Abstract
Copper homeostasis within the cell is established and preserved by different mechanisms. Changes in gene expression constitute a way of maintaining this homeostasis. In Schizosaccharomyces pombe, the Cuf1 transcription factor is critical for the activation of copper transport gene expression under conditions of copper starvation. However, in the presence of elevated intracellular levels of copper, the mechanism of Cuf1 inactivation to turn off gene expression remains unclear. In this study, we provide evidence that inactivation of copper transport gene expression by Cuf1 is achieved through a copper-dependent, cytosolic retention of Cuf1. We identify a minimal nuclear localization sequence (NLS) between amino acids 11 to 53 within the Cuf1 N terminus. Deletion of this region and specific mutation of the Lys13, Arg16, Arg19, Lys24, Arg28, Lys45, Arg47, Arg50, and Arg53 residues to alanine within this putative NLS is sufficient to abrogate nuclear targeting of Cuf1. Under conditions of copper starvation, Cuf1 resides in the nucleus. However, in the presence of excess copper as well as silver ions, Cuf1 is sequestered in the cytoplasm, a process which requires the putative copper binding motif, 328Cys-X-Cys-X3-Cys-X-Cys-X2-Cys-X2-His342 (designated C-rich), within the C-terminal region of Cuf1. Deletion of this region and mutation of the Cys residues within the C-rich motif result in constitutive nuclear localization of Cuf1. By coexpressing the Cuf1 N terminus with its C terminus in trans and by using a two-hybrid assay, we show that these domains physically interact with each other in a copper-dependent manner. We propose a model wherein copper induces conformational changes in Cuf1 that promote a physical interaction between the Cuf1 N terminus and the C-rich motif in the C terminus that masks the NLS. Cuf1 is thereby sequestered in the cytosol under conditions of copper excess, thereby extinguishing copper transport gene expression.
As a donor (Cu1+) and acceptor (Cu2+) of electron, copper is an important catalytic cofactor utilized by a variety of enzymes, making it essential for all living organisms (47, 50). The same chemistry that makes copper indispensable, however, also makes it a potent cytotoxin when present in excess. Due to its proclivity to change redox state within the cell, copper (Cu1+) can react with hydrogen peroxide (H2O2) to produce the highly destructive hydroxyl radical (18). Therefore, copper concentrations must remain within homeostatic boundaries to allow sufficient copper to serve as an electron transfer intermediate for enzymes and yet prevent its accumulation to cytotoxic levels.
In fungi, a common response of cells to fluctuations in copper levels is to reprogram the transcription of genes that are important for its uptake and sequestration (33, 72). Metallosensing regulatory proteins play key roles in controlling the homeostatic processes that regulate cell responses to both essential and toxic levels of copper ions. In general, under conditions of copper deprivation, specialized pathways are positively regulated to allow efficient transport of the metal from the environment or intracellular stores. Conversely, when copper is in excess of physiological requirements, components that function in copper acquisition are down-regulated and protective proteins are up-regulated to safeguard cells against metal toxicity (53).
Studies in Saccharomyces cerevisiae have led to the identification of many components of the copper homeostatic machinery (13, 51). Genetic screens and postgenomic analyses have uncovered genes that are responsible for copper uptake under low environmental copper conditions, copper distribution to appropriate target proteins or compartments, and detoxification upon exposure to high environmental copper levels (35, 52). For high-affinity transport into cells, copper is reduced from Cu2+ to Cu1+ by the Fre cell surface reductases (11, 17, 19, 36). Simultaneously or subsequent to reduction, copper is transported across the plasma membrane via two distinct membrane-associated copper transporters, Ctr1 and Ctr3 (12, 30, 48, 49). Under copper-limiting conditions, mRNA levels of the FRE1/7, CTR1, and CTR3 genes increase, while their expression is repressed under copper-replete conditions (34, 36, 74). The Mac1 transcriptional activator is directly responsible for this copper-dependent regulation (26). It binds to the copper response element (5′-TTTGC(T/G)C(A/G)-3′) located in tandem or inverted repeats at the promoters of its target genes (22, 34, 36). Mac1 resides constitutively in the nucleus (25). Two regions in Mac1, one in the N terminus encompassing residues 70 to 287 and one in the C terminus encompassing residues 289 to 417, are required for its nuclear localization (56). The N-terminal 159 amino acids harbor its DNA binding domain (24). Within this region, the first 40 amino acids display sequence identity to the Ace1 copper-detoxifying factor, which includes a zinc binding domain and a conserved (R/K)GRP sequence motif that are essential for binding to the minor groove of DNA (31, 65). Near its C terminus, Mac1 has two Cys-His repeats (termed C1 and C2, respectively) that contain the metal binding motif Cys-X-Cys-X4-Cys-X-Cys-X2-Cys-X2-His (28, 56). All the dominant mutations thus far identified in MAC1 lie in the C1 motif, suggesting that the two repeats are not functionally redundant with respect to copper sensing (28, 77). The C1 motif has been shown to bind four Cu1+ ions (25). When C1 is disrupted, cells exhibit elevated basal CTR1/3 mRNA levels that are unresponsive to copper. While the C2 motif can also coordinate with four Cu1+ atoms, its partial or complete disruption alters the ability of Mac1 to trans-activate target gene expression, rather than its ability to sense copper (77). Based on these observations, it was proposed that the apo form of Mac1 binds to copper response elements to activate transcription of copper transport genes. At elevated levels, copper induces intramolecular conformational changes, possibly at the C1 motif of Mac1, that inactivate its DNA binding activity and consequently its ability to trans-activate copper transport gene expression (25).
Resistance to copper toxicity in S. cerevisiae is mediated by the Cup1 and Crs5 metallothioneins (MTs), which protect cells by chelating copper, as well as the antioxidant enzyme copper-zinc superoxide dismutase (SOD1) (8-10). The transcription of these copper-detoxifying genes is induced by the presence of excess copper and is regulated by the transcription factor Ace1 (62). Ace1 is constitutively expressed and exists in an inactive form in the nucleus (61). Upon exposure to copper, Ace1 cooperatively binds Cu1+ to form a polycopper cluster through specific Cys residues within its N-terminal DNA-binding domain. Copper binding leads to a conformational switch in this domain that results in specific binding of Ace1 to the metal-regulatory elements, resulting in increased expression of the genes encoding proteins involved in copper sequestration and protection against copper toxicity.
Previously, we have identified a nutritional copper-sensing transcription factor in the fission yeast Schizosaccharomyces pombe named Cuf1 (32). Cuf1 is functionally similar to Mac1 from S. cerevisiae. When copper is scarce, Cuf1 activates the transport of copper at the cell surface and mobilizes vacuolar copper by up-regulating the expression of the Ctr4-Ctr5 two-component copper transporting complex at the plasma membrane and the Ctr6 vacuolar efflux system, respectively (3, 4, 6, 32, 76). Structurally, the similarity between the Cuf1 and Ace1 proteins extends beyond the similarity observed between the Mac1 and Ace1 N-terminal DNA binding domains. This is supported by the finding that Cuf1 can activate the expression of Ace1 target genes when introduced into an ace1 null S. cerevisiae strain (3). Cuf1 binds specifically to DNA sequences similar to those of the Ace1 protein. We previously showed that Cuf1 binds to the consensus sequence 5′-D(T/A)DDHGCTGD-3′ (D = A, G, or T; H = A, C, or T) termed CuSE (copper signaling element) that is closely related to the metal-regulatory element (3). The Cuf1 C terminus harbors a Cys-His element (named C-rich motif) that is absent in Ace1 but found duplicated in Mac1. Mutations that altered all of the five Cys residues of the C-rich motif in Cuf1 generated a protein that is unable to repress target gene expression in response to copper, suggesting that this motif likely serves a copper-sensing function to modulate expression of the copper transport genes (4).
Because Cuf1 is too large to diffuse through the nuclear pore complex, the accumulation of Cuf1 in the nucleus must be an active facilitated process. Proteins destined for the nucleus contain a nuclear localization sequence (NLS) that is recognized by importin β receptors (37, 71). NLSs are broadly divided into classical and nonclassical types (23). The former requires an adapter molecule (importin α) to bridge the interaction between the cargo protein and the importin β receptor, while the latter involves direct binding of the importin β receptor to its cargo protein (21). A number of transcription factors have been shown to be regulated by mechanisms that affect their nucleocytoplasmic transport. Examples include the mammalian transcription factors NF-κB (2) and NF-AT (59) and the sterol-regulatory binding proteins (70) as well as the budding yeast transcription factors Swi5 (40) and Pho4 (45). In all these cases, the regulation appears to be mediated by relocalization of the transcriptional activator, either by the masking or unmasking of an NLS or by the disruption of protein-protein interactions that serve to anchor the regulatory protein in the cytoplasmic compartment.
In this study, we characterize the NLS of Cuf1 and show that a functional Cuf1-green fluorescent protein (GFP) is localized in the nucleus in the presence of only low copper concentrations. When cells were grown in the presence of excess copper, Cuf1-GFP is retained in the cytoplasm. More importantly, we demonstrate that the cytoplasmic accumulation of Cuf1 results from a copper-mediated intramolecular interaction between the N- and C-terminal regions of Cuf1.
MATERIALS AND METHODS
Yeast strains and growth conditions.
The S. pombe strains used in this study were the wild-type FY435 (h+ his7-366 leu1-32 ura4-Δ18 ade6-M210) (7) and the cuf1Δ disruption mutants JSY17 (h+ his7-366 leu1-32 ura4-Δ18 ade6-M210 cuf1Δ::ura4+) and JSY8 (h+ his7-366 leu1-32 ura4-Δ18 ade6-M210 cuf1Δ::hisG). The cuf1Δ strain JSY8 was created by replacing the coding region of cuf1+ with a hisG-ura4+-hisG cassette through homologous recombination as described previously (38). Under nonselective conditions, S. pombe cells were grown in a yeast extract plus supplements (YES) medium that contains 3% glucose (5). The nonfermentable carbon source medium YES-glycerol was prepared by replacing the glucose in YES with 3% glycerol. When plasmid maintenance was required, cells were grown in Edinburgh minimal medium (EMM) (1) with the appropriate amino acids (225 mg/liter adenine, histidine, and uracil, unless otherwise stated); unsupplemented EMM contains 160 nM copper. When the wild-type or mutant cuf1 alleles were expressed under the control of the nmt1+ promoter, cells expressing these alleles were induced by the removal of thiamine from the medium. In contrast, to prevent expression of the cuf1 alleles, cells were grown in the presence of 5 μM thiamine. The S. cerevisiae strain L40 (Mata his3Δ200 trp1-901 leu2-3, 112 ade2 LYS2::(lexAop)4-HIS3 URA3::(lexAop)8-lacZ) (69) was used for two-hybrid analysis. S. cerevisiae cells were grown either in yeast extract/peptone/dextrose medium or in the appropriate drop-out synthetic medium (58).
Plasmids.
The gfp gene derived from pSF1-GP1 (29) was isolated by PCR by using primers designed to generate XmaI and SstI sites at the 5′ and 3′ termini of the open reading frame. The resulting DNA fragment was used to fuse the gfp gene in frame with the cuf1+ open reading frame, in which XmaI and SstI restriction sites were previously engineered by PCR and placed immediately before the stop codon. The pSP1 (3) derivative was denoted pSP1JBcuf1+-GFP. Subsequently, the S. pombe cuf1+ promoter up to position −2283 from the start codon of the cuf1+ gene was isolated by PCR from the S. pombe FY435 genomic DNA. Once generated, the DNA fragment was digested with ApaI and PstI and the fragment containing the cuf1+ promoter was cloned immediately upstream of the cuf1+-GFP fusion gene, creating the pSP1JB-2283cuf1+-GFP plasmid. Using primers designed to introduce PstI and SstI restriction sites at the start and stop regions of the gfp open reading frame, gfp was reisolated by PCR. Subsequently, this PstI-SstI DNA fragment was exchanged with the PstI-SstI DNA fragment in plasmid pSP1JB-2283cuf1+-GFP to generate pSP1JB-2283GFP, which expresses the GFP alone under the control of the cuf1+ promoter. The wild-type cuf1+ codons 1 to 20, 1 to 30, 1 to 40, 1 to 53, 1 to 61, 11 to 53, 21 to 53, 62 to 410, and 71 to 410 were isolated by PCR using primers that contained PstI and XmaI sites. The PCR products obtained were digested with PstI and XmaI and swapped into the corresponding sites of pSP1JB-2283cuf1+-GFP, generating a series of plasmids bearing deletions within different regions of cuf1+. These plasmids were termed pSP1JB-2283cuf1+1-20-GFP, pSP1JB-2283cuf1+1-30-GFP, pSP1JB-2283cuf1+1-40-GFP, pSP1JB-2283cuf1+1-53-GFP, pSP1JB-2283cuf1+1-61-GFP, pSP1JB-2283cuf1+11-53-GFP, pSP1JB-2283cuf1+21-53-GFP, pSP1JB-2283cuf1+62-410-GFP, and pSP1JB-2283cuf1+71-410-GFP. Importantly, for all of these constructs except the last two, a second copy of the gfp gene was cloned into the XmaI and SstI sites to generate recombinant plasmids containing tandemly repeated GFP coding sequences. The pSP1JB-2283cuf1Δ11-46-GFP plasmid was created by PCR using primers that contained PstI and XmaI sites. The PCR product was obtained from the cuf1Δ11-46 allele (4) and exchanged with the PstI-XmaI DNA fragment in plasmid pSP1JB-2283cuf1+-GFP. When the cuf1+ gene was not expressed from its own promoter, the regulatable nmt1+ promoter system was used for cuf1+ function analysis. The nmt1+ promoter region from position −1178 to position −1 with respect to the A of the initiator codon was isolated from pREP3X (14) by PCR. This PCR product was purified and subcloned to replace the cuf1+ promoter by using the ApaI and PstI restriction sites. To test whether the Cuf1 C-rich motif can act in trans with the N-terminal portion of Cuf1, we coexpressed 1Cuf161-GFP and 62Cuf1410-FLAG2 as separate molecules. Under the control of a nmt1+ promoter, an ApaI-SacII PCR-amplified fragment containing the first 61 codons of the cuf1+ open reading frame fused to the entire gfp open reading frame was isolated from the pSP1JB-1178cuf1+1-61-GFP plasmid. The ApaI-SacII DNA fragment was inserted into the ApaI-SacII-cut pJK210 plasmid (27). The resulting plasmid, denoted pJK210JB-1178cuf1+1-61-GFP, was then used to integrate, in single copy, the 1cuf161-GFP gene at the ura4+ locus of the JSY8 strain. To integrate the fusion gene into the genome by homologous recombination, we digested the plasmid with StuI. Likewise, an ApaI-SacII PCR-amplified fragment containing the last 350 codons of cuf1+ fused to FLAG2 was isolated from the pSP1Cuf1-FLAG2 plasmid (4). The purified DNA fragment was cloned into pJR1-3XL (41) to create pJR1-cuf1+62-410-FLAG2.
Two-hybrid analysis.
To examine if the Cuf1 C terminus can interact with its N-terminal region, we constructed bait and prey plasmids as follows. The lexA gene of Escherichia coli was isolated from pLex-a (68) by PCR using primers that contained PstI and SalI restriction sites. The fragment was digested and cloned into the p424GPD vector (44). The resulting plasmid, p424GPDLexA, was digested with SpeI and PstI and a XbaI-PstI DNA containing the synthetic codons of SV40 NLS optimized for yeast (S. cerevisiae) was cloned into the SpeI (which produces compatible ends to that of XbaI) and PstI sites of the p424GPDLexA vector. The resulting bait plasmid was named p424GPD-NLexA. The VP16 acidic activation domain was obtained by PCR using the plasmid pVP16 (20) as a template. The purified fragment was digested with PstI and SalI and subsequently cloned into the corresponding sites in plasmid p425GPD (44). As described above, an XbaI-PstI DNA fragment containing the synthetic codons of SV40 NLS was placed upstream of and in frame to the VP16 gene, creating the p425GPD-NVP16 plasmid. The p424GPD1Cuf161-NLexA plasmid was created by subcloning a BamHI-PstI DNA fragment (containing the first 61 codons of cuf1+) into vector p424GPD-NLexA. To construct a prey plasmid that has the last 350 codons of cuf1+ fused to VP16, BamHI and PstI sites were created by PCR mutagenesis at the termini of the upstream and downstream DNA fragment, and used to insert the fragment into p425GPD-NVP16 vector. The L40 strain was used as the recipient for the indicated bait and prey plasmids. Cells were grown in low-copper synthetic media (34) to mid-logarithmic phase (A600 of ∼1.0) and then treated with the addition of either 100 μM bathocuproinedisulfonic acid (BCS) or 100 μM CuSO4. After a 1-h incubation at 30°C, cells were harvested, washed with sterile water, and resuspend in 700 μl of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, pH 7.0, 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol). The cells were permeabilized by adding 50 μl of chloroform and 50 μl of 0.1% sodium dodecyl sulfate. After the cell suspension was vortex-mixed for 10 s, 200 μl of 4 mg/ml o-nitrophenyl-β-d-galactopyranoside was added to each sample. Following a 10-min incubation at 30°C, 350 μl of 1 M Na2CO3 was added to stop the reactions. After clarification by centrifugation at 4°C, the A420 was measured within the linear response range and expressed in standard units (39). Values shown are the average of triplicate assays of three independent transformants.
Fluorescence microscopy and biochemical techniques.
For localization of Cuf1, JSY17 or JSY8 cells that were transformed with plasmids harboring the cuf1+-GFP fusion gene or its mutant derivatives were grown in liquid EMM plus 225 mg/liter adenine, histidine, and uracil and 5 μM thiamine, unless otherwise stated. Cells were grown to A600 of ∼1.0. At this mid-logarithmic phase, cells were washed twice to remove thiamine and diluted 100-fold in EMM with adenine, histidine, and uracil. At this point, cells were grown in the presence of CuSO4 (100 μM), AgNO3 (2 μM), ZnSO4 (1 mM), FeCl3 (100 μM), CdCl2 (25 μM), or BCS (100 μM). After 4 h treatment, direct fluorescence microscopy was carried out as described previously (46), except that 4′,6′-diamidino-2-phenylindole (DAPI) (Sigma; D-8417) was used for DNA staining instead of Hoechst 33342. Cell fields shown in this study are representative of experiments repeated at least five times. Procedures for RNase protection assays have been described previously (4). For Western blotting experiments, the following primary antisera were used: monoclonal anti-GFP antibody B-2 (Santa Cruz Biotechnology, Santa Cruz, CA), monoclonal anti-FLAG antibody M2 (Sigma, Saint-Louis, MO), monoclonal anti-PCNA antibody PC10 (Sigma, St. Louis, MO), monoclonal anti-LexA antibody 2-12, monoclonal anti-VP16 antibody 1-21 (both from Santa Cruz Biotechnology, Santa Cruz, CA), and monoclonal anti-PGK antibody (Molecular Probes, Eugene, OR). After a 2-h incubation with the appropriate primary antibody, membranes were washed in Tris-buffered saline (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% bovine serum albumin) and incubated for 1 h with the sheep anti-mouse immunoglobulin G-horseradish peroxidase-conjugated secondary antibody (Amersham Biosciences, Uppsala, Sweden). The proteins were detected by chemiluminescence (Perkin Elmer Life Sciences, Boston, MA).
RESULTS
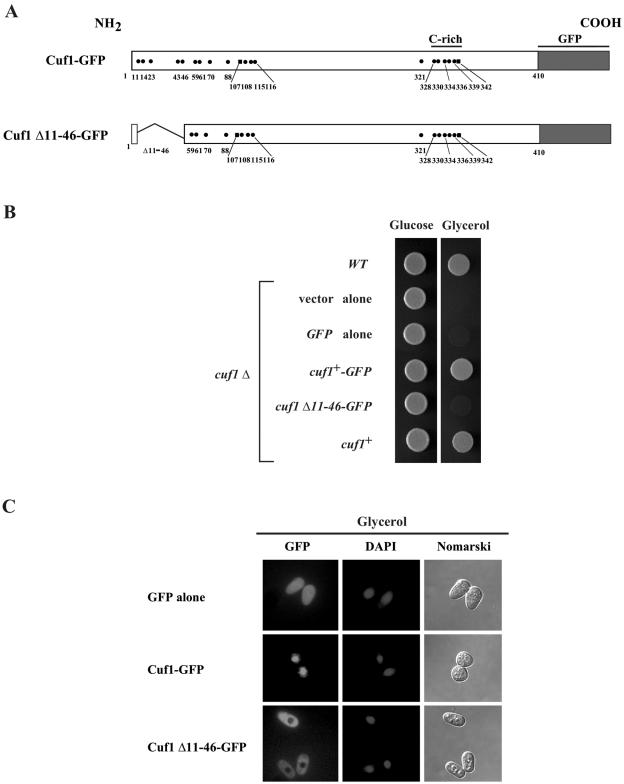
The N-terminal region of Cuf1 from residues 11 to 46 is required for its nuclear localization.
We have previously shown that the deletion of 36 amino acids from the N-terminal region of Cuf1, including residues 11 to 46 (Cuf1Δ11-46), results in the inactivation of Cuf1 (4). To determine the role of these amino acids in Cuf1 function, we analyzed the intracellular location of the Cuf1Δ11-46 mutant by fluorescence microscopy (Fig. 1). To facilitate this analysis, a cuf1+-GFP fusion gene was constructed, transformed, and expressed in a cuf1Δ strain. As shown in Fig. 1B, a plasmid-borne copy expressing the Cuf1-GFP fusion protein fully complements the cuf1Δ growth defect in glycerol. To analyze the Cuf1Δ11-46-GFP mutant protein, a plasmid expressing its corresponding allele was transformed into a cuf1Δ strain and tested for its ability to confer growth in glycerol compared to that of cuf1+-GFP, cuf1+, GFP alone, or the empty vector. Cells expressing Cuf1Δ11-46-GFP were unable to grow on a respiratory carbon source (Fig. 1B). Furthermore, when Cuf1Δ11-46-GFP was expressed, most of the mutant protein was localized in the cytoplasm of cells (Fig. 1C). In contrast, Cuf1-GFP, which functionally complements the respiratory deficiency of a cuf1Δ strain in a manner indistinguishable from that of the Cuf1 wild-type protein, was localized in the nucleus (Fig. 1C). Presumably due to its small size (∼27 kDa), GFP alone diffuses across the nuclear envelope and its localization was seen throughout cytoplasm and nucleus (Fig. 1C). These results suggest that the N-terminal residues 11 to 46 of Cuf1 contain amino acids that are required for Cuf1 nuclear localization. Accumulation of Cuf1Δ11-46-GFP in the cytoplasm may result from the loss of a putative NLS contained within these residues and consequently inhibit its import into the nucleus.
FIG. 1.
The N-terminal region of Cuf1 between residues 11 to 46 is required for nuclear localization when cells are grown in a nonfermentable carbon source. (A) Schematic representation of the Cuf1 and Cuf1 Δ11-46 fused to the green fluorescent protein. The dots and squares depict positions of cysteine and histidine residues, respectively. (B) The Cuf1-GFP fusion protein complements the cuf1Δ growth defect on a nonfermentable carbon source (glycerol) in a manner indistinguishable from the unadulterated Cuf1 protein. Cultures grown to identical optical densities were spotted (3,000 cells/5 μl) onto YES-glucose and YES-glycerol media and incubated at 30°C for 3 and 8 days, respectively. (C) Shown are representative cells expressing GFP alone, Cuf1-GFP, and Cuf1 Δ11-46-GFP proteins, respectively. Cells were grown in respiratory growth media to exponential phase and visualized by fluorescence microscopy. DAPI staining visualized nuclear DNA and Nomarski microscopy was used to examine cell morphology.
Cuf1 11-53 contains a noncanonical NLS with several basic amino acid residues.
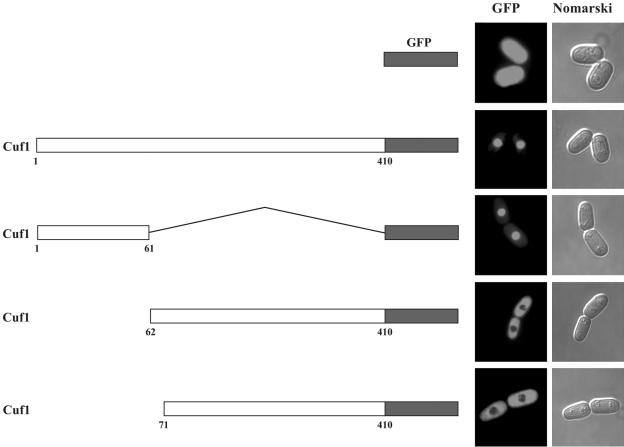
The N terminus of Cuf1 harbors stretches of basic amino acid residues characteristic of sequences found in nuclear localization signals. Together with the observation that Cuf1Δ11-46-GFP was predominantly localized in the cytoplasm, we ascertained the ability of the Cuf1 N-terminal region, designated 1Cuf161, to direct nuclear localization when fused to a cytoplasmic GFP protein. We created three chimeric proteins. The first contained 1Cuf161 fused to GFP, while the second and third chimeric proteins harbored the last 349 and 340 amino acids of Cuf1 fused to GFP, designated 62Cuf1410-GFP and 71Cuf1410-GFP, respectively. As shown in Fig. 2, fusion of 1Cuf161 to GFP triggered nuclear targeting of the chimeric protein. In contrast, 62Cuf1410-GFP and 71Cuf1410-GFP, which contained fusions of the C-terminal regions of Cuf1 to GFP, were mainly localized in the cytoplasm (Fig. 2). Under the same conditions, GFP alone was cytoplasmic as well as nuclear. These results suggest that the N-terminal 61-amino-acid region of Cuf1 harbors a nuclear localization sequence.
FIG. 2.
The chimeric protein bearing the N-terminal 61-amino-acid segment of Cuf1 fused to GFP localizes to the nucleus. JSY17 cells were transformed with the indicated constructs and grown in EMM containing 100 μM BCS. The left panel shows a schematic representation of the GFP alone, wild-type Cuf1, 1Cuf161, 62Cuf1410, and 71Cuf1410 fused to the green fluorescent protein. The amino acid sequence of the Cuf1-GFP fusion protein is numbered relative to its initiator codon. Fluorescence microscopy (middle and right panels) was used to visualize the cellular location of GFP fusions and GFP alone, and cell morphology was examined using Nomarski optics.
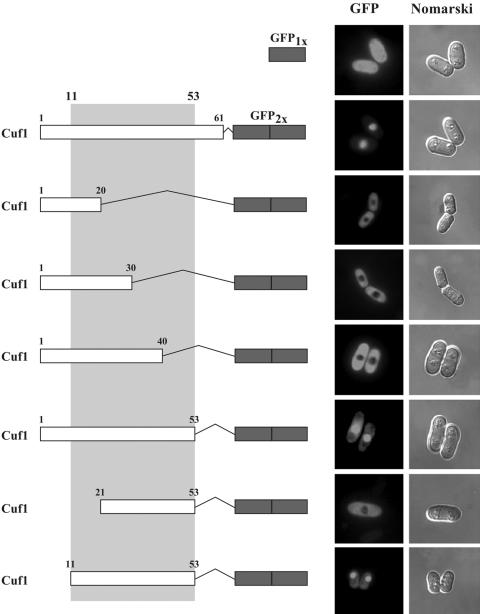
To delineate the specific region within 1Cuf161 that directs nuclear localization, we generated a series of N- and C-terminal deletions within this region (Fig. 3) and fused two copies of the GFP protein (GFP2x) to the C terminus of each truncated protein. The tandem duplication of GFP protein is suitable for sufficiency experiments (54) when fused to small peptides (Fig. 3). Plasmids expressing the truncated proteins fused to GFP2x, shown in Fig. 3, were transformed into a cuf1Δ strain of S. pombe and analyzed by fluorescence microscopy to identify which mutants localized to the nucleus. The results show that 1Cuf120-GFP2x, 1Cuf130-GFP2x, and 1Cuf140-GFP2x, which contain deletions of the last 41, 31, and 21 amino acids, respectively, in 1Cuf161 were exclusively cytosolic (Fig. 3). However, 1Cuf153-GFP2x, which was truncated at residue 53, was efficiently targeted to the nucleus (Fig. 3). We then generated N-terminal deletions of 1Cuf153-GFP2x. The deletion of amino acids 1 to 10 from the N terminus to generate 11Cuf153-GFP2x did not affect nuclear localization of the fusion protein. However, further deletion of 10 amino acids in 21Cuf153-GFP2x abrogated its ability to localize in the nucleus. Together, these data reveal that the N-terminal segment of Cuf1 from amino acids 11 to 53 constitutes a minimal region which is sufficient for nuclear targeting.
FIG. 3.
Minimal region in 1Cuf161 required for translocation to the nucleus. The left panel shows schematic illustration of 1Cuf161 and its N-terminal and C-terminal deletions fused to tandemly repeated copies of the GFP coding sequence (GFP2x). The middle panel represents subcellular locations of the fusion proteins determined by fluorescence microscopy. The right panel represents cells viewed by Nomarski microscopy to examine cell morphology. A single copy of the GFP protein (GFP1x) was expressed from the cuf1+ promoter as a control for GFP localization. Cells were grown in EMM containing 100 μM BCS.
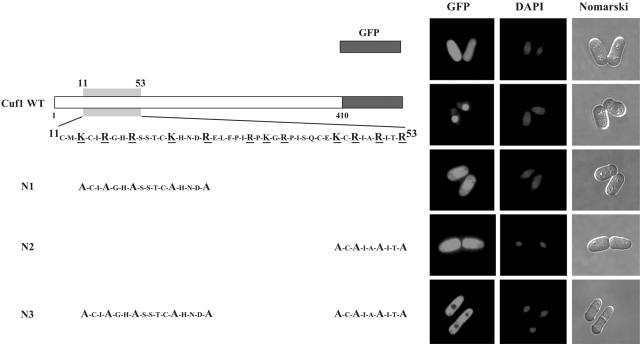
To identify the critical amino acids that constitute the NLS in Cuf1, we converted five positively charged amino acids found in 11Cuf153, Lys13, Arg16, Arg19, Lys24, and Arg28, to Ala in full-length Cuf1 to generate Cuf1-N1. We also examined the effect of mutating Lys45, Arg47, Arg50, and Arg53 to Ala on the ability of Cuf1 to localize to the nucleus. This mutant was designated Cuf1-N2. Furthermore, we combined the mutated residues within Cuf1-N2 with the Lys13→Ala, Arg16→Ala, Arg19→Ala, Lys24→Ala, and Arg28→Ala mutations in Cuf1-N1 to generate the Cuf1-N3 mutant (Fig. 4). Analysis by fluorescence microscopy showed that cells harboring the mutant cuf1-N1 and cuf1-N2 alleles appeared to have less nuclear accumulation compared to that of the wild-type protein. Furthermore, like GFP alone, these Cuf1-N1 and -N2 mutant proteins were also present throughout the cells, suggesting that they are localized to both the cytosol and nucleus (Fig. 4). On the other hand, cells harboring Cuf1-N3 exhibited no obvious nuclear accumulation. Taken together, these data suggest that the basic residues Lys13, Arg16, Arg19, Lys24, Arg28, Lys45, Arg47, Arg50, and Arg53 or at least some subset of these residues is important for targeting Cuf1 to the nucleus.
FIG. 4.
Cuf1 11-53 contains basic amino acid residues that are required for Cuf1 nuclear import. The left side shows a schematic representation of mutant versions of the Cuf1-GFP fusion protein that were expressed in a cuf1Δ mutant strain of S. pombe under copper-limiting conditions (100 μM BCS). The substitutions in N1, N2, and N3 mutants corresponding to the residues in the wild-type (WT) Cuf1 protein are indicated. (Right side) Cells were visualized for GFP by fluorescence microscopy. DAPI was used to stain nuclei, and Nomarski optics were utilized to monitor cell morphology. JSY17 cells transformed with GFP alone are shown as a control.
Elevated copper levels result in cytoplasmic localization of Cuf1-GFP, while low-copper conditions direct the nuclear localization of Cuf1-GFP.
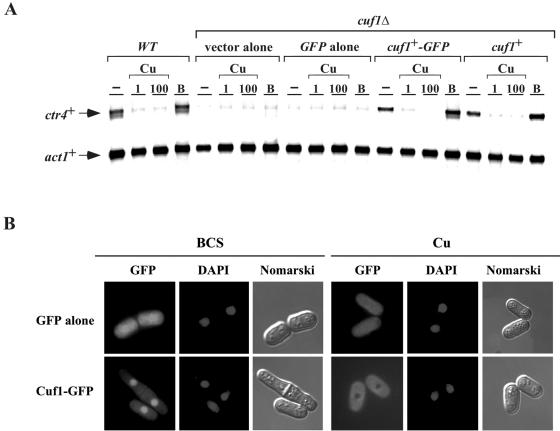
In S. pombe, the genes encoding proteins that are members of the copper transporter family are regulated at the level of gene transcription. These genes are induced under conditions of copper starvation and turned off under conditions of copper repletion. This copper-regulated gene expression is dependent on a functional cuf1+ gene. As shown in Fig. 5A, ctr4+ mRNA levels in a cuf1Δ strain expressing the wild-type Cuf1 protein was down-regulated (approximately fivefold) in the presence of both low (1 μM) and elevated (100 μM) copper concentrations. Conversely, in the presence of 100 μM BCS, a copper chelator, ctr4+ mRNA levels were induced (approximately threefold) over basal levels. In the absence of Cuf1, ctr4+ mRNA was barely detectable under all conditions. Importantly, copper-dependent regulation of ctr4+ gene expression was restored by returning a cuf1+-GFP fusion allele expressed from its own promoter, indicating that the fusion retained wild-type function (Fig. 5A). In previous studies, we demonstrated that the cuf1+ mRNA and protein levels are stable and unaffected by copper deprivation or repletion (4, 32). To further investigate the mechanism by which Cuf1 activity is regulated, we examined the localization of Cuf1 in response to changes in copper levels. As shown in Fig. 5B, under conditions of copper starvation in the presence of BCS, Cuf1-GFP localizes to the nucleus. Consistently, Cuf1-GFP fluorescence colocalized with the DNA-staining dye DAPI, which was used as a marker for nuclear staining. However, when cells were grown in the presence of exogenous copper, Cuf1-GFP is exclusively localized in the cytoplasm of cells (Fig. 5B). Furthermore, the copper-dependent cytoplasmic sequestration of Cuf1 was also observed when a cuf1+-GFP fusion allele was returned in cuf1Δ cells by integration (J. Beaudoin and S. Labbé, unpublished data). When minimal medium without copper or BCS was used, we observed that Cuf1-GFP localizes to the nucleus in a proportion of ∼75%, while ∼25% of the fusion protein is present in the cytoplasm (Beaudoin and Labbé, unpublished). Therefore, the copper-dependent cytoplasmic sequestration of Cuf1 may serve as an important mechanism to abrogate the expression of copper transport genes under conditions of copper excess, thereby preventing copper toxicity.
FIG. 5.
Effect of copper on the subcellular localization of a functional Cuf1-GFP fusion protein. (A) An S. pombe strain bearing a disrupted cuf1Δ allele was transformed with an empty vector (vector alone), GFP alone, a functional cuf1+-GFP allele, or a wild-type (WT) cuf1+ allele. Total RNA from control (−), CuSO4 (1 and 100 μM), or BCS (B) (100 μM) cultures was isolated. Shown is a representative RNase protection assay of ctr4+ and act1+ mRNA steady-state levels. Wild-type strain FY435 (cuf1+) was used as a control. (B) Localization of a functional Cuf1-GFP fusion protein in S. pombe. Cells expressing the Cuf1-GFP fusion protein were grown to mid-logarithmic phase and then treated with either BCS (100 μM) or CuSO4 (Cu) (100 μM) for 4 h (GFP). Cells with green fluorescent proteins were treated with DAPI for nuclear DNA staining (DAPI). Nomarski optics was used to examine cell morphology (Nomarski). Cells transformed with GFP alone distributed throughout the cells (cytosol and nucleus) are shown as control.
Copper- and silver-induced cytoplasmic accumulation of Cuf1.
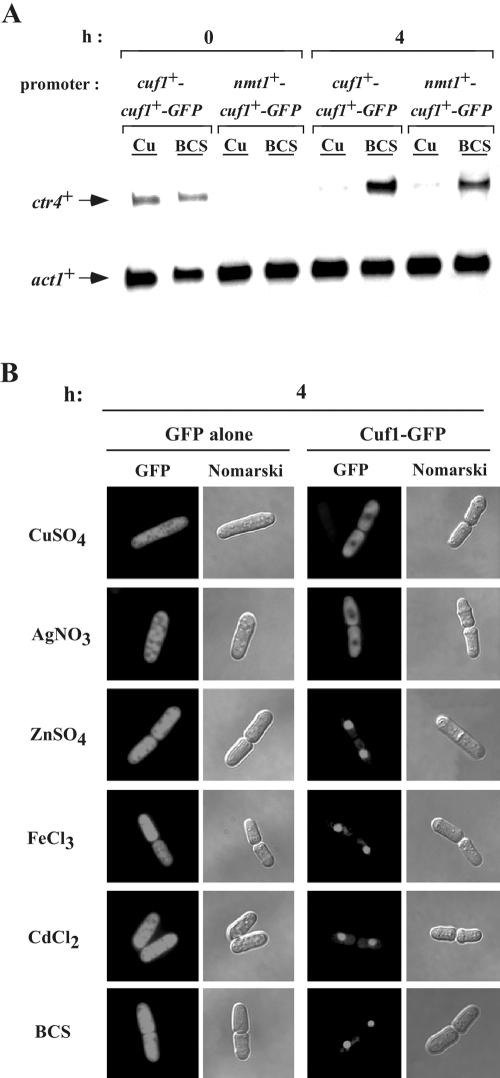
To determine the specificity of the cytoplasmic sequestration of Cuf1 following exposure of cells to a variety of metals, the cuf1+-GFP allele was expressed under the control of the thiamine-regulatable promoter. This system allowed us to induce the synthesis of Cuf1-GFP in the presence of well-defined metal ion concentrations in the media. First, we ascertained whether Cuf1-GFP can be expressed from this inducible expression system without affecting its function. After induction for 4 h, we analyzed the ability of Cuf1-GFP to activate and direct copper-dependent regulation of copper transporter gene expression. As shown in Fig. 6A, we analyzed ctr4+ steady-state mRNA levels over time in response to either copper repletion or starvation. Prior to nmt1+ induction at the zero time point, only basal levels of ctr4+ mRNA were observed in cells expressing cuf1+-GFP under the control of its native promoter (cuf1+ promoter-cuf1+-GFP). ctr4+ mRNA was not seen in the cells expressing cuf1+-GFP under the control of the nmt1+ promoter. After induction and treatment for 4 h with either 100 μM CuSO4 or 100 μM BCS, the ctr4+ mRNA levels in a strain expressing the cuf1+-GFP gene under the control of the nmt1+ promoter were regulated in a copper-dependent manner similar to that of cuf1+-GFP under its own promoter: repressed by copper and induced by copper deprivation. Furthermore, the nmt1+ 41X promoter gave regulatable levels of ctr4+ mRNA comparable to those observed with the strongest nmt1+ 3X promoter (Beaudoin and Labbé, unpublished). Based on these results, we utilized the nmt1+ inducible promoter system to examine the abilities of other heavy metals with chemical properties similar to those of copper to induce the cytoplasmic accumulation of Cuf1-GFP. The metal concentrations used were those which allowed 100% survival of S. pombe cells. Among the five different metal ions tested, CuSO4 (100 μM), AgNO3 (2 μM), ZnSO4 (1 mM), FeCl3 (100 μM), and CdCl2 (25 μM), only copper and silver, a metal that is electronically analogous to the reduced form of Cu2+, triggered the cytoplasmic sequestration of Cuf1-GFP (Fig. 6B). For each metal tested, increasing the incubation times up to 8 h or using concentrations which allowed 50% cell survival did not alter the Cuf1-GFP localization patterns from those shown in Fig. 6B (data not shown). Furthermore, we found that neither low nor high doses of hydrogen peroxide trigger the cytoplasmic retention of Cuf1-GFP (data not shown). Under all the metal conditions examined, GFP alone was localized to the cytosol and nucleus. The ability of silver ions to efficiently sequester Cuf1-GFP in the cytoplasm and the electronic similarity of Ag1+ to Cu1+ but not Cu2+ suggest that cuprous [Cu1+] might be the active form of copper that is required for cytosolic retention of Cuf1-GFP.
FIG. 6.
Metal specificity of cytoplasmic sequestration of Cuf1-GFP. (A) A cuf1Δ strain of S. pombe which was transformed with either pSP1JB-2283cuf1+-GFP or pJBnmt1+-cuf1+-GFP was grown to mid-logarithmic phase in EMM containing 5 μM thiamine. After two washes, the cells were incubated during the indicated time (0 and 4 h) in EMM without thiamine and in the presence of CuSO4 (Cu) (100 μM) or BCS (100 μM). Total RNA was prepared and analyzed from culture aliquots. Arrows indicate signals corresponding to ctr4+ and act1+ mRNA steady-state levels. Results shown are representative of three independent experiments. (B) Strain JSY17 expressing Cuf1-GFP was grown to mid-logarithmic phase in the presence of 5 μM thiamine. Cells were washed twice and then incubated in the presence of CuSO4 (100 μM), AgNO3 (2 μM), ZnSO4 (1 mM), FeCl3 (100 μM), CdCl2 (25 μM), or BCS (100 μM). After 4 h treatment, the full-length Cuf1-GFP protein was viewed by direct fluorescence microscopy (GFP). GFP alone expressed under the control of the nmt1+ promoter was also examined by direct fluorescence. Corresponding Nomarski images are shown after each GFP panel.
Cuf1-GFP requires the C-rich motif for copper-induced cytoplasmic sequestration.
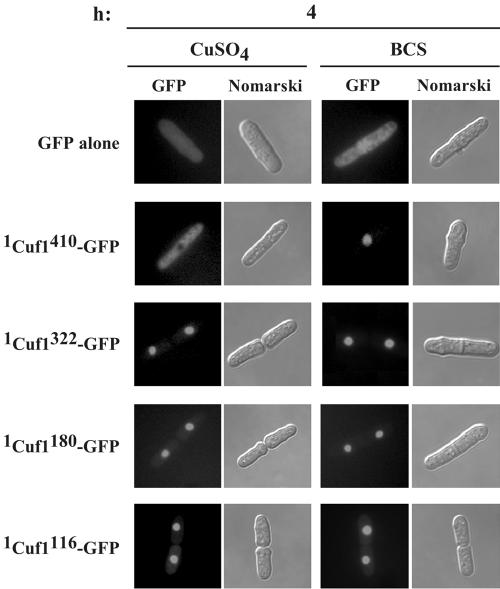
We have previously determined that the C-rich motif (residues 328 to 342) within the C-terminal region of Cuf1 constitutes a copper-sensing module that serves to inactivate Cuf1 function when cells are grown under copper-replete conditions (4). In the present study, our results reveal that a NLS exists within the N-terminal region of Cuf1. We hypothesized that metallation of Cuf1, possibly within the C-rich motif, induces conformational changes that mask the Cuf1 NLS and consequently block Cuf1 nuclear import. To test this hypothesis, we created a series of C-terminal deletions aimed at identifying the regions required for copper-dependent masking of the NLS and fused the truncated proteins to GFP. All mutant alleles were expressed under the control of the inducible nmt1+ promoter and transformed into a cuf1Δ strain. Analysis of transformed cells after induction showed that truncation of the Cuf1 C-terminal region at amino acid residue 322, which eliminates the 328Cys-X-Cys-X3-Cys-X-Cys-X2-Cys-X2-His342 motif, produced a mutant protein that was efficiently imported into the nucleus under both low and high copper concentrations (Fig. 7). When further deletions up to amino acid residues 180 or 116 were created, cells expressing these mutants and analyzed by fluorescence microscopy revealed that, like 1Cuf1322-GFP, these mutant proteins were localized exclusively in the nucleus. Furthermore, their localization was unaffected by changes in intracellular copper levels (Fig. 7). It should be noted that even though the high-affinity copper transport machinery is possibly inactive, copper ions (100 μM) are transported via the low-affinity copper transport system. Therefore, the nuclear localization of mutant proteins is not due to the lack of copper assimilation in cells with cuf1Δ alleles.
FIG. 7.
C terminus of Cuf1 is required for copper-dependent cytoplasmic localization. The cuf1Δ strain of S. pombe JSY17 was transformed with plasmids that encode GFP, 1Cuf1410-GFP, 1Cuf1322-GFP, 1Cuf1180-GFP, or 1Cuf1116-GFP proteins. Cells were grown to mid-logarithmic phase in medium supplemented with 5 μM thiamine. Thiamine was withdrawn from cell cultures, and then cells were grown for 4 h in CuSO4 (100 μM) or BCS (100 μM). Cells were analyzed by fluorescence microscopy for GFP (GFP). Cells were also examined by Nomarski microscopy for cell morphology (Nomarski).
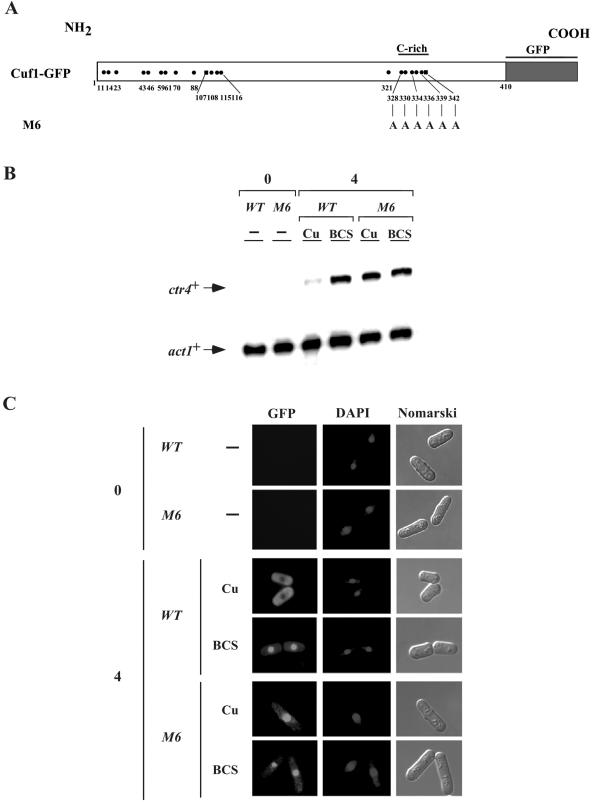
To further investigate the role of the C-rich motif in the copper-dependent cytoplasmic localization of Cuf1, we created a mutant allele, termed cuf1-M6-GFP, in which all of the five Cys residues as well as His342 were mutated to alanines (Fig. 8A). Like the wild-type cuf1+-GFP allele, the mutant allele was expressed under the control of the nmt1+ promoter (Fig. 8B). As shown in Fig. 8C, after 4 h of induction in the presence of exogenous copper, the wild-type Cuf1-GFP fusion protein was localized in the cytosol. Consistent with its absence from the nucleus, virtually no expression of ctr4+ mRNA was observed under these conditions (Fig. 8B). Under the conditions of copper starvation, wild-type Cuf1 was localized in the nucleus and a concomitant high level of ctr4+ mRNA expression was observed (Fig. 8B and C). In contrast, upon induction, the Cuf1-M6-GFP mutant protein was localized within the nucleus under both copper-replete and copper starvation conditions (Fig. 8C). Analysis of ctr4+ mRNA levels in a strain expressing the M6 mutant protein showed an elevated and sustained expression of ctr4+ mRNA even in the presence of high copper concentrations, consistent with its presence in the nucleus (Fig. 8B and C). Furthermore, cells expressing the cuf1-M6-GFP allele exhibited high basal levels of ctr5+ and ctr6+ mRNAs and a lack of copper-dependent repression of ctr5+ and ctr6+ (Beaudoin and Labbé, unpublished). Taken together, these data indicate that the presence of the C-rich motif in wild-type Cuf1-GFP is responsible for determining its cytoplasmic localization in the presence of exogenous copper. Removing the C-rich motif, as in Cuf1-M6-GFP, possibly unmasks the NLS, resulting in its constitutive localization in the nucleus, leading to constitutive elevated levels of ctr4+ mRNA.
FIG. 8.
Cuf1-GFP requires the 328Cys-X-Cys-X3-Cys-X-Cys-X2-Cys-X2-His342 motif within its C-terminal region for copper-dependent cytoplasmic localization. (A) Schematic representation of the Cuf1-GFP and Cuf1-M6-GFP proteins. The mutations in Cuf1-M6-GFP corresponding to the Cys and His residues in the wild-type Cuf1-GFP protein are depicted. (B) cuf1Δ cells from S. pombe, transformed with the wild-type (WT) cuf1+-GFP or cuf1-M6-GFP as indicated, were grown in selective media in the presence of thiamine (5 μM) to mid-logarithmic phase. The cultures were washed twice to remove thiamine, and Cuf1-GFP or Cuf1-M6-GFP expression was induced. Total RNA was prepared from culture aliquots after 0 or 4 h induction. Where indicated, cells were untreated (−) or treated with CuSO4 (Cu) (100 μM) or BCS (100 μM). ctr4+ and act1+ mRNA levels (arrows) were detected using a RNase protection assay. Data illustrated are representative of three independent experiments. (C) Aliquots of cultures described above for panel B were analyzed by microscopy. GFP fluorescence (left panels), DAPI staining (middle panels), and Nomarski phase contrast (right panels) images of cells expressing Cuf1-GFP or Cuf1-M6-GFP from the nmt1+ promoter are shown.
The association between the N and C termini of Cuf1 inhibits its nuclear import.
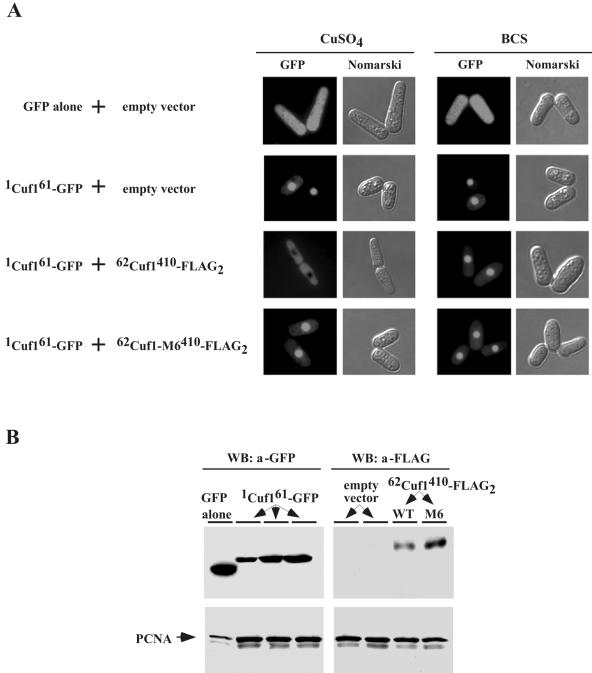
To further test our hypothetical model, we examined the possibility that the Cuf1 C terminus containing the C-rich motif physically interacts with the N terminus harboring the NLS to inhibit its nuclear import. First, we determined the ability of the Cuf1 C terminus to act in trans with its N terminus and its effect on Cuf1 intracellular location. We used a chimeric protein containing the N-terminal 61 amino acids of Cuf1 fused to the GFP protein, 1Cuf161-GFP. In separate molecules, we fused two copies of the FLAG epitope to the C-terminal residues 62 to 410 of Cuf1 or Cuf1-M6 to generate 62Cuf1410-FLAG2 or 62Cuf1-M6410-FLAG2, respectively. We coexpressed 1Cuf161-GFP with 62Cuf1410-FLAG2 or 62Cuf1-M6410-FLAG2 in the yeast strain JSY8 (cuf1Δ) and analyzed the localization of 1Cuf161-GFP. As shown in Fig. 9A, when 1Cuf161-GFP was expressed with the empty vector, it was localized in the nucleus under both copper-replete and copper starvation conditions. However, coexpression of 1Cuf161-GFP with 62Cuf1410-FLAG2 inhibited the nuclear import of 1Cuf161-GFP in the presence of copper. Interestingly, this inhibitory effect was completely reversed upon treatment with the copper chelator BCS or by coexpressing 1Cuf161-GFP with the mutant 62Cuf1-M6410-FLAG2, which lacks the C-rich motif, instead of 62Cuf1410-FLAG2 (Fig. 9A). To ensure that the chimeric proteins were expressed in the cotransformed cells, immunoblot analyses of protein extracts were performed using anti-GFP and anti-FLAG antibodies (Fig. 9B). These results suggest that copper promotes an interaction between the Cuf1 N terminus, 1Cuf161, and its C terminus, 62Cuf1410. Furthermore, we conclude that all or at least a subset of the amino acid residues Cys328, Cys330, Cys334, Cys336, Cys339, and His342 within the C-rich motif in the C terminus is required for this interaction that leads to inhibition of the nuclear import of 1Cuf161-GFP.
FIG. 9.
Cu-mediated inhibition of nuclear import of Cuf1 by its C-rich motif. (A) S. pombe JSY8 cells were cotransformed with an empty control plasmid and GFP alone, an empty vector and 1cuf161-GFP, 1cuf161-GFP and 62cuf1410-FLAG2, or 1cuf161-GFP and 62cuf1-M6410-FLAG2. To examine GFP fluorescence, the cotransformed cells specified above were grown to A600 of 1.0 in the presence of 5 μM thiamine, at which step the cultures were washed twice. Cultures were divided for their respective treatments (100 μM CuSO4 or 100 μM BCS) and grown in selective media lacking thiamine to induce protein synthesis. After 4 h, cells were subjected to fluorescence microscopy to visualize the 1Cuf161-GFP fusion protein (GFP). Cell morphology was also examined through Nomarski optics (Nomarski). (B) Whole-cell extracts were prepared from aliquots of cultures described above for panel A and analyzed by immunoblotting using either anti-GFP or anti-FLAG antibody (a). For simplicity, results shown are samples that were analyzed from copper-replete cells since the expression level of proteins detected from copper-deficient cells were virtually identical. As a control, total extract preparations were probed with anti-PCNA antibody. WB, Western blot.
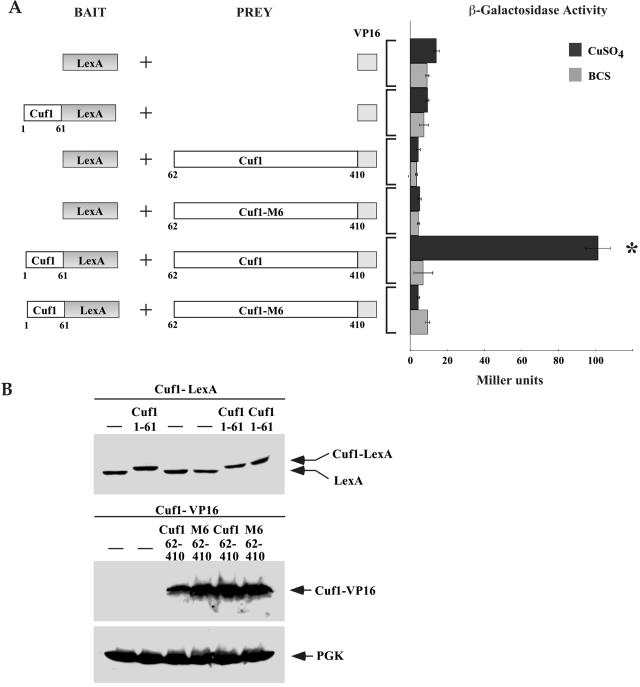
We next determined whether 1Cuf161 physically interacts with 62Cuf1410 using a yeast two-hybrid analysis. We fused 1Cuf161 upstream of the E. coli LexA DNA binding domain as bait. The prey vectors consisted of either 62Cuf1410 or 62Cuf1-M6410 fused to the VP16 acidic activation domain. The bait plasmid expressing the 1Cuf161-LexA or the empty bait plasmid was cotransformed into S. cerevisiae strain L40 with prey plasmids encoding 62Cuf1410-VP16, 62Cuf1-M6410-VP16, or the empty VP16 vector, and β-galactosidase activity was measured in cells grown in the presence or absence of copper. The results show that coexpression of 1Cuf161-LexA and 62Cuf1410-VP16 fusion proteins in copper-treated cells produced elevated levels of β-galactosidase activity (∼100 Miller units) (Fig. 10A). However, in the presence of the copper chelator BCS, no significant β-galactosidase activity was measured. When 1Cuf161-LexA and 62Cuf1-M6410-VP16 were coexpressed, no meaningful β-galactosidase activity was measured in cells grown in the presence of either copper or BCS, suggesting that these proteins fail to physically interact with each other (Fig. 10A). As shown in Fig. 10B, the loss of protein-protein interactions was not due to the lack of protein expression because all the fusion proteins tested for two-hybrid interactions were synthesized as confirmed by immunoblot analyses. The exception was the VP16 polypeptide alone that we were unable to detect, perhaps owing to its low predicted molecular mass of ∼8 kDa. Based on these data, we conclude that the Cuf1 C terminus physically interacts with the N terminus in a copper-dependent manner. The C-rich motif at the C terminus is absolutely required, as its removal in 62Cuf1-M6410-VP16 abrogates the physical interaction between these two domains.
FIG. 10.
The C-terminal C-rich domain of Cuf1 is required for interaction with Cuf1 N terminus in a copper-dependent manner by two-hybrid assay. (A) Schematic diagram of the LexA DNA binding domain alone or fused downstream of and in frame to the Cuf1 coding region that corresponds to codons 1 through 61. Bait molecules were coexpressed with the VP16 activation domain or two distinct 62Cuf1410-VP16 fusion derivatives (PREY). The amino acid sequences of Cuf1 are numbered relative to its first initiator codon. Cotransformed cells were grown under copper-deficient conditions (100 μM BCS) or with excess copper (100 μM CuSO4). Protein-protein interactions were detected by liquid β-galactosidase assays and indicated in Miller units (right side). Error bars indicate the standard deviations of samples analyzed in triplicate. The asterisk indicates a positive interaction detected between the 1Cuf161-LexA and 62Cuf1410-VP16 fusion proteins when coexpressed in copper-replete cells. (B) Protein extracts were prepared from aliquots of cultures described above for panel A and then analyzed by immunoblotting using either anti-LexA, anti-VP16, or anti-PGK (as an internal control) antibody.
DISCUSSION
In this study, we provide evidence that the subcellular localization of the fission yeast transcription factor Cuf1 is regulated as a function of copper availability. We show that Cuf1 localizes to the nucleus in copper-deficient cells but is retained in the cytoplasm when cells are grown under copper-replete conditions. The nuclear localization of Cuf1 is directed by an N-terminal NLS, which is nonclassical (23). The NLS sequence is relatively large, encompassing residues 11 to 53. Furthermore, this nuclear targeting segment does not contain any strict consensus cluster of basic amino acids. Instead, it contains several lysines and arginines that are interrupted by single or few nonbasic residues. Nonetheless, the Cuf1 segment containing the NLS is necessary and sufficient to promote nuclear translocation of a cytoplasmic reporter protein, GFP, indicating that its activity is transferable. We also found that multiple mutations within the basic segments of the NLS were needed to impair its activity since single alanine substitutions within these positively charged residues were ineffective (Beaudoin and Labbé, unpublished). This suggests that proper folding may be necessary in order for the NLS to be recognized by an importin β receptor. Interestingly, in the case of many transcription factors, including Cuf1, the NLS is embedded or located next to the DNA binding domain (67). It is possible that association between an import receptor and the NLS of these transcription factors might block their DNA binding domains and inhibit DNA binding activity until these factors reach an appropriate DNA binding site. Subsequently, the binding of the transcription factor to the DNA binding site might accelerate the dissociation of the receptor-cargo interaction possibly in a Ran-GTP-mediated manner (55).
Cuf1 joins three other metalloregulatory transcription factors, Aft1, Crf1, and MTF-1, that are regulated through nuclear localization (16, 54, 60, 75). The S. cerevisiae Aft1 protein is an iron-sensing transcriptional activator that regulates the expression of genes involved in the acquisition of iron (57, 73). Under iron-replete conditions, Aft1 is cytosolic but, following iron depletion, it translocates to the nucleus (75). The nuclear import of Aft1 is dependent on Pse1, a member of the importin β family of nuclear receptors (66). There is no sequence homology between the NLS of Cuf1 and that of Aft1, except that the basic amino acid residues responsible for nuclear import are found within relatively large regions for both proteins. Because, like Aft1, Cuf1 enters the nucleus upon metal starvation, it is tempting to speculate that the putative S. pombe homologue of Pse1, named Sal3, may be involved in the nuclear import of Cuf1 in fission yeast. In Yarrowia lipolytica, Crf1 (16) resembles the Ace1 protein from S. cerevisiae (15, 62). Although Ace1 protects cells from copper toxicity via induction of MT gene transcription, it has been shown that Crf1 mounts a protective response against copper overload by regulating the expression of a yet-unidentified target gene(s) which is not MT (16). An Ace1-β-galactosidase fusion protein was shown to be constitutively localized in the nucleus (61). Conversely, it has been shown that a Crf1-β-galactosidase fusion protein responds to changes in copper levels (16). Crf1-β-galactosidase is cytosolic when copper is limited and translocates to the nucleus when copper is replete (16). The potential mechanism(s) of copper-dependent Crf1 nuclear import in Y. lipolytica cells, however, has not been ascertained. MTF-1, which plays a role in zinc homeostasis, cadmium detoxification, and protection from reactive free radicals in mammals, is transported in and out of the nucleus (54, 60). Under nonstress conditions in the absence of metals, MTF-1 is located predominantly in the cytoplasm. However, upon the addition of zinc or cadmium, MTF-1 is concentrated in the nucleus. Inhibition of the export factor exportin 1 (Crm1) by the drug leptomycin B causes a nuclear accumulation of MTF-1, indicating that nuclear MTF-1 export occurs via Crm1 (54). However, information on the import process remains limited.
Although the cuf1+ gene of S. pombe encodes a copper sensor orthologous to S. cerevisiae Mac1, there are differences between Cuf1 and Mac1 intracellular localization in response to changes in copper concentrations and their nuclear localization sequences. Once synthesized, Mac1 is constitutively transported into the nucleus and its nuclear localization is unaffected by cellular copper status (25). In contrast to the Cuf1 NLS which is found between residues 11 to 53, the N-terminal 70-amino-acid segment of Mac1 expressed as a chimeric protein with three copies of the hemagglutinin (HA) epitope was observed in the cytoplasm (56). Two separate regions in Mac1 were capable of localizing to the nucleus, 1Mac1287-HA3 and 289Mac1417-HA3 (56). Because 1Mac170-HA3 was cytoplasmic, two NLS in Mac1 were suggested, one between amino acids 70 to 287 and the other between amino acids 289 to 417 (56). Within these latter regions, two potential sequences encompassing residues 155 to 177 (155K-K-X-R-X-R160-X13-K-K-X-K177) and 356 to 368 (356R-X2-R-K-X3-K-X-K-X-K368) were proposed to be the putative elements for nuclear targeting in Mac1. However, experimental evidence supporting a role for these amino acids in Mac1 nuclear localization is currently lacking. Importantly, the basic amino acids encompassed within residues 155 to 177 and 356 to 368 in Mac1 are absent in Cuf1. Furthermore, many of the basic amino acid residues that are found in Cuf1 11-53, such as Lys13, Arg47, Arg50, and Arg53, are not present in Mac1. Taken together, these differences may explain the distinct modes by which the subcellular localization and nuclear targeting of these proteins are controlled.
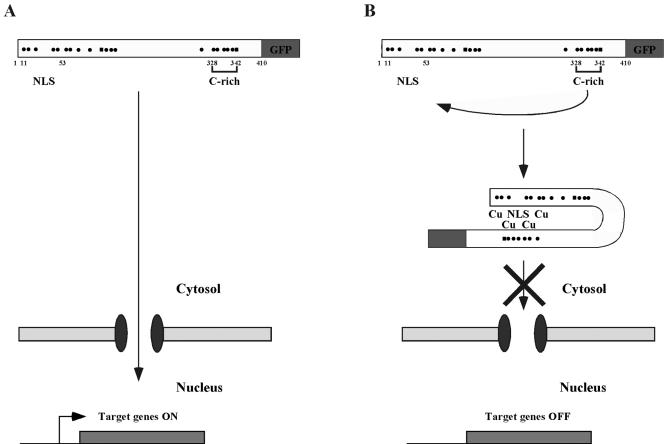
Cuf1 accumulates in the nucleus in response to low levels of copper. Conversely, as the concentration of copper increases, Cuf1 is inhibited from entering the nucleus. The requirement of the C-rich motif in the copper-dependent regulation of Cuf1 nuclear localization was confirmed by three experimental results. First, deletion analysis or point mutations designed to disrupt the Cuf1 C-rich motif resulted in a constitutive nuclear protein. Second, copper induced cytoplasmic retention of 1Cuf161 when this peptide was coexpressed as a separate molecule with the Cuf1 C-terminal domain containing the C-rich motif. Third, using a yeast two-hybrid assay, we showed that 1Cuf161 and 62Cuf1410, which harbors the wild-type C-rich motif, physically interact with each other in a copper-dependent manner. Based on these observations, we propose a model for the regulation of Cuf1 subcellular localization as a function of copper availability (Fig. 11). Under conditions of copper starvation, Cuf1 associates with an importin β via an accessible N-terminal NLS. Cuf1 is imported into the nucleus with a concomitant increase in Cuf1 transcriptional activity. Conversely, under conditions of copper excess, Cuf1 binds copper ions through specific Cys residues found in both the N terminus and the C terminus, particularly the C-rich motif, and adopts an inhibitory conformation that masks the NLS. Lack of access to the NLS allows Cuf1 to accumulate in the cytosol, since the interaction between Cuf1 and importin β is precluded.
FIG. 11.
A model for copper-dependent cytosolic retention of the Cuf1 protein. (A) Under low-copper conditions, Cuf1 interacts with an importin β receptor via association with its accessible N-terminal NLS. Cuf1 is localized to the nucleus with a concomitant increase in Cuf1 transcriptional activity. (B) In the presence of elevated copper concentrations, metallation of Cuf1 induces intramolecular conformational changes that mask its NLS. Cuf1 is retained in the cytosol, and target gene expression is extinguished. The dots and squares depict positions of cysteine and histidine residues, respectively.
The existence of a mechanism for the regulation of Cuf1 subcellular localization raises a number of questions. For example, it would be interesting to examine how the nuclear pool of Cuf1 protein is inactivated during a shift from low to high copper. Does the shut off of this pool require nucleocytoplasmic export? Herein, our experimental design served specifically to test the effect of metal on the nuclear accumulation of Cuf1. However, whether a mechanism exists for nuclear export of Cuf1 will require additional studies. It is interesting to note that our data on the regulation of Cuf1 nuclear localization through a C-terminal C-rich domain are reminiscent of those observed in the S. pombe Pap1 protein (42, 63, 64). Pap1 nuclear localization is regulated by oxidative stress (63). Pap1 is homologous to Yap1 in S. cerevisiae (43). These two proteins are often referred to as yeast AP-1-like proteins (yAP-1). yAP-1 proteins have two C-rich domains called the N-terminal and C-terminal C-rich domains, respectively (n-CRD and c-CRD) (42). During unstressed conditions, a model predicts that the export of Yap1 and Pap1 is more efficient than their import, resulting in a majority of the yAP-1 proteins being cytoplasmic. Upon oxidative stress, disulfide bonds are formed between the n-CRD and c-CRD of Yap1 or Pap1, masking its nuclear export sequence leading to nuclear accumulation of the transcription factor. While this oxidant-regulated mechanism occurs in the nucleus and involves inhibition of the interaction between Yap1 or Pap1 and the Crm1 nuclear export factor, one can envision the possibility that copper-induced conformational changes in Cuf1, through its C-rich domain, can inactivate its nuclear translocation by masking its NLS and inhibiting the interaction between Cuf1 and importin β. Experimental evidence to support this mechanism for the regulation of Cuf1 nuclear localization is the subject of ongoing studies.
Acknowledgments
We are indebted to Maria M. O. Peña for critical reading of the manuscript.
This work was supported by the CIHR of Canada grant MOP-36450 to S.L. Infrastructure equipment that was essential for conducting this investigation was obtained in part by the Canada Foundation for Innovation grant NOF-3754 and the NSERC Research Tools and Instruments grant 299851-04 to S.L. S.L. is a New Investigator Scholar from the Canadian Institutes of Health Research.
REFERENCES
- 1.Alfa, C., P. Fantes, J. Hyams, M. McLeod, and E. Warbrick. 1993. Experiments with fission yeast: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 2.Baeuerle, P. A., and T. Henkel. 1994. Function and activation of NF-κ B in the immune system. Annu. Rev. Immunol. 12:141-179. [DOI] [PubMed] [Google Scholar]
- 3.Beaudoin, J., and S. Labbé. 2001. The fission yeast copper-sensing transcription factor Cuf1 regulates the copper transporter gene expression through an Ace1/Amt1-like recognition sequence. J. Biol. Chem. 276:15472-15480. [DOI] [PubMed] [Google Scholar]
- 4.Beaudoin, J., A. Mercier, R. Langlois, and S. Labbé. 2003. The Schizosaccharomyces pombe Cuf1 is composed of functional modules from two distinct classes of copper metalloregulatory transcription factors. J. Biol. Chem. 278:14565-14577. [DOI] [PubMed] [Google Scholar]
- 5.Bellemare, D. R., M. Sanschagrin, J. Beaudoin, and S. Labbé. 2001. A novel copper-regulated promoter system for expression of heterologous proteins in Schizosaccharomyces pombe. Gene 273:191-198. [DOI] [PubMed] [Google Scholar]
- 6.Bellemare, D. R., L. Shaner, K. A. Morano, J. Beaudoin, J. Langlois, and S. Labbé. 2002. Ctr6, a vacuolar membrane copper transporter in Schizosaccharomyces pombe. J. Biol. Chem. 277:46676-46686. [DOI] [PubMed] [Google Scholar]
- 7.Bezanilla, M., S. L. Forsburg, and T. D. Pollard. 1997. Identification of a second myosin-II in Schizosaccharomyces pombe: Myp2p is conditionally required for cytokinesis. Mol. Biol. Cell 8:2693-2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butt, T. R., E. J. Sternberg, J. A. Gorman, P. Clark, D. Hamer, M. Rosenberg, and S. T. Crooke. 1984. Copper metallothionein of yeast, structure of the gene, and regulation of expression. Proc. Natl. Acad. Sci. USA 81:3332-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Culotta, V. C., W. R. Howard, and X. F. Liu. 1994. CRS5 encodes a metallothionein-like protein in Saccharomyces cerevisiae. J. Biol. Chem. 269:25295-25302. [PubMed] [Google Scholar]
- 10.Culotta, V. C., H. D. Joh, S. J. Lin, K. H. Slekar, and J. Strain. 1995. A physiological role for Saccharomyces cerevisiae copper/zinc superoxide dismutase in copper buffering. J. Biol. Chem. 270:29991-29997. [DOI] [PubMed] [Google Scholar]
- 11.Dancis, A., D. G. Roman, G. J. Anderson, A. G. Hinnebusch, and R. D. Klausner. 1992. Ferric reductase of Saccharomyces cerevisiae: molecular characterization, role in iron uptake, and transcriptional control by iron. Proc. Natl. Acad. Sci. USA 89:3869-3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dancis, A., D. S. Yuan, D. Haile, C. Askwith, D. Eide, C. Moehle, J. Kaplan, and R. D. Klausner. 1994. Molecular characterization of a copper transport protein in S. cerevisiae: an unexpected role for copper in iron transport. Cell 76:393-402. [DOI] [PubMed] [Google Scholar]
- 13.Eide, D. J. 1998. The molecular biology of metal ion transport in Saccharomyces cerevisiae. Annu. Rev. Nutr. 18:441-469. [DOI] [PubMed] [Google Scholar]
- 14.Forsburg, S. L. 1993. Comparison of Schizosaccharomyces pombe expression systems. Nucleic Acids Res. 21:2955-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furst, P., S. Hu, R. Hackett, and D. Hamer. 1988. Copper activates metallothionein gene transcription by altering the conformation of a specific DNA binding protein. Cell 55:705-717. [DOI] [PubMed] [Google Scholar]
- 16.Garcia, S., M. Prado, R. Degano, and A. Dominguez. 2002. A copper-responsive transcription factor, CRF1, mediates copper and cadmium resistance in Yarrowia lipolytica. J. Biol. Chem. 277:37359-37368. [DOI] [PubMed] [Google Scholar]
- 17.Georgatsou, E., L. A. Mavrogiannis, G. S. Fragiadakis, and D. Alexandraki. 1997. The yeast Fre1p/Fre2p cupric reductases facilitate copper uptake and are regulated by the copper-modulated Mac1p activator. J. Biol. Chem. 272:13786-13792. [DOI] [PubMed] [Google Scholar]
- 18.Halliwell, B., and J. M. Gutteridge. 1992. Biologically relevant metal ion-dependent hydroxyl radical generation. FEBS Lett. 307:108-112. [DOI] [PubMed] [Google Scholar]
- 19.Hassett, R., and D. J. Kosman. 1995. Evidence for Cu(II) reduction as a component of copper uptake by Saccharomyces cerevisiae. J. Biol. Chem. 270:128-134. [DOI] [PubMed] [Google Scholar]
- 20.Hollenberg, S. M., R. Sternglanz, P. F. Cheng, and H. Weintraub. 1995. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol. Cell. Biol. 15:3813-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hood, J. K., and P. A. Silver. 1999. In or out? Regulating nuclear transport. Curr. Opin. Cell Biol. 11:241-247. [DOI] [PubMed] [Google Scholar]
- 22.Jamison McDaniels, C. P., L. T. Jensen, C. Srinivasan, D. R. Winge, and T. D. Tullius. 1999. The yeast transcription factor Mac1 binds to DNA in a modular fashion. J. Biol. Chem. 274:26962-26967. [DOI] [PubMed] [Google Scholar]
- 23.Jans, D. A., C.-Y. Xiao, and M. H. C. Lam. 2000. Nuclear targeting signal recognition: a key control point in nuclear transport? BioEssays 22:532-544. [DOI] [PubMed] [Google Scholar]
- 24.Jensen, L. T., M. C. Posewitz, C. Srinivasan, and D. R. Winge. 1998. Mapping of the DNA binding domain of the copper-responsive transcription factor Mac1 from Saccharomyces cerevisiae. J. Biol. Chem. 273:23805-23811. [DOI] [PubMed] [Google Scholar]
- 25.Jensen, L. T., and D. R. Winge. 1998. Identification of a copper-induced intramolecular interaction in the transcription factor Mac1 from Saccharomyces cerevisiae. EMBO J. 17:5400-5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jungmann, J., H. A. Reins, J. Lee, A. Romeo, R. Hassett, D. Kosman, and S. Jentsch. 1993. MAC1, a nuclear regulatory protein related to Cu-dependent transcription factors is involved in Cu/Fe utilization and stress resistance in yeast. EMBO J. 12:5051-5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keeney, J. B., and J. D. Boeke. 1994. Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics 136:849-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keller, G., C. Gross, M. Kelleher, and D. R. Winge. 2000. Functional independence of the two cysteine-rich activation domains in the yeast Mac1 transcription factor. J. Biol. Chem. 275:29193-29199. [DOI] [PubMed] [Google Scholar]
- 29.Kim, J., and J. P. Hirsch. 1998. A nucleolar protein that affects mating efficiency in Saccharomyces cerevisiae by altering the morphological response to pheromone. Genetics 149:795-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knight, S. A., S. Labbé, L. F. Kwon, D. J. Kosman, and D. J. Thiele. 1996. A widespread transposable element masks expression of a yeast copper transport gene. Genes Dev. 10:1917-1929. [DOI] [PubMed] [Google Scholar]
- 31.Koch, K. A., and D. J. Thiele. 1996. Autoactivation by a Candida glabrata copper metalloregulatory transcription factor requires critical minor groove interactions. Mol. Cell. Biol. 16:724-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Labbé, S., M. M. O. Peña, A. R. Fernandes, and D. J. Thiele. 1999. A copper-sensing transcription factor regulates iron uptake genes in Schizosaccharomyces pombe. J. Biol. Chem. 274:36252-36260. [DOI] [PubMed] [Google Scholar]
- 33.Labbé, S., and D. J. Thiele. 1999. Pipes and wiring: the regulation of copper uptake and distribution in yeast. Trends Microbiol. 7:500-505. [DOI] [PubMed] [Google Scholar]
- 34.Labbé, S., Z. Zhu, and D. J. Thiele. 1997. Copper-specific transcriptional repression of yeast genes encoding critical components in the copper transport pathway. J. Biol. Chem. 272:15951-15958. [DOI] [PubMed] [Google Scholar]
- 35.Luk, E., L. T. Jensen, and V. C. Culotta. 2003. The many highways for intracellular trafficking of metals. J. Biol. Inorg. Chem. 8:803-809. [DOI] [PubMed] [Google Scholar]
- 36.Martins, L. J., L. T. Jensen, J. R. Simons, G. L. Keller, and D. R. Winge. 1998. Metalloregulation of FRE1 and FRE2 homologs in Saccharomyces cerevisiae. J. Biol. Chem. 273:23716-23721. [DOI] [PubMed] [Google Scholar]
- 37.Mattaj, I. W., and L. Englmeier. 1998. Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 67:265-306. [DOI] [PubMed] [Google Scholar]
- 38.McNabb, D. S., S. M. Pak, and L. Guarente. 1997. Cassette for the generation of sequential gene disruptions in the yeast Schizosaccharomyces pombe. BioTechniques. 22:1134-1139. [DOI] [PubMed] [Google Scholar]
- 39.Miller, J. H. 1972. Experiments in molecular genetics, p 352-355. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Moll, T., G. Tebb, U. Surana, H. Robitsch, and K. Nasmyth. 1991. The role of phosphorylation and the CDC28 protein kinase in cell cycle-regulated nuclear import of the S. cerevisiae transcription factor SWI5. Cell 66:743-758. [DOI] [PubMed] [Google Scholar]
- 41.Moreno, M. B., A. Duran, and J. C. Ribas. 2000. A family of multifunctional thiamine-repressible expression vectors for fission yeast. Yeast 16:861-872. [DOI] [PubMed] [Google Scholar]
- 42.Moye-Rowley, W. S. 2003. Regulation of the transcriptional response to oxidative stress in fungi: similarities and differences. Eukaryot. Cell 2:381-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moye-Rowley, W. S., K. D. Harshman, and C. S. Parker. 1989. Yeast YAP1 encodes a novel form of the jun family of transcriptional activator proteins. Genes Dev. 3:283-292. [DOI] [PubMed] [Google Scholar]
- 44.Mumberg, D., R. Muller, and M. Funk. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156:119-122. [DOI] [PubMed] [Google Scholar]
- 45.O'Neill, E. M., A. Kaffman, E. R. Jolly, and E. K. O'Shea. 1996. Regulation of PHO4 nuclear localization by the PHO80-PHO85 cyclin-CDK complex. Science 271:209-212. [DOI] [PubMed] [Google Scholar]
- 46.Pelletier, B., A. Trott, K. A. Morano, and S. Labbé. 2005. Functional characterization of the iron-regulatory transcription factor Fep1 from Schizosaccharomyces pombe. J. Biol. Chem. 280:25146-25161. [DOI] [PubMed] [Google Scholar]
- 47.Peña, M. M. O., J. Lee, and D. J. Thiele. 1999. A delicate balance: homeostatic control of copper uptake and distribution. J. Nutr. 129:1251-1260. [DOI] [PubMed] [Google Scholar]
- 48.Peña, M. M. O., S. Puig, and D. J. Thiele. 2000. Characterization of the Saccharomyces cerevisiae high-affinity copper transporter Ctr3. J. Biol. Chem. 275:33244-33251. [DOI] [PubMed] [Google Scholar]
- 49.Puig, S., J. Lee, M. Lau, and D. J. Thiele. 2002. Biochemical and genetic analyses of yeast and human high-affinity copper transporters suggest a conserved mechanism for copper uptake. J. Biol. Chem. 277:26021-26030. [DOI] [PubMed] [Google Scholar]
- 50.Puig, S., and D. J. Thiele. 2002. Molecular mechanisms of copper uptake and distribution. Curr. Opin. Chem. Biol. 6:171-180. [DOI] [PubMed] [Google Scholar]
- 51.Radisky, D., and J. Kaplan. 1999. Regulation of transition metal transport across the yeast plasma membrane. J. Biol. Chem. 274:4481-4484. [DOI] [PubMed] [Google Scholar]
- 52.Rees, E. M., and D. J. Thiele. 2004. From aging to virulence: forging connections through the study of copper homeostasis in eukaryotic microorganisms. Curr. Opin. Microbiol. 7:175-184. [DOI] [PubMed] [Google Scholar]
- 53.Rutherford, J. C., and A. J. Bird. 2004. Metal-responsive transcription factors that regulate iron, zinc, and copper homeostasis in eukaryotic cells. Eukaryot. Cell 3:1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saydam, N., O. Georgiev, M. Y. Nakano, U. F. Greber, and W. Schaffner. 2001. Nucleo-cytoplasmic trafficking of metal-regulatory transcription factor 1 is regulated by diverse stress signals. J. Biol. Chem. 276:25487-25495. [DOI] [PubMed] [Google Scholar]
- 55.Seedorf, M., M. Damelin, J. Kahana, T. Taura, and P. A. Silver. 1999. Interactions between a nuclear transporter and a subset of nuclear pore complex proteins depend on Ran GTPase. Mol. Cell. Biol. 19:1547-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Serpe, M., A. Joshi, and D. J. Kosman. 1999. Structure-function analysis of the protein-binding domains of Mac1p, a copper-dependent transcriptional activator of copper uptake in Saccharomyces cerevisiae. J. Biol. Chem. 274:29211-29219. [DOI] [PubMed] [Google Scholar]
- 57.Shakoury-Elizeh, M., J. Tiedeman, J. Rashford, T. Ferea, J. Demeter, E. Garcia, R. Rolfes, P. O. Brown, D. Botstein, and C. C. Philpott. 2004. Transcriptional remodeling in response to iron deprivation in Saccharomyces cerevisiae. Mol. Biol. Cell 15:1233-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 59.Shibasaki, F., E. R. Price, D. Milan, and F. McKeon. 1996. Role of kinases and the phosphatase calcineurin in the nuclear shuttling of transcription factor NF-AT4. Nature 382:370-373. [DOI] [PubMed] [Google Scholar]
- 60.Smirnova, I. V., D. C. Bittel, R. Ravindra, H. Jiang, and G. K. Andrews. 2000. Zinc and cadmium can promote rapid nuclear translocation of metal response element-binding transcription factor-1. J. Biol. Chem. 275:9377-9384. [DOI] [PubMed] [Google Scholar]
- 61.Szczypka, M. S., and D. J. Thiele. 1989. A cysteine-rich nuclear protein activates yeast metallothionein gene transcription. Mol. Cell. Biol. 9:421-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thiele, D. J. 1988. ACE1 regulates expression of the Saccharomyces cerevisiae metallothionein gene. Mol. Cell. Biol. 8:2745-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Toone, M. W., S. Kuge, M. Samuels, B. A. Morgan, T. Toda, and N. Jones. 1998. Regulation of the fission yeast transcription factor Pap1 by oxidative stress: requirement for the nuclear export factor Crm1 (Exportin) and the stress-activated MAP kinase Sty1/Spc1. Genes Dev. 12:1453-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Toone, M. W., B. A. Morgan, and N. Jones. 2001. Redox control of AP-1-like factors in yeast and beyond. Oncogene 20:2336-2346. [DOI] [PubMed] [Google Scholar]
- 65.Turner, R. B., D. L. Smith, M. E. Zawrotny, M. F. Summers, M. C. Posewitz, and D. R. Winge. 1998. Solution structure of a zinc domain conserved in yeast copper-regulated transcription factors. Nat. Struct. Biol. 5:551-555. [DOI] [PubMed] [Google Scholar]
- 66.Ueta, R., A. Kukunaka, and Y. Yamaguchi-Iwai. 2003. Pse1p mediates the nuclear import of the iron-responsive transcription factor Aft1p in Saccharomyces cerevisiae. J. Biol. Chem. 278:50120-50127. [DOI] [PubMed] [Google Scholar]
- 67.Vandromme, M., C. Gauthier-Rouviere, N. Lamb, and A Fernandez. 1996. Regulation of transcription factor localization: fine-tuning of gene expression. Trends Biochem. Sci. 21:59-64. [PubMed] [Google Scholar]
- 68.Vojtek, A. B., J. A. Cooper, and S. M. Hollenberg. 1997. Searching for interacting proteins with the two-hybrid system II, p. 29-42. In P. Bartel and S. Fields (ed.), The yeast two-hybrid system. Oxford University Press, New York, N.Y.
- 69.Vojtek, A. B., S. M. Hollenberg, and J. A. Cooper. 1993. Mammalian Ras interacts directly with the serine threonine kinase Raf. Cell 74:205-214. [DOI] [PubMed] [Google Scholar]
- 70.Wang, X., R. Sato, M. S. Brown, X. Hua, and J. L. Goldstein. 1994. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell 77:53-62. [DOI] [PubMed] [Google Scholar]
- 71.Weis, K. 1998. Importins and exportins: how to get in and out of the nucleus. Trends Biochem. Sci. 23:185-189. [DOI] [PubMed] [Google Scholar]
- 72.Winge, D. R. 1998. Copper-regulatory domain involved in gene expression. Prog. Nucleic Acid Res. Mol. Biol. 58:165-195. [DOI] [PubMed] [Google Scholar]
- 73.Yamaguchi-Iwai, Y., A. Dancis, and R. D. Klausner. 1995. AFT1: a mediator of iron regulated transcriptional control in Saccharomyces cerevisiae. EMBO J. 14:1231-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yamaguchi-Iwai, Y., M. Serpe, D. Haile, W. Yang, D. J. Kosman, R. D. Klausner, and A. Dancis. 1997. Homeostatic regulation of copper uptake in yeast via direct binding of MAC1 protein to upstream regulatory sequences of FRE1 and CTR1. J. Biol. Chem. 272:17711-17718. [DOI] [PubMed] [Google Scholar]
- 75.Yamaguchi-Iwai, Y., R. Ueta, A. Fukunaka, and R. Sasaki. 2002. Subcellular localization of Aft1 transcription factor responds to iron status in Saccharomyces cerevisiae. J. Biol. Chem. 277:18914-18918. [DOI] [PubMed] [Google Scholar]
- 76.Zhou, H., and D. J. Thiele. 2001. Identification of a novel high-affinity copper transport complex in the fission yeast Schizosaccharomyces pombe. J. Biol. Chem. 276:20529-20535. [DOI] [PubMed] [Google Scholar]
- 77.Zhu, Z., S. Labbé, M. M. O. Peña, and D. J. Thiele. 1998. Copper differentially regulates the activity and degradation of yeast Mac1 transcription factor. J. Biol. Chem. 273:1277-1280. [DOI] [PubMed] [Google Scholar]