When HA-Ebs1p was immunoprecipitated, Myc-Dcp1p was detected by Western blotting with the post-IP lysate (Fig. 6C). Since Ebs1p contains a functional RRM (51), the protein lysate from the HA-Ebs1p IP was incubated with RNase A prior to IP. After RNase treatment, no Dcp1-cMyc was detected in the post-IP lysate, indicating that the copurification of Ebs1p and Dcp1p could be due to an indirect interaction since it requires the presence of RNA.
Genetic interactions detected in ebs1 dcp1 double mutants.
Genetic interactions in strains carrying mutations in ebs1 and dcp1 were assessed using a previously described growth assay with strains compromised for both 5′→3′ and 3′→5′ decay pathways (2). Strains defective in both pathways are inviable, but dcp1-2 ski8Δ and dcp2-7 ski3Δ double mutants grow conditionally at 24°C while failing to grow at 33°C or 37°C, respectively (2, 12). Genes coding for Dcp2p and the decapping enhancers Edc1p, Edc2p, and Edc3p suppress temperature-sensitive lethality in the double mutants when overexpressed (11, 12, 25). The growth of a dcp1-2 ski8Δ double mutant was assayed in strain backgrounds carrying ebs1-Δ or expressing EBS1 from a 2μm plasmid (Fig. 6D). The dcp1-2 ski8Δ ebs1-Δ triple mutant failed to grow at 30°C and 37°C. When EBS1 was expressed from a CEN plasmid, growth was slightly improved. When EBS1 was expressed from a 2μm plasmid, growth was restored at 30°C and 33°C (not shown) but not at 37°C. These phenotypes resemble the growth suppression conferred when EDC genes are overexpressed, suggesting that Ebs1p enhances decapping in double mutants impaired for decapping and 3′ decay.
We also analyzed the effects of deleting or overexpressing EBS1 in strains carrying dcp1-2 or a dcp1-Δ null allele in the absence of mutations in the SKI genes. Strains that carry dcp1-Δ or both dcp1-Δ and ebs1-Δ were slightly growth-impaired at 30°C, and both single and double mutants failed to grow at 37°C (Fig. 6D, middle panels). When EBS1 was overexpressed from a 2μm plasmid, growth was completely restored at both 30°C and 37°C, suggesting that excess Ebs1p compensates for a complete loss of Dcp1p function. Similar results were observed in strains carrying dcp1-2. Interestingly, a suppression of growth was not observed in a dcp2-7 ski3-Δ double mutant or in a dcp2-7 single mutant coding for a partially impaired catalytic subunit of the decapping complex (data not shown).
Changes in EBS1 expression have little to no effect on decapping of MFA2 mRNA.
To determine whether the physical and genetic interactions described above were indicative of an effect of EBS1 on the rate of decapping, we used a modified MFA2 gene coding for an mRNA that contains a 3′-proximal G loop (12). When the cap is removed, decay from the 5′ end halts at the G loop, leaving a relatively stable decay product that can be detected by Northern blotting. The ratio of capped full-length MFA2 mRNA to the uncapped degradation product provides a relative measure of the decay rate. Both the dcp1-2 and dcp2-7 mutations impair decapping, as indicated by decreased ratios of the decay products to the full-length mRNA (Fig. 6E). Expression of EBS1 from a 2μm plasmid had little to no effect on the ratio compared to expression from a CEN plasmid in strains carrying dcp1-2 and a small, statistically marginal effect in strains carrying dcp2-7. These results indicate that Ebs1p does not modulate decapping as the underlying basis for the physical and genetic interactions describe above.
Upf proteins control EBS1 mRNA abundance.
The Upf proteins affect the abundance of hundreds of mRNAs (18, 28). Some of the changes in abundance detected on DNA arrays correlate with changes in the mRNA half-lives, while others are uncorrelated, typically with no change in half-life. EBS1 mRNA increases in abundance in strains carrying mutations in UPF1, UPF2, or UPF3 and in a strain carrying mutations in all three genes (28).
EBS1 mRNA was first analyzed by Northern blotting with UPF1+ and upf1− strains. Consistent with the results from microarray analysis (28), a threefold increase was observed for the upf1-Δ mutant (Fig. 7A). However, the EBS1 mRNA half-life was the same (12 to 15 min) in upf1-Δ mutant and UPF1+ strains (Fig. 7B and C). This result indicates that the difference in abundance was most likely due to a change in the rate of transcription of the EBS1 gene and that an unidentified regulator of EBS1 expression may be the direct target of Upf-mediated mRNA decay.
FIG. 7.
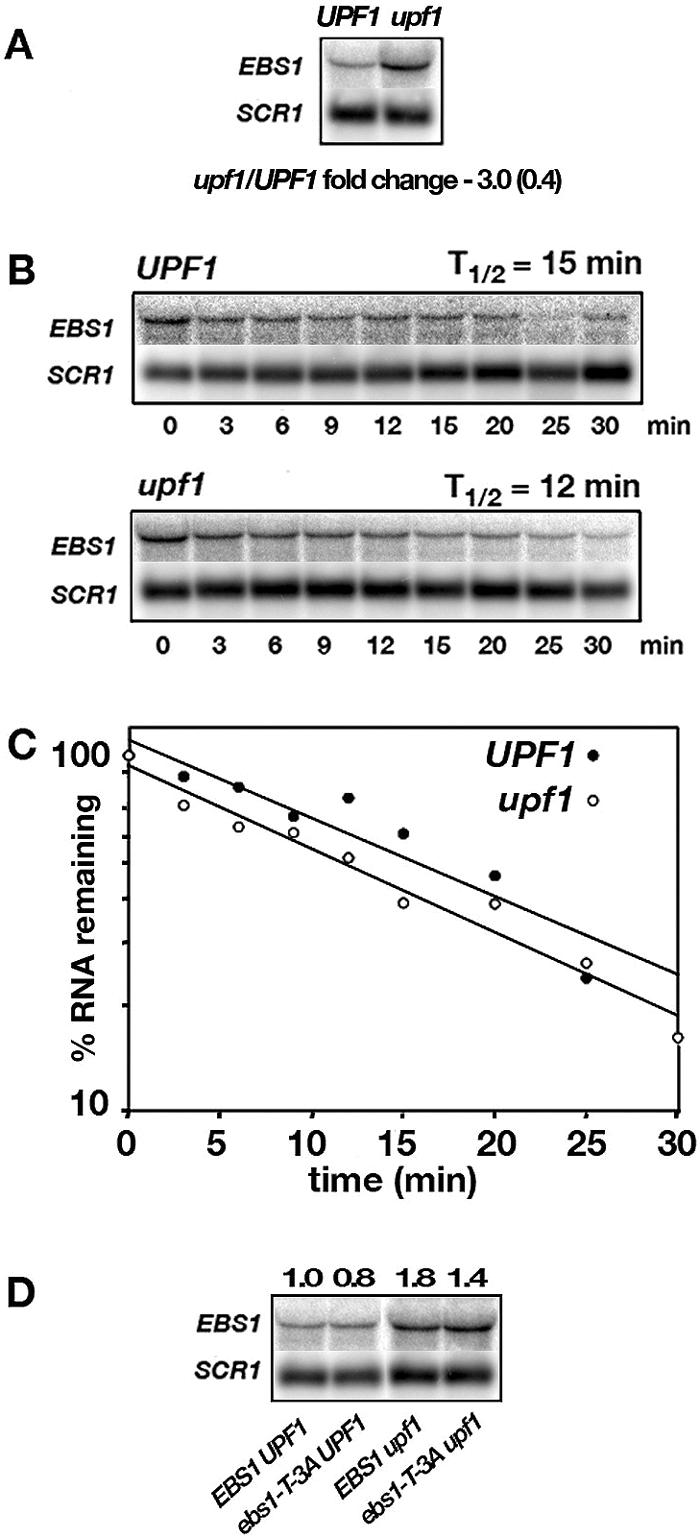
EBS1 mRNA accumulation depends on NMD. (A) EBS1 mRNA expressed from the chromosomal EBS1 gene in UPF1 and upf1-Δ mutant strains was detected by Northern blotting. The difference in expression was calculated after normalizing the signals to the SCR1 RNA signal. (B and C) The EBS1 mRNA half-life was determined by Northern blotting following the addition of 25 μg/ml thiolutin to inhibit transcription. Half-lives were determined from four experiments. (D) Strain QGY37 (upf1-Δ ebs1-Δ) was cotransformed with combinations of CEN plasmids carrying EBS1, UPF1, or ebs1-T-3A or with an empty vector. RNA accumulation levels were determined by Northern blotting.
EBS1 mRNA was considered a candidate for leaky scanning, which occurs in the SPT10 transcript when ribosomes bypass the first AUG and initiate at a downstream AUG in an alternate reading frame (50). Termination at an out-of-frame stop codon triggers a Upf-dependent change in the mRNA decay rate. In upf− mutants, the SPT10 transcript is stabilized. The frequency of ribosome bypass of the first AUG depends on the context for initiation, which can be evaluated by the codon adaptation index for the initiating AUG (AUGCAI), which reflects the extent of deviation from the optimal consensus (A/U)A(A/C)AA(A/C)AUGUC(U/C) (33). A low score within the AUGCAI range of 0 to 1 indicates fewer matches to the optimal context and thus a higher probability of inefficient translation initiation. Purines at the −3 position have the greatest influence in favoring efficient initiation. For SPT10 mRNA, AUGCAI = 0.25; for EBS1 mRNA, AUGCAI = 0.41.
To test whether EBS1 mRNA is subject to leaky scanning, a T-to-A mutation at the −3 position was made, which improves the context from an AUGCAI of 0.41 to an AUGCAI of 0.61. Compared to a reference set of highly expressed genes, an index score of 0.61 corresponds to a good context for initiation (unpublished observation). Strain QGY37 (upf1-Δ ebs1-Δ) was transformed with an empty vector or with CEN plasmids carrying either EBS1 or ebs1-T-3A (Fig. 7D). If leaky scanning triggered UPF-dependent decay of EBS1 mRNA, then the mutant ebs1-T-3A transcript should have been more stable and more abundant than EBS1 mRNA in an Nmd+ strain. However, Northern blotting indicated that the abundances of ebs1-T-3A and EBS1 mRNAs were similar in the Nmd+ strain. This result indicates that leaky scanning plays no role in targeting EBS1 for NMD, consistent with the absence of an altered EBS1 mRNA half-life in upf− strains.
DISCUSSION
Cosuppressors of the missense mutation ctf13-30 and the frameshift mutation his4-38 were selected in a strain carrying duplications of UPF1, UPF2, and UPF3. This paper focuses on mutations in the EBS1 gene that were recovered in the merodiploid selection. The ebs1-1 allele is a +1 insertion in the middle of an RNA recognition motif in the coding region. In addition to suppressing the ctf-13-30 and his4-38 alleles, ebs1-1 suppressed the nonsense mutations leu2-2 (UGA) and tyr7-1 (UAG). The null allele ebs1-Δ suppressed the same mutations to the same extent as ebs1-1 but had no effect on growth in rich medium, indicating that ebs1-1 causes a complete loss of function of a nonessential gene.
The loss of EBS1 function had no effect on the steady-state levels of three NMD-sensitive RNAs, namely, CYH2 pre-mRNA, his4-38 mRNA, and leu2-2 mRNA. The mRNA levels of the nonsense transcripts were initially normalized to the levels of the PGK1 and HIS4 transcripts, respectively. However, to guard against the possibility that EBS1 affects the abundance of the control transcripts, the RNA levels were also normalized to SCR1 RNA, a structural component of the signal recognition particle (16) that is not translated and therefore not likely to be affected by Ebs1p. The results indicate that suppression of the his4-38 and leu2-2 mutations occurs without any change in transcript accumulation. Mutations in EBS1 are therefore distinguished from UPF mutations, which cause suppression partly through changes in mRNA accumulation and partly through increased readthrough of premature translation termination codons. EBS1 is not a fourth UPF gene.
The overexpression of wild-type EBS1 alters resistances to 3-AT, copper, and canavanine, indicating reduced expression of HIS3, CUP1, and CAN1, respectively. Using noncoding SCR1 RNA as the loading control for Northern blots, no changes were detected in the abundance of the HIS3, CUP1, and CAN1 transcripts, suggesting that altered resistances result from changes in gene expression that do not involve changes in rates of transcription or mRNA decay. The results indicate that disrupting EBS1 function increased gene expression and overexpressing EBS1 reduced gene expression without commensurate changes in transcript abundance for seven wild-type genes, including PGK1, HIS4, PET18, HIS3, CUP1, CAN1, and CYH2. Our results support the model that Ebs1p inhibits the expression of many yeast genes, but we cannot be certain that it inhibits the expression of all genes. However, a previous study showed that the expression of genes controlling telomere length is affected by the expression of EBS1, supporting a possible global regulatory role for the Ebs1 protein (51).
To establish a role for Ebs1p in translation, a lacZ reporter was assayed to compare the amounts of β-galactosidase activity produced per transcript in strains where EBS1 was deleted or overexpressed. The results indicate that the enzyme activity per transcript was highest in an ebs1-Δ mutant strain but was reduced to an extent proportional to increased dosage of the wild-type EBS1 gene. These results are consistent with a role for Ebs1p as an inhibitor of translation.
No changes in readthrough efficiencies at premature stop codons were observed with a dual reporter system (3). To explain ebs1-mediated suppression, a requirement of the translation model is that natural readthrough rates for premature termination codons must be high enough that increased rates of translation lead to a sufficient accumulation of full-length readthrough proteins to cause suppression. Of the three stop codons, UAA is the most efficient and has the lowest readthrough rate, but the context surrounding termination at any premature stop codon influences the frequency of readthrough (3, 24). Premature UAA codons in a good context for termination might therefore be the least likely to be suppressed. This may be why ebs1-Δ fails to suppress two UAA nonsense mutations, namely, can1-100 and leu2-1. Interestingly, the can1-100 mutation is suppressed by ebs1-Δ in upf1-Δ mutant strains. The likely reason is that the can1-100 mutation is in a good context for termination and because the upf1-Δ mutation may increase the rate of readthrough. A combination of increased translation initiation caused by ebs1-Δ and increased readthrough caused by upf1-Δ could allow greater accumulation of full-length Can1p, leading to suppression.
Ebs1p contains consensus motifs suggesting binding to the cytoplasmic cap binding protein eIF-4E (30). Although Ebs1p and eIF-4E did not appear to interact well in the two-hybrid system, eIF-4E copurified with Ebs1p in IP experiments, even in the presence of RNase A. The results suggest that the two proteins are not merely tethered to common RNAs, but rather that the interaction is a direct protein-protein interaction. The relatively small proportion of input eIF-4E that copurified with Ebs1p could be due to a lower total abundance of Ebs1p than of eIF-4E. No interactions were detected between Ebs1p and the nuclear cap binding protein Cbc1p or the translation initiation factors eIF-4A and eIF-4G. If Ebs1p inhibits translation through the interaction with eIF-4E, the effect is most likely at the level of translation initiation. In addition, effects on elongation are not likely because the elongation rate is preset by kinetic proofreading and not readily altered without negative consequences for growth and because deleting EBS1 does not impair growth.
Two other eIF-4E binding proteins have been described, namely, Caf20p and Eap1 (5). These proteins function in the TOR kinase signaling pathway that controls the rate of translation initiation during the cell cycle (10). TOR signaling is sensitive to rapamycin, and mutations that impair the function of some proteins that function in the TOR pathway, such as Eap1p, confer resistance to rapamycin (5). We found that the ebs1-Δ mutation confers increased resistance to rapamycin. Although more data will be needed to interpret this result, the rapamycin-resistant phenotype is another indicator that Ebs1p may function in a complex network of proteins that influence translation initiation.
Ebs1p and the decapping subunit Dcp1p interact in the two-hybrid system (47) and copurify during immunoprecipitation. However, copurification was abolished by pretreatment with RNase A, suggesting that Ebs1p and Dcp1p associate only when both proteins are bound to mRNA. The Ebs1p-Dcp1p interaction might therefore be indirect and might not involve direct protein-protein contacts. A surprising finding was that overexpression of Ebs1p suppressed the temperature-sensitive growth defect caused by a complete deletion of DCP1, indicating that overexpressed Ebs1p compensates for the absence of Dcp1p. Overexpressed Ebs1p also suppressed growth defects that occur in dcp1 ski8 double mutants impaired for both the 5′ and 3′ decay pathways. Three other proteins, Edc1p, Edc2p, and Edc3p, confer a similar phenotype when overexpressed. However, these proteins influence the rate of decapping (2, 12). Ebs1p differs from the Edc proteins because Ebs1p did not affect the decapping rate in an assay that measures decapping of MFA2 mRNA. The lack of an effect on decapping is consistent with the absence of changes in mRNA abundance when the expression level of EBS1 is altered. Although the basis for the genetic interactions observed in strains carrying mutations in ebs1 and dcp1 are not yet understood, they do not appear to be indicative of a direct role for EBS1 in decapping.
We also uncovered an interesting circuit that connects the function of Ebs1p with NMD. In wild-type strains, NMD controls EBS1 expression by reducing the abundance of the EBS1 transcript severalfold compared to that in a mutant that is unable to support NMD. This occurs without a change in the EBS1 mRNA decay rate, indicating that EBS1 is member of a larger class of indirect targets of NMD that may be regulated by NMD-sensitive transcription factors (28). Also, our results show that Ebs1p and the NMD factor Upf1p are physically proximal through interactions with Dcp1p, which participates along with Dcp2p and Xrn1p in the 5′ decay pathway triggered by NMD. It was shown previously that NMD increases the translation efficiency of nonsense transcripts (35). This might occur through down-regulation of Ebs1p, which could reduce translation inhibition, leading to increased translation initiation during NMD.
Recently, it was shown that NMD targets both Cbc- and eIF-4E-bound transcripts for degradation (14). NMD-sensitive transcripts that escape decay during pioneer translation of Cbc-bound transcripts are prone to NMD in subsequent rounds when bound to eIF-4E. If the interactions we have described for Ebs1p are indicative of a function in bulk translation, then translation inhibition by Ebs1p could be limited to eIF-4E-bound transcripts. What, then, is the net result of placing EBS1 expression under the control of NMD? Since NMD reduces EBS1 expression, the inhibition of translation of eIF-4E-bound NMD-sensitive transcripts that escape NMD during pioneer translation would be reduced, causing increased translation of the NMD-sensitive transcripts during the multiple rounds of translation that take place in polyribosomes. Further studies will be required to test the validity of these ideas.
Acknowledgments
This work was supported by the University of Wisconsin College of Agricultural and Life Sciences, the University of Wisconsin Medical School, and NIH grant GM65172 (M.R.C.).
We thank Cynthia Spring and Natalie Barszcz for technical assistance.
Footnotes
This is University of Wisconsin Laboratory of Genetics paper no. 3625.
REFERENCES
- 1.Anders, K. R., A. Grimson, and P. Anderson. 2002. SMG-5, required for nonsense-mediated mRNA decay, associates with SMG-2 and protein phosphatase 2A. EMBO J. 22:641-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, J. S., and R. Parker. 1998. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA exonucleases of the exosome complex. EMBO J. 17:1497-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bidou, L., G. Stahl, I. Hatin, O. Namy, J. P. Rousset, and P. J. Farabaugh. 2000. Nonsense-mediated decay mutants do not affect programmed −1 frameshifting. RNA 6:952-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cali, B. M., S. L. Kuchma, J. Latham, and P. Anderson. 1999. smg-7 is required for mRNA surveillance in Caenorhabditis elegans. Genetics 151:605-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cosentino, G. P., T. Schmezle, A. Haghighat, S. B. Helliwell, M. N. Hall, and N. Sonenberg. 2000. Eap1p, a novel eukaryotic translation initiation factor 4E-associated protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:4604-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Culbertson, M. R., and P. F. Leeds. 2003. Looking at mRNA decay pathways through the window of molecular evolution. Curr. Opin. Genet. Dev. 13:207-214. [DOI] [PubMed] [Google Scholar]
- 7.Culbertson, M. R., and E. Neeno-Eckwall. 2005. Transcript selection and the recruitment of mRNA decay factors for NMD in Saccharomyces cerevisiae. RNA 11:1333-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Culbertson, M. R., K. M. Underbrink, and G. R. Fink. 1980. Frameshift suppression in Saccharomyces cerevisiae. II. Genetic properties of group II suppressors. Genetics 95:833-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dahlseid, J. N., J. Puziss, R. L. Shirley, A. L. Atkin, P. Hieter, and M. R. Culbertson. 1998. Accumulation of mRNA coding for the Ctf13p kinetochore subunit of Saccharomyces cerevisiae depends on the same factors that promote rapid decay of nonsense mRNAs. Genetics 150:1019-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De La Cruz, J., I. Iost, D. Kressler, and P. Linder. 1997. The p20 and Ded1 proteins have antagonistic roles in eIF4E-dependent translation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94:5201-5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunckley, T., and R. Parker. 1999. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 18:5411-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunckley, T., M. Tucker, and R. Parker. 2001. Two related proteins, Edc1p and Edc2p, stimulate mRNA decapping in Saccharomyces cerevisiae. Genetics 157:27-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051-1060. [DOI] [PubMed] [Google Scholar]
- 14.Gao, Q., B. Das, F. Sherman, and L. Maquat. 2005. Cap-binding protein 1-mediated and eukaryotic translation initiation factor 4E-mediated pioneer rounds of translation in yeast. Proc. Natl. Acad. Sci. USA 102:4258-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grimson, A., S. O'Connor, C. L. Newman, and P. Anderson. 2004. SMG-1 is a phosphatidylinositol kinase-related protein kinase required for nonsense-mediated mRNA decay in Caenorhabditis elegans. Mol. Cell. Biol. 24:7483-7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hann, B. C., and P. Walter. 1991. The signal recognition particle in S. cerevisiae. Cell 67:131-144. [DOI] [PubMed] [Google Scholar]
- 17.He, F., and A. Jacobson. 1995. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev. 9:437-454. [DOI] [PubMed] [Google Scholar]
- 18.He, F., X. Li, P. Spatrick, R. Casillo, S. Dong, and A. Jacobson. 2003. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol. Cell 12:1439-1452. [DOI] [PubMed] [Google Scholar]
- 19.He, F., S. W. Peltz, J. L. Donahue, M. Rosbash, and A. Jacobson. 1993. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1− mutant. Proc. Natl. Acad. Sci. USA 90:7034-7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Izaurralde, E., and S. Adam. 1998. Transport of macromolecules between the nucleus and the cytoplasm. RNA 4:351-364. [PMC free article] [PubMed] [Google Scholar]
- 21.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaufer, N. F., H. M. Fried, W. F. Schwindinger, M. Jasin, and J. R. Warner. 1983. Cycloheximide resistance in yeast: the gene and its protein. Nucleic Acids Res. 11:3123-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kebaara, B., T. Nazarenus, R. Taylor, and A. L. Atkin. 2003. Genetic background affects relative nonsense mRNA accumulation in wild-type and upf mutant yeast strains. Curr. Genet. 43:171-177. [DOI] [PubMed] [Google Scholar]
- 24.Keeling, K., J. Lanier, M. Du, J. Salas-Marco, L. Gao, A. Kaenjak-Anageletti, and D. Bedwell. 2004. Leaky termination at premature stop codons antagonizes nonsense mediated mRNA decay in S. cerevisiae. RNA 10:691-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kshirsagar, M., and R. Parker. 2004. Identification of Edc3p as an enhancer of mRNA decapping in Saccharomyces cerevisiae. Genetics 166:729-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leeds, P., S. W. Peltz, A. Jacobson, and M. R. Culbertson. 1991. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 5:2303-2314. [DOI] [PubMed] [Google Scholar]
- 27.Leeds, P., J. M. Wood, B. S. Lee, and M. R. Culbertson. 1992. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol. Cell. Biol. 12:2165-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lelivelt, M. J., and M. R. Culbertson. 1999. Yeast Upf proteins required for RNA surveillance affect the global expression of the yeast transcriptome. Mol. Cell. Biol. 19:6710-6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lesser, C. F., and C. Guthrie. 1993. Mutational analysis of pre-mRNA splicing in Saccharomyces cerevisiae using a sensitive new reporter gene, CUP1. Genetics 133:851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mader, S., H. Lee, A. Pause, and N. Sonnenberg. 1995. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol. Cell. Biol. 15:4990-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maderazo, A. B., F. He, D. A. Mangus, and A. Jacobson. 2000. Upf1p control of nonsense mRNA translation is regulated by Nmd2p and Upf3p. Mol. Cell. Biol. 20:4591-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitchell, P., and D. Tollervey. 2003. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′→5′ degradation. Mol. Cell 11:1405-1413. [DOI] [PubMed] [Google Scholar]
- 33.Miyasaka, H. 1999. The positive relationship between codon usage bias and translation initiation AUG context in Saccharomyces cerevisiae. Yeast 15:633-637. [DOI] [PubMed] [Google Scholar]
- 34.Morrissey, J. P., J. A. Deardorff, C. Hebron, and A. Sachs. 1999. Decapping of stabilized mRNA in yeast pab1 mutants. Yeast 15:687-702. [DOI] [PubMed] [Google Scholar]
- 35.Muhlrad, D., and R. Parker. 1999. Aberrant mRNAs with extended 3′ UTRs are substrates for rapid degradation by mRNA surveillance. RNA 5:1299-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muhlrad, D., and R. Parker. 1994. Premature translational termination triggers mRNA decapping. Nature 370:578-581. [DOI] [PubMed] [Google Scholar]
- 37.Neville, M., F. Stutz, L. Lee, L. I. Davis, and M. Rosbash. 1997. The importin-β family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr. Biol. 7:767-775. [DOI] [PubMed] [Google Scholar]
- 38.Nguyen, V. T., M. Moragne, and O. Bensuade. 1988. Firefly luciferase luminescence assays using scintillation counters for quantitation in transfected mammalian cells. Anal. Biochem. 171:404-408. [DOI] [PubMed] [Google Scholar]
- 39.Page, M. F., B. Carr, K. R. Anders, A. Grimson, and P. Anderson. 1999. SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p in yeast. Mol. Cell. Biol. 19:5943-5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pulak, R., and P. Anderson. 1993. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 7:1885-1897. [DOI] [PubMed] [Google Scholar]
- 41.Shirley, R. L., A. S. Ford, R. Richards, M. Albertini, and M. R. Culbertson. 2002. Nuclear import of Upf3p is mediated by importin-α/β and export to the cytoplasm is required for a functional nonsense-mediated mRNA decay pathway in yeast. Genetics 161:1465-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shirley, R. L., L. R. Schenkman, M. J. Lelivelt, J. N. Dahlseid, and M. R. Culbertson. 1998. A factor required for nonsense-mediated mRNA decay in yeast is exported from the nucleus to the cytoplasm by a nuclear export signal sequence. J. Cell Sci. 14:3129-3143. [DOI] [PubMed] [Google Scholar]
- 43.Stade, K., C. S. Ford, C. Guthrie, and K. Weis. 1997. Xeportin 1 (Crm1p) is an essential nuclear export factor. Cell 90:1041-1050. [DOI] [PubMed] [Google Scholar]
- 44.Stahl, G., L. Bidou, J. P. Rousset, and M. Cassan. 1995. Versatile vectors to study recoding: conservation of rules between yeast and mammalian cells. Nucleic Acids Res. 23:1557-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takahashi, S., Y. Araki, T. Sakuno, and T. Katada. 2003. Interaction between Ski7p and Upf1p is required for nonsense-mediated 3′-to-5′ mRNA decay in yeast. EMBO J. 22:3951-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tharun, S., and R. Parker. 1999. Analysis of mutations in the yeast mRNA decapping enzyme. Genetics 151:1273-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, et al. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403:623-627. [DOI] [PubMed] [Google Scholar]
- 48.Ursic, D., K. Chinchilla, J. S. Finkel, and M. R. Culbertson. 2004. Multiple protein/protein and protein/RNA interactions suggest roles for yeast DNA/RNA helicase Sen1p in transcription, transcription-coupled DNA repair and RNA processing. Nucleic Acids Res. 32:2441-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, W., K. Czaplinski, Y. Rao, and S. W. Peltz. 2001. The role of Upf proteins in modulating the translation readthrough of nonsense-containing transcripts. EMBO J. 20:880-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Welch, E. M., and A. Jacobson. 1999. An internal open reading frame triggers nonsense-mediated decay of yeast SPT10 mRNA. EMBO J. 18:6134-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou, J., K. Hidaka, and B. Futcher. 2000. The Est1 subunit of yeast telomerase binds the Tlc1 telomerase RNA. Mol. Cell. Biol. 20:1947-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]