Abstract
A common property of G protein-coupled receptors is that they become less responsive with prolonged stimulation. Regulators of G protein signaling (RGS proteins) are well known to accelerate G protein GTPase activity and do so by stabilizing the transition state conformation of the G protein α subunit. In the yeast Saccharomyces cerevisiae there are four RGS-homologous proteins (Sst2, Rgs2, Rax1, and Mdm1) and two Gα proteins (Gpa1 and Gpa2). We show that Sst2 is the only RGS protein that binds selectively to the transition state conformation of Gpa1. The other RGS proteins also bind Gpa1 and modulate pheromone signaling, but to a lesser extent and in a manner clearly distinct from Sst2. To identify other candidate pathway regulators, we compared pheromone responses in 4,349 gene deletion mutants representing nearly all nonessential genes in yeast. A number of mutants produced an increase (sst2, bar1, asc1, and ygl024w) or decrease (cla4) in pheromone sensitivity or resulted in pheromone-independent signaling (sst2, pbs2, gas1, and ygl024w). These findings suggest that Sst2 is the principal regulator of Gpa1-mediated signaling in vivo but that other proteins also contribute in distinct ways to pathway regulation.
G protein-coupled receptors respond to a vast array of chemical and sensory signals, including hormones, neurotransmitters, odors, and light. Approximately one-third of all drugs act by binding directly to receptors of this class (64). Upon agonist stimulation of the receptor, a cognate G protein α subunit will exchange GDP for GTP and undergo dissociation from the G protein βγ subunit dimer. The dissociated subunits bind to effector enzymes, which in turn activate protein kinases, trigger new gene transcription, and ultimately produce programmed changes in cell homeostasis or differentiation (90). Regulators of G protein signaling (RGS proteins) function as GTPase-accelerating proteins (GAPs) and, in this manner, promote rapid inactivation or desensitization of the signal (89).
Whereas mammalian genome analysis has revealed at least 16 Gα- and ∼40 RGS-encoding genes (89, 106), a similar analysis in the yeast Saccharomyces cerevisiae reveals only two Gα subunits but at least four RGS protein homologues. Gpa1 mediates cellular responses to mating pheromones. These pheromones, called a-factor and α-factor, are produced by haploid a and α cells and bind to G protein-coupled receptors on cells of the opposite mating type. Upon activation of pheromone receptors, Gpa1 binds to GTP and dissociates from the Gβγ dimer Ste4/Ste18, and the dissociated subunits activate a multitude of downstream effectors leading to cell fusion (mating) to form an a/α diploid (36, 50). Prominent among the known effectors are components of a MAP (mitogen-activated protein) kinase cascade comprised of Ste20, Ste11, Ste7, and Fus3. A parallel signaling pathway responds to glucose stimulation, leading to activation of a distinct receptor (Gpr1) (66, 73, 76, 99, 124), a distinct G protein α subunit (Gpa2), and an atypical G protein βγ complex comprised of Gpb1 or Gpb2 and Gpg1 (4, 54).
Among the RGS proteins in yeast, Sst2 is by far the best characterized. The gene was originally identified through a screen for negative regulators of the pheromone response (15, 16). Subsequent analyses revealed that Sst2 interacts genetically (33, 37) and physically (39) with Gpa1 and can accelerate Gpa1 GTPase activity (2, 133). A second yeast RGS protein, Rgs2, was identified as a multicopy suppressor of Gpa2-dependent loss of heat shock resistance in stationary-phase cells and was also shown to accelerate Gpa2 GTP binding and hydrolysis (123).
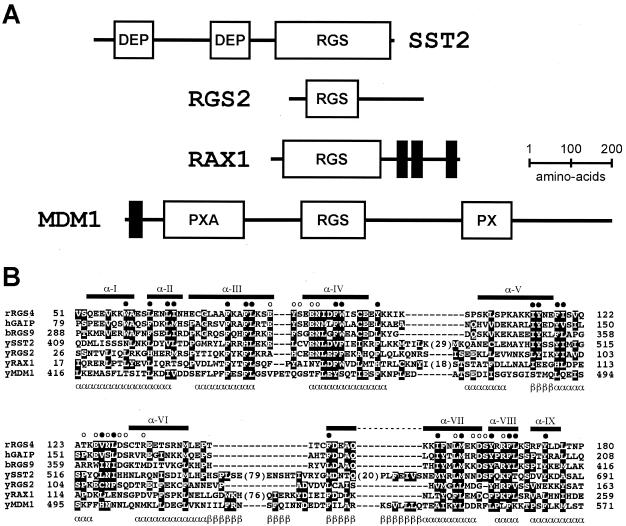
Two additional RGS protein homologues in yeast have not been implicated previously in G protein signaling events (Fig. 1). RAX1 (revert to axial) was identified as a suppressor of axl1/ste22 mutations (46). Axl1 is a haploid-specific endoprotease required for maturation of a-factor mating pheromone and for the normal axial budding pattern of haploid cells (23, 26, 75, 91). Conversely, diploid rax1/rax1 mutants exhibit a random or axial-like budding pattern normally found only in haploid cells (46, 91). These findings suggest a role for Rax1 in the establishment and maintenance of cell polarity. A fourth RGS protein is Mdm1. This is the least conserved member of the RGS family in yeast, but the one most similar to human RGS-PX1 (also known as SNX13) (137). Both Mdm1 and RGS-PX1 have a Phox (PX) domain (137), which binds to SH3 domains and phosphoinositides and contributes to membrane localization in vivo (20, 58, 134). Overexpression of human RGS-PX1 inhibits transport of epidermal growth factor receptors from endosomes to lysosomes, thereby enhancing the growth factor receptor signal (137). Yeast Mdm1 is expressed predominantly in late G1 to early S phase of the cell cycle and appears to be required for proper nuclear and mitochondrial inheritance in cells grown at elevated temperatures (45, 80).
FIG. 1.
Architecture of the four RGS proteins in yeast. (A) Schematic of the multiple domains of Sst2, Rgs2, Rax1, and Mdm1. DEP, Dishevelled/EGL-10/pleckstrin homology domain; RGS, regulator of G-protein signaling domain; PXA, PX-associated domain; PX, p40phox and p47phox homology domain. Putative transmembrane regions are denoted by solid vertical bars. (B) Multiple sequence alignment of the RGS box regions of rat (r) RGS4, human (h) GAIP/RGS19, bovine (b) RGS9, and the four yeast (y) RGS box proteins. Boxed residues are conserved amino acids identified using Clustal-X (120). Numbers in parentheses within Sst2 and Rax1 protein sequences denote the number of residues not shown in each insert region. The nine α-helices observed within the nuclear magnetic resonance solution structure of human GAIP are numbered in roman numerals and marked with horizontal bars above the sequences (29). Closed circles (•) denote conserved residues forming the RGS box hydrophobic core. Open circles (○) highlight conserved residues making direct contacts with Gα in the RGS4/Gαi1 crystal structure (119). Predicted α-helical (α) and β-strand (β) secondary structure within the Mdm1 RGS-box, based on the PSI-Pred algorithm (81), is denoted underneath the Mdm1 sequence. Primary sequences in the alignment are derived from rat RGS4 (SwissProt accession no. P49799), human GAIP/RGS19 (SwissProt accession no. P49795), bovine RGS9 (SwissProt accession no. O46469), yeast Sst2 (SwissProt accession no. P11972), yeast Rgs2 (GenBank accession no. NP_014750), yeast Rax1 (GenBank accession no. NP_014945), and yeast Mdm1 (GenBank accession no. NP_013603).
While the GAP function of RGS proteins is well established, not all RGS proteins exhibit this activity. Two prominent examples are Axin and the G protein-coupled receptor kinase GRK2 (13, 101, 113). Even when GAP activity has been documented, the physiological function of most RGS protein family members remains poorly understood. Moreover, there is growing evidence in mammalian cells that RGS proteins regulate specific Gα subunits in vivo, even when such specificity is absent in vitro. For example, RGS4 and Gα-interacting protein (GAIP) behave similarly towards Giα and Goα in vitro (7) yet have dissimilar effects in cultured neuronal cells (35). Likewise, in yeast it is not known if more than one RGS protein specifically regulates Gpa1 (or Gpa2) signaling. The promiscuity of RGS-Gα coupling observed in vitro highlights the need to analyze the specificity of RGS-Gα interactions in vivo, as well as in vitro.
Here we present a comprehensive analysis of G protein signal regulation in yeast. Through systematic overexpression and disruption of all four RGS genes, we establish the ability of each to regulate the well-characterized Gpa1 signaling pathway. We show that all four RGS proteins regulate signaling to some extent, but by distinct mechanisms, and with Sst2 as by far the most important contributor. To determine whether additional factors regulate Gpa1 signaling, we present results of pheromone dose-response profiles for nearly all viable gene deletion mutants in yeast. This analysis reveals several additional components that regulate the pheromone signaling pathway, either by restricting basal activity or by pheromone-induced activation of the G protein.
MATERIALS AND METHODS
Strains.
Standard methods for the growth, maintenance, and transformation of yeast and bacteria, the preparation of growth medium, and the manipulation of DNA were used throughout (3). The yeast Saccharomyces cerevisiae strains used in this study were DC17 (MATα his1) (from J. Thorner, University of California—Berkeley), BJ2168 (MATa ura3-52 leu2-Δ1 trp1-Δ63 prb1-1,122 prc1-407 pep4-3) (from E. Jones, Carnegie Mellon University), YPH499 (MATa ura3-52 lys2-801amber ade2-101ochre trp1-Δ63 his3-Δ200 leu2-Δ1), BY4741 (MATa his3Δ leu2Δ met15Δ ura3Δ), and commercially available gene deletion strains in BY4741 (Research Genetics, Huntsville, AL).
Multiple RGS deletion strains were made in the BY4741 background and designated BY4741-sst2/rgs2 (sst2Δ::G418R rgs2Δ::hisG), BY4741-sst2/rax1 (sst2Δ::G418R rax1Δ::hisG), BY4741-sst2/mdm1 (sst2Δ::G418R mdm1Δ::hisG), BY4741-rgs2/rax1 (rgs2Δ::G418R rax1Δ::hisG), BY4741-rgs2/mdm1 (rgs2Δ::G418R mdm1Δ::hisG), BY4741-rax1/mdm1 (rax1Δ::G418R mdm1Δ::hisG), and BY4741-sst2/rgs2/rax1/mdm1 (sst2Δ::G418R rgs2Δ::hisG rax1Δ::hisG mdm1Δ::hisG). Multiple gene disruptions were constructed by replacing a portion of the indicated gene with the hisG-URA3-hisG cassette from pUC19-hisG-URA3, containing the hisG-URA3-hisG BamHI-BglII digestion product from pNK51 (1) ligated into the BamHI site of pUC19. To this end, RGS2 was PCR amplified using primers 5′-CTGTC ATTTG CCTTA CTGTT-3′ and 5′-TGAGC CACAG ACATT GGGTC-3′, cloned into pYES2.1/V5 TOPO (Invitrogen), digested with SacI, blunt-ended with T4 polymerase, and digested with EcoRI. The deleted portion of the gene was replaced with hisG-URA3-hisG digested in the same manner. RAX1 was PCR amplified using primers 5′-CGAAT GGGAG TTATT TGGCC-3′ and 5′-CGGTA GGAAG GATCA GTCAT-3′, digested with XhoI (blunt ended) and SacI, and ligated with hisG-URA3-hisG digested with SalI (blunt ended) and AscII. MDM1 was PCR amplified using primers 5′-CCACT ATCTG TTGCA ATACA-3′ and 5′-GCTGC GCCAA TTAAT TGTCC-3′, digested with HindIII (blunt ended) and BamHI, and ligated to hisG-URA3-hisG digested with SalI (blunt ended) and BamHI. Cells transformed with the hisG-URA3-hisG mutants were grown in 5-fluoroorotic acid to select against URA3. The integrity of the mutants was confirmed by PCR amplification of the disrupted gene.
Constitutively active mutants (group VI) were introduced into YPH499 and YPH499-based sterile mutants YDK101 (ste2::HIS3, from J. Thorner, University of California), YTG4 (ste4::hisG) (47), JTY2556 (ste7::ADE2, from J. Thorner), and YDK12/JDY3 (ste12::LEU2) (28). Primers designed to amplify the G418R selection gene module from genomic DNA derived from the deletion strains of interest were either purchased (GenePairs PCR primers; Research Genetics) (for SST2 and YGL024W) or custom designed for PBS2 (5′-GCAAA GGTCT AGATT TCTTG C-3′, 5′-GGTAA TTCTA GACTG TTTTC C-3′) and GAS1 (5′-GAATC TTCCG AGCTC ACAAC C-3′, 5′-CCTAA CGTGA GCTCG TACAC G-3′). The amplification products were gel purified, transformed, and plated on yeast extract-peptone-dextrose medium containing 300 μg/ml G418. Proper integration of the gene deletion cassette was confirmed by PCR.
Plasmids.
Previously described expression plasmids used in this study were pRS316-ADH (CEN, ampR, URA3, ADH1 promoter/terminator) (112), pAB27 (2μm, ampR, URA3, leu2-d, GAP1 promoter and terminator; from Anthony Brake, University of California—San Francisco), pAD4M (2μm, ampR, LEU2, ADH1 promoter/terminator; from Peter McCabe, Onyx Pharmaceutical), pAD4M-GPA1Q323L (39), pAD4M-GPA1-GST, pAD4M-GST (112), and HOG1 in pRS426 (from M. Gustin, Rice University) (11). pAD4M-GPA2-GST was constructed as described previously for pAD4M-GPA1-GST (112), using PCR primers 5′-GCGGT CGACA TGGGT CTCTG CGCAT CTTCA-3′ and 5′-GCGGA GCTCT TGTAA CACTC CAGAG TCTTT-3′. The boldface print indicates added SalI or SacI restriction sites used to digest the product prior to ligation to the corresponding sites in pAD4M.
Overexpression of RGS proteins was achieved by subcloning each gene into the blunt-ended BglII site of pAB27. For construction of pAB27-RGS2 the RGS2 gene was PCR amplified using primers 5′-CGCGG ATCCG CCAGA TGGCG AGTGT ACCAA GTCTA-3′ and 5′-CGCGG ATCCC TATCT TTGTT GATGA CTGTT-3′. For pAB27-RAX1 the gene was PCR amplified using primers 5′-CGCGG ATCCT CATCA TGAAG GAAGA GCTCA GCAAA-3′ and 5′-CGCGG ATCCGAATTCT CATAC ACGAC GGCCG GGAAC-3′. For pAB27-MDM1 the gene was PCR amplified using primers 5′-GACGC GTCGA CGCCT TCTGA TATGA TATCG T-3′ and 5′-GACGC GTCGA CTAGA TTGTT CGGTA CTTAG T-3′. The boldface print indicates added BamHI, EcoRI, or SalI sites, which were used to digest the product prior to blunt-ending and ligation to pAB27. Construction of a similar plasmid used to overexpress SST2 (pAB23-SST2) was described previously (39).
Constitutive expression of RGS proteins was achieved by subcloning PCR amplification products into pRS316-ADH. For construction of pRS316-ADH-RGS2, RGS2 was amplified using primers 5′-CCGGT CGACG CCAGA TGGCG AGTGT ACCAA GTCA-3′ and 5′-CAGGA TCCCT ATCTT TGTTG ATGAC TGTT-3′. The boldface print indicates added SalI or BamHI sites, used to subclone the PCR product into the corresponding sites in pRS316-ADH. pRS316-ADH-RAX1 was constructed by PCR amplification of RAX1 using the same primers as for pAB27-RAX1 and subcloned as a BamHI-EcoRI fragment. pRS316-ADH-MDM1 was constructed by PCR amplification of MDM1 using the same primers as for pAB27-MDM1 and subcloned as a SalI fragment. Construction of pRS316-ADH-SST2 was described previously (61).
Constitutive expression of epitope-tagged RGS proteins was achieved by subcloning PCR amplification products containing the primer-encoded Myc epitope into pRS316-ADH. pRS316-ADH-RGS2-Myc was constructed by PCR amplification of RGS2 using primers 5′-CCGGT CGACG CCAGA TGGCG AGTGT ACCAA GTCA-3′ and 5′-CGCGG ATCCT TACCT CTTCC TGAGG AGGTC CTCTT CGCTG ATTAA TTTCT GCTCC TCGAG TCTTT GTTGA TGACT GTTTT GTCCT TTCAA-3′ and subcloned as a SalI-BamHI fragment into pRS316-ADH. pRS316-ADH-MDM1-Myc was constructed by PCR amplification of MDM1 using primers 5′-ACGCG TCGAC CTTCT GATAT GATAT CGTAT G-3′ and 5′-GCGAG CTCTT ACCTC TTCCT GAGGA GGTCC TCTTC GCTGA TTAAT TTCTG CTCCT CGAGG TCATT ACATA TTATG TCCAA TAAAA TTGC-3′ and subcloned as a SalI-SacI fragment. pRS316-ADH-RAX1-Myc was constructed by amplification of RAX1 using primers 5′-CGCGG ATCCT CATCA TGAAG GAAGA GCTCA GCAAA-3′ and 5′-CCGGA ATTCT TACCT CTTCC TGAGG AGGTC CTCTT CGCTG ATTAA TTTCT GCTCC TCGAG TACAC GACGG CCGGG AACAC AGCTG AAAA-3′ and subcloned as a BamHI-EcoRI fragment. pRS316-ADH-SST2-Myc was constructed by subcloning SST2-Myc as a BamHI digestion product from pBS-SST2-Myc (39) into pRS316-ADH. Added BamHI, SalI, EcoRI, or SacI sites are in boldface print, and the Myc epitope is underlined.
A slightly different strategy was used to clone non-RGS regulators. GenePairs PCR primers (Research Genetics) were used to PCR amplify group I, group II, and group III genes. Custom primers were used to amplify YAL047c (5′-GATCG AATTC ACCAC CATGG TACGT CGATG GATTC GTAG-3′ and 5′-GATCC CCGGG AATTG CCATG TTAGG GATTG TTGAT TGAT-3′) and YDR462w (5′-GATCG AATTC ACCAC CATGC TGGCA CAAAC ATTCA AAAA-3′ and 5′-GATCC CCGGG AATTG CCATG TCATT TTCGG AAGTT ATAAT GCCAT AACG-3′). The resulting PCR products were subcloned directly into pYES2.1/V5 TOPO (Invitrogen). The integrity of BAR1, SST2, YGL024W, ASC1, UBC4, and CLA4 genes was confirmed by DNA sequencing.
To determine which, if any, group III mutants were truly sterile, we PCR amplified the wild-type genes of all open reading frames not previously reported to be sterile and expressed each clone in the corresponding deletion strain. All remained unresponsive to α-factor, indicating they were likely false positives (data not shown). These mutants were likely to have switched mating type or to contain contaminating diploid cells, as reported previously for some strains in the collection, and would therefore be unresponsive to α-factor. Note that the sir mutants are likely to cause a loss of silencing at the HMLα locus in a cells, resulting in expression of a- and α-type genes, creating a pseudodiploid state that is unresponsive to α-factor (79).
Assay of RGS binding to Gα proteins.
Binding assays were performed as described previously for monitoring the interaction of Gpa1 and Ste4/Ste18, with minor modifications (112). Briefly, BJ2168 cells were transformed with the pRS316-ADH plasmids containing Myc epitope-tagged RGS proteins and either pAD4M-GST, pAD4M-GPA1-GST, or pAD4M-GPA2-GST. Cells were grown to an A600 of ∼1.0 in selective medium and transferred to ice, and growth was stopped by addition of NaN3 to 10 mM. All subsequent manipulations were carried out at 0 °C to 4°C.
Thirty A600 units of cells (per treatment) was harvested by centrifugation at 1,000 × g for 10 min, washed once with 10 mM NaN3, and washed once with lysis buffer (40 mM triethanolamine [pH 7.2], 2 mM EDTA, 150 mM NaCl, 2 mM dithiothreitol, 1 mM benzamidine, protease inhibitor cocktail [Sigma no. P8215], 3 mM MgCl2, 10 μM GDP) (condition 1) or the same buffer plus 60 μM AlCl3 and 20 mM NaF (condition 2). Cells were subjected to glass bead vortex homogenization for 8 × 1 min, with 1 min on ice in between. The lysate was centrifuged twice at 1,000 × g for 10 min, and the resulting supernatant was supplemented with 1.5% Triton X-100 (final concentration) for 1 h to solubilize membrane proteins. The insoluble material was removed by centrifugation at 1,000 × g for 10 min. The “applied” samples contain 30 μl of the solubilized supernatant mixed with an equal volume of 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer and boiled for 10 min. The glutathione S-transferase-purified (“GST-purified”) sample was obtained by mixing a 40% slurry of 100 μl glutathione-Sepharose 4B resin in lysis buffer with the remainder of the solubilized lysate for 12 h. The resin was then washed five times with phosphate-buffered saline containing 1% Triton X-100 supplemented with 300 mM NaCl. Bound proteins were eluted by boiling in 30 μl of 2× SDS-PAGE sample buffer for 10 min. Samples were resolved by SDS-10% PAGE, transferred to nitrocellulose, and probed with 9E10 anti-Myc mouse monoclonal antibodies (ascites) at 1:1,000 to detect the RGS proteins (44) or anti-GST rabbit polyclonal antibodies at 1:1,500 to confirm equal expression and recovery of the GST proteins (from J. Steitz, Yale University). Immunoreactive species were visualized by enhanced chemiluminescence detection (Pierce) of horseradish peroxidase-conjugated anti-mouse or anti-rabbit immunoglobulin G (Bio-Rad).
For immunoblot detection of whole-cell lysates, cell growth was stopped by the addition of 10 mM NaN3 and transfer to an ice bath. Cells were washed and resuspended directly in SDS-PAGE sample buffer, boiled for 10 min, subjected to glass bead homogenization for 2 min, and clarified by microcentrifugation for 2 min at maximum speed. Following SDS-PAGE and transfer to nitrocellulose, the membrane was probed with antibodies to phospho-p44/p42 MAP kinase at 1:500 (Cell Signaling Technology no. 9101L), phospho-Tyr (4G10; Upstate Cell Signaling Solutions no. 05-321), phospho-p38 MAP kinase at 1:500 (Cell Signaling Technologies no. 9211L), Gpa1 at 1:1,000 (38), Ste4 at 1:2,000 (from D. Jenness, University of Massachusetts), Sst2 at 1:2,000 (39), and Pgk1 at 1:75,000 (from J. Thorner) as indicated. Specificity of antibody detection was established using diploid cells (which do not express the G protein or RGS protein) or the corresponding gene deletion mutants.
Growth, transcription, and mating bioassays.
Analysis of RGS gene deletion and overexpression using the reporter-transcription assay was carried out as described previously (60). The methods used to screen the yeast deletion strain array and to analyze data have been published previously (18). Briefly, gene deletion mutants in the parental strain BY4741 were arrayed in 96-well plates provided by Research Genetics and transformed with the pheromone response reporter plasmid pRS423-FUS1-lacZ (60). β-Galactosidase activity assays were performed at mid-log-phase growth (corresponding to an A600 of 0.420 to 0.9) (60). β-Galactosidase activity values were normalized to an A600 of 0.8 and used to calculate the change in activity (n-fold) over that of the wild type, determined for the parent wild-type strain present on each plate. Dose-response curves (variable slope) were plotted and the 50% effective concentration (or concentration necessary to achieve 50% of the maximum response) (EC50) was calculated for each data set using the GraphPad Prism software package (GraphPad Software, San Diego, CA). Selected mutants were arrayed, retransformed, and reanalyzed at a wider range of α-factor concentrations as described above (60).
The quantitative mating assay was described previously (114). Briefly, 1 × 107 to 1.5 × 107 DC17 “tester” cells were mixed with 2 × 106 to 5 × 106 exponentially growing a cells containing an RGS gene deletion or overexpression plasmid (pAB27). The starting concentration of input α cells was measured by counting the number of viable colonies formed after serial dilution of the cultures. The mixtures were collected using a 0.45-μm 25-mm nitrocellulose filter (HAWP02500; Millipore Corp.) and SWINNEX filter cartridge (SX0002500; Millipore Corp.) and were placed on solid yeast extract-peptone-dextrose medium for 12 h. The filters were resuspended in sterile distilled water, serially diluted, and plated onto synthetic medium lacking uracil and all amino acids to select for diploids. Mating frequency for deletion mutants was expressed as the ratio of diploid colonies obtained for each mutant strain divided by the number of colonies obtained for the isogenic wild-type control strain (BY4741). Mating frequency with RGS gene overexpression was compared to that for the same wild-type strain containing the parent vector.
RESULTS
Multiple RGS proteins in yeast.
In the yeast Saccharomyces cerevisiae, Sst2 and Rgs2 have been identified previously as RGS proteins. Sst2 regulates the pheromone signaling pathway through mating factor receptors and the G protein α subunit Gpa1 (36). Rgs2 regulates dextrose signaling through a distinct receptor and Gα protein Gpa2 (123). We searched the yeast genome sequence database and identified two additional RGS homologues, Rax1 and Mdm1 (Fig. 1). Neither Rax1 nor Mdm1 has been characterized previously with respect to G protein signaling. Thus, we employed methods established for Sst2 to determine whether any of the other RGS proteins modulate Gpa1 signaling in vivo.
Initially, we tested the ability of each RGS protein to bind to Gpa1. Most RGS proteins act as GTPase-accelerating proteins and exert this function by stabilizing the transition state conformation of the G protein α subunit. RGS proteins that function as GAPs bind with low affinity to Gα in the GTP- or GDP-bound state but with high affinity to Gα in the presence of GDP-AlF4− (6, 119, 126), which functions as a transition state mimic (111, 117). Thus, preferential binding of an RGS protein to Gα-GDP-AlF4− correlates with GAP activity, as documented previously for Sst2 and Gpa1 as well as for Rgs2 and Gpa2 (2, 123).
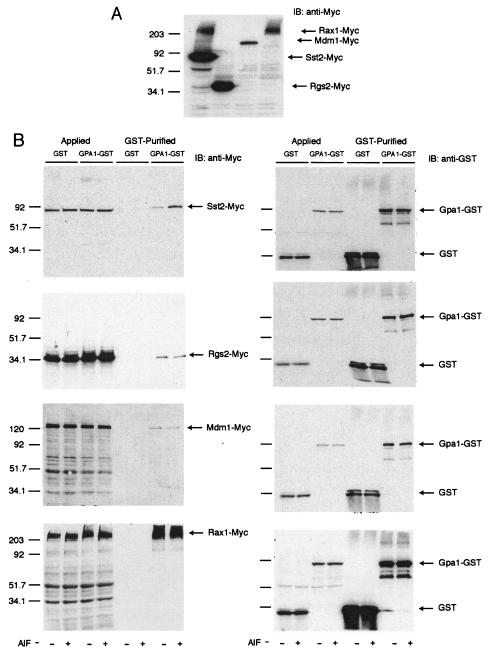
In order to determine which RGS proteins interact with Gpa1 in a guanine nucleotide-dependent manner, we fused full-length Sst2 (positive control), Rgs2, Rax1, and Mdm1 to the Myc epitope tag and fused Gpa1 to GST. The resulting RGS-Myc and Gpa1-GST fusions were coexpressed in yeast, and the GST fusion proteins were purified by glutathione-Sepharose affinity chromatography. GST alone was purified as a negative control, to detect nonspecific binding. The purified samples (Fig. 2, lanes labeled “GST-Purified”), as well as the starting material applied to each column (Fig. 2, lanes labeled “Applied”), were resolved by SDS-PAGE. Each RGS protein was detected by immunoblotting using the anti-Myc antibodies, and equal loading of each lane was confirmed using anti-GST antibodies. For unknown reasons, the majority of cellular Rax1 migrated well above the predicted molecular weight of 50,160 (Fig. 2A, lane 4). A similar (though less prominent) higher-molecular-weight form of Sst2 was also detected (Fig. 2A, lane 1) and has been demonstrated previously to result from Sst2 ubiquitination (53).
FIG. 2.
Binding of RGS proteins to Gpa1. Plasmid pRS316-ADH, containing Myc epitope-tagged SST2, RGS2, RAX1, or MDM1, was cotransformed with plasmid pAD4M, containing GPA1-GST or GST, into strain BJ2168. (A) Whole-cell lysates were resolved by 10% SDS-PAGE and detected by immunoblotting (IB) with anti-Myc antibodies. Note that the Rax1 sample ran anomalously whether or not the sample was boiled. (B) Cells were grown to mid-log phase and then lysed in the presence of GDP (−) or GDP+AlF4− (+) as indicated. Detergent-solubilized and clarified extracts were immobilized on glutathione-Sepharose, washed, and eluted with SDS-PAGE sample buffer. Total cell lysate (“Applied”) and retained protein (“GST-Purified”) were subjected to immunoblotting and probed with anti-Myc antibodies (left panels) or anti-GST antibodies (right panels) (to ensure equal loading), as indicated. The Applied sample represents ∼8% of the total cell lysate in every case. The arrows indicate bands specifically recognized by each antibody. Numbers at left of gels indicate molecular weights (in thousands). AlF, AlF4−.
As shown in Fig. 2B, all four RGS proteins bound detectably to Gpa1-GST. Whereas Sst2 binding was enhanced by the addition of GDP-AlF4− (2), the other RGS proteins exhibited equal binding in the presence of either GDP or GDP-AlF4−. Binding in the presence of GTP or GTP analogs was not tested because it is limited by the slow dissociation of GDP, whereas AlF4− binds to the GDP-occupied form of the G protein without the need for exchange (132). In no case was binding to GST alone detected. Thus, it appears that all four RGS proteins can bind to Gpa1, but Sst2 alone binds preferentially to the transition state conformation of this G protein.
As an additional control, we tested the ability of each RGS protein to bind Gpa2. Again, all four RGS proteins bound detectably to Gpa2-GST, but not to GST alone. In this case, Rgs2 binding was enhanced by the addition of GDP-AlF4− (123), while the other proteins exhibited equal binding in the presence of GDP or GDP-AlF4− (data not shown). Taken together, our data confirm that Sst2 binds the transition state conformation of Gpa1 and Rgs2 binds the transition state conformation of Gpa2. In addition, we show these are the only RGS-Gα pairs in yeast that distinguish between the transition state and the inactive state of either G protein. Other RGS proteins bind detectably to Gpa1 and Gpa2, but not in the manner of the canonical GAPs.
Multiple RGS proteins regulate Gpa1.
Having established that all four RGS proteins bind to Gpa1, we then examined whether any RGS proteins other than Sst2 influence pheromone signaling. Initially we measured mating efficiency in cells lacking individual RGS genes. Each mutant (as the a mating type) was mixed with an excess of α cells and then plated on medium to select for the growth of a/α diploids. Mating efficiency was calculated as the percentage of a cells that formed a viable diploid. The sst2Δ-mutant strain mated with about half the frequency of wild-type cells (Table 1), as reported previously (15). In contrast, deletion of RGS2, RAX1, or MDM1 each had a moderate enhancing effect on mating efficiency.
TABLE 1.
Effects of RGS protein expression on mating efficiencya
| Strain | Plasmid | Mating % (SEM %) |
|---|---|---|
| WT | 100 (4.4) | |
| sst2Δ | 55 (7) | |
| rgs2Δ | 120 (1.1) | |
| rax1Δ | 130 (1.5) | |
| mdm1Δ | 120 (9) | |
| WT | pAB | 100 (6) |
| WT | pAB-SST2 | 86 (10) |
| WT | pAB-RGS2 | 90 (5) |
| WT | pAB-RAX1 | 110 (8) |
| WT | pAB-MDM1 | 90 (7) |
Mating, mating efficiency was determined as the mating frequency of the mutant divided by the mating frequency of the WT control; control was defined as 100%. SEM is expressed as a percentage. Data are representative of assays performed three times in triplicate.
A second measure of pheromone response is the growth inhibition plate assay (halo assay). In this method a nascent lawn of cells is exposed to a point source of α-factor, and the resulting zone of growth inhibition, or halo, provides a measure of pheromone sensitivity over several days. The SST2-deficient strain is considerably more sensitive to pheromones, producing much larger zones of growth inhibition surrounding the source of α-factor, as reported previously (15, 16). In contrast, and in accord with the mating assay, the RGS2-, RAX1-, and MDM1-deficient strains all produced halos very similar to those of the wild-type strain (data not shown).
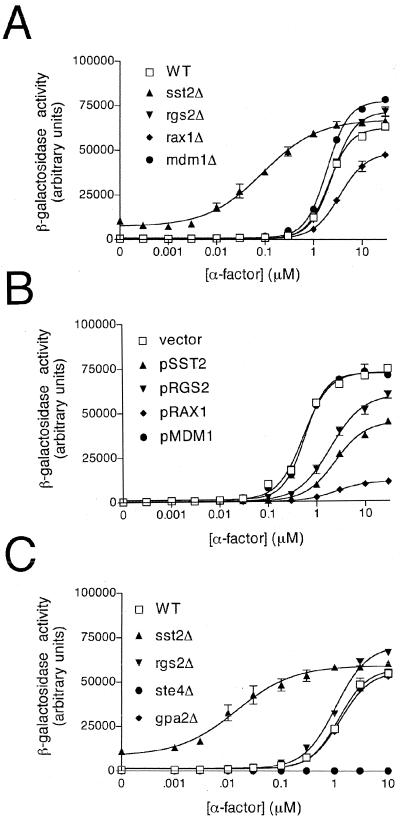
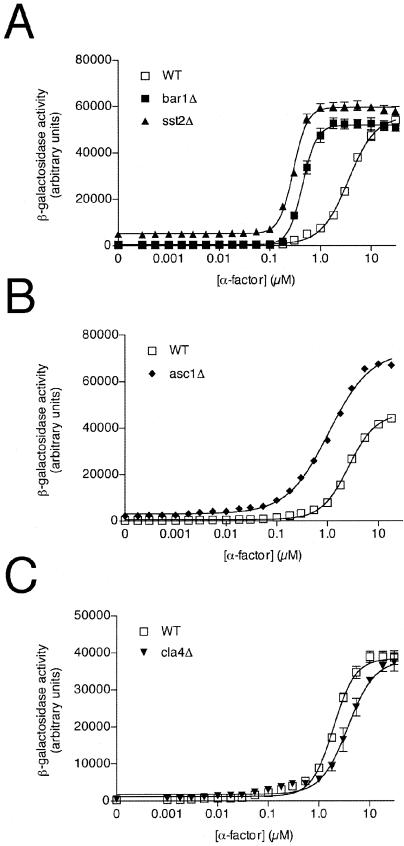
A third measure of pheromone signaling is the reporter transcription assay, which provides quantitative information about pheromone sensitivity over a relatively short time. For this we used the pheromone-inducible FUS1 promoter and lacZ (β-galactosidase) reporter gene. As shown in Fig. 3A and in Fig. 4, the mdm1Δ and rgs2Δ mutants produced a modest increase in maximum responsiveness (efficacy), while the rax1Δ mutant showed a significant reduction in this activity. The rax1Δ strain was also unique in that it exhibited an increase in the EC50 for pheromone (i.e., decreased potency). All of these effects were in striking contrast to the SST2-deletion mutant, which exhibited a >10-fold decrease in the EC50 as well as an increase in basal activity (activity in the absence of added pheromone). These data reveal a minor role for Mdm1, Rgs2, and Rax1 in pheromone-dependent gene regulation. Moreover, the phenotypes exhibited by the three mutants are distinct from that of the sst2Δ strain, suggesting that these other RGS proteins function in a manner different from Sst2.
FIG. 3.
Pheromone response after deletion or overexpression of RGS proteins. (A) Wild-type (WT) cells and the indicated mutant cells were transformed with a plasmid containing the pheromone-inducible FUS1 promoter and lacZ reporter gene (pRS423-FUS1-lacZ), grown to mid-log phase, and treated with the indicated concentration of α-factor for 90 min. β-Galactosidase activity was measured spectrofluorometrically and expressed as arbitrary fluorescence units. (B) Wild-type cells that overexpress the indicated RGS protein (in plasmids pAB27 or pAB23), cotransformed with the FUS1-lacZ reporter, were assayed for β-galactosidase activity as described above. (C) Transcription reporter assay control experiments. Wild-type cells and the indicated gene deletion mutants were assayed for β-galactosidase activity as described above. Data shown are typical of three independent experiments performed in triplicate. Error bars indicate standard errors of the means (SEM).
FIG. 4.
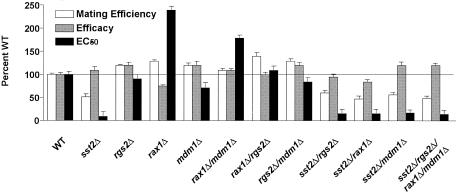
Pheromone response after deletion of RGS proteins alone or in combination. WT cells and the indicated mutant cells were assayed for mating efficiency and also transformed with the FUS1-lacZ reporter and assayed for β-galactosidase activity as described in the legend to Fig. 3. Mating efficiency as well as pheromone efficacy and potency was calculated, as described in Materials and Methods. Data shown are expressed as a percentage of WT strain activity and are typical of three independent experiments performed in triplicate. Error bars indicate SEM. The horizontal line indicates 100% of wild type.
The data presented above indicate that all four RGS proteins regulate Gpa1 signaling activity. However, unlike Sst2, the other RGS proteins (i) do not distinguish between the transition state and inactive conformations of the G protein, (ii) increase rather than decrease mating efficiency, (iii) do not alter the growth arrest response, (iv) do not alter basal transcriptional activity, and (v) exhibit unique changes in pheromone potency and/or efficacy. If the mechanisms of action are indeed different, we would predict that deletion of multiple RGS proteins should produce nonoverlapping or additive effects on signaling. To test this model we constructed all possible combinations of double mutants, as well as a quadruple mutant lacking all four RGS-encoding genes, and then measured pheromone responses for each. Any multigene deletion mutant lacking SST2 exhibited a larger halo, decreased EC50, and an elevated basal response, as seen with sst2Δ alone (Fig. 4 and data not shown). Thus, in every case where SST2 was deleted, the robust mutant phenotype prevailed over that of any other deletion, and significantly quantifiable differences were not observed. For the other multigene deletions, however, approximately additive effects were observed in the mating efficiency and reporter transcription assays (Fig. 4). These data further support our model of distinct modes of regulation by each of the four RGS proteins, but with Sst2 as the principal regulator of the pathway.
We then examined whether high-level expression of any RGS protein affects pheromone signaling. Overexpression of a gene product typically results in a phenotype opposite that of the gene disruption but can sometimes reveal additional functions not evident from simple knockout mutations. We first examined whether RGS overexpression affects mating efficiency, and we observed no appreciable differences for any of the genes, including SST2 (Table 1). We then examined the effects of RGS overexpression, using the growth inhibition halo assay. Overexpression of SST2 led to a reduced arrest response as previously reported (37), while overexpression of the other RGS genes had no effect (data not shown). Finally, we measured pheromone response using the reporter-transcription assay. In this case, overexpression of Sst2, Rgs2, or Rax1 diminished pheromone potency and, most dramatically, pheromone efficacy, while overexpression of Mdm1 had no effect (Fig. 3B). The effects of Rax1 were especially strong, equaling or exceeding even those of Sst2 (it is worth noting that of the three “atypical” RGS proteins tested, Rax1 appeared to bind with the highest affinity to Gpa1 [Fig. 2]). Taking these results together, it appears that deletion of SST2 or RGS2 can enhance the maximum transcription response slightly, while overexpression of these proteins diminishes the response. Deletion of MDM1 promotes signaling slightly, while overexpression has no effect. Either deletion or overexpression of RAX1 diminishes the pheromone signal.
The results described above reveal that all four RGS proteins can—in distinct ways—alter Gpa1-mediated signaling events. These effects are best detected using the reporter transcription assay, which is more quantitative, more sensitive, and, presumably, more selective than the longer-term growth arrest or mating efficiency assays. To confirm that the transcription reporter assay is indeed specific for the Gpa1 pathway (and not that of Gpa2), we performed several control experiments using activated or inactive mutants of each G protein and monitored signaling using the reporter transcription assay. First, we examined the transcription response in a gpa1Δ mutant. Deletion of GPA1 ordinarily results in constitutive growth arrest (through uncontrolled release of Gβγ) but can be maintained by placing a downstream effector kinase under the control of an inducible promoter (83, 88). The gpa1Δ mutation resulted in an increase in FUS1-lacZ activity similar to that observed in wild-type cells treated with pheromone (50) (data not shown). Conversely, ste4Δ-mutant cells, which lack the Gβ subunit associated with Gpa1, were completely unresponsive to pheromone (129) (Fig. 3C). Finally, cells lacking the only other Gα subunit in yeast (GPA2) responded normally, in a manner indistinguishable from wild-type cells and in striking contrast to the RGS2-deficient cells (Fig. 3C). These results strongly suggest that the changes in FUS1-lacZ expression reflect the activation state of Gpa1 and not Gpa2.
Additional regulators of G protein signaling.
Having established that Sst2 is the principal RGS regulator of Gpa1, we sought to determine whether other proteins regulate the same signaling pathway, perhaps in a manner distinct from Sst2. Indeed, in mammalian cells, G protein signaling is regulated by a number of non-RGS proteins. For example, phospholipase C-β1 is a downstream effector of Gq but also serves to regulate pathway activation by accelerating Gqα GTPase activity (9). Likewise, GRK2 (G protein-coupled receptor kinase 2) can downregulate G protein signaling by phosphorylation and inactivation of the receptor as well as binding to Gα in competition with effectors (13, 32). Considering the precedent of multiple alternative modes of G protein signal modulation, we anticipated that additional proteins might also modulate G protein signaling in yeast.
To this end, we measured the pheromone response in each of 4,847 available yeast gene deletion mutants, representing nearly all nonessential genes in the genome. To conduct this analysis on several thousand strains, we adapted the reporter-transcription assay described above to a 96-well microplate format and employed a robotic liquid handling system (18). Mutant strains in microplates were transformed with the FUS1-lacZ reporter and then exposed to five different concentrations of α-factor pheromone or left untreated, in triplicate. From the resulting β-galactosidase activity, we tabulated for each mutant the EC50 and the maximum response to pheromone, as well as the basal activity. Any mutant whose results differed from the wild type by more than twofold and at least 2 standard deviation units in either the EC50 or the maximum response, or by more than fivefold in basal activity, was re-arrayed and retested. (See the supplemental material for a complete list of the 90 mutants that showed a reproducibly altered signaling phenotype after two rounds of testing in the 96-well format.) A subset of these mutants were validated further as described below and ultimately classified into six functional groups. Group I consists of five validated strains that exhibited a decrease in the EC50 for α-factor. These include known pathway components sst2Δ, bar1Δ (also known as sst1Δ), and ubc4Δ, as well as two novel mutants with a similar phenotype, ygl024wΔ and asc1Δ. Group II consists of one validated mutant, cla4Δ, that exhibited an increase in the EC50 (Table 2). Group III mutants exhibited no pheromone response whatsoever. These include mutants shown previously to block or diminish the α-factor response (ste2Δ, ste4Δ, ste5Δ, ste7Δ, ste8Δ/sir3Δ, ste9Δ/sir4Δ, ste11Δ, ste50Δ, and sir2Δ) (Table 3). Mutants with several additional known-sterile genes were not present in the strain collection (ste12Δ, ste18Δ) or failed to reach an appropriate cell density and were not tested (ste20Δ). Group IV consists of 38 mutants that exhibited a >50% decrease in maximum responsiveness, compared with the wild-type strain. Group V contains six mutants with a more than twofold increase in maximum responsiveness. Group IV and V mutants were shown to have reproducible phenotypes, but they were not validated or characterized further (see Table S1 in the supplemental material). Finally, group VI contains four mutants with a more than fivefold elevation in basal activity; of these, sst2Δ and gas1Δ were previously shown to exhibit pheromone-independent signaling (25, 105) (Table 4). The identification of known sterile, supersensitive, and constitutively active mutants confirmed the integrity of the strain collection and the validity of the screening method used.
TABLE 2.
Group I and II mutants alter pheromone potencya
| Group | Strain | EC50 (fold vs WT) | Efficacy (fold vs WT) | Halo size | Plasmid rescue? |
|---|---|---|---|---|---|
| I | yil015w (bar1Δ) | 7.9 | 0.93 | ++ | Yes |
| I | ylr452c (sst2Δ) | 12.0 | 1.1 | ++ | Yes |
| I | ygl024w (ygl024wΔ) | 3.3 | 1.3 | + | Yes |
| I | ymr116c (asc1Δ) | 2.5 | 1.6 | + | Yes |
| I | ybr082c (ubc4Δ) | 1.9 | 1.3 | + | ND |
| II | ynl298w (cla4Δ) | 0.52 | 1.0 | = | Yes |
Group I (lower EC50) and group II (higher EC50) mutants retested over a wide range of α-factor concentrations to quantitate EC50 and maximum response (“Efficacy”). EC50 and efficacy are expressed as the ratio of mutant and WT activities, where the activity of the fully stimulated wild type equals 1. Halo size is expressed as larger (+) or smaller (−) than the wild type. ND, not done.
TABLE 3.
Group III mutants are pheromone-unresponsivea
| Gene (strain) | ID? | Role |
|---|---|---|
| STE2 (YFL026W) | YES | α-factor receptor |
| STE3 (YKL178C) | NA | a-factor receptor |
| STE4 (YOR212W) | YES | G protein β subunit |
| STE5 (YDR103W) | YES | Kinase scaffold |
| STE6 (YKL209C) | NT | a-factor export |
| STE7 (YDL159W) | YES | MAP kinase kinase |
| STE8/SIR3 (YLR442C) | YES | Transcription silencing |
| STE9/SIR4 (YDR227W) | YES | Transcription silencing |
| STE11 (YLR362W) | YES | MAP kinase kinase kinase |
| STE12 (YHR084W) | NT | Transcription factor |
| STE13 (YOR219C) | NA | α-factor processing |
| STE14 (YDR410C) | ND | a-factor processing |
| STE16/RAM1 (YDL090C) | NA | a-factor processing |
| STE18 (YJR086W) | NT | G protein γ subunit |
| STE20 (YHL007C) | ND | MAP kinase kinase kinase |
| STE21/MSN5 (YDR335W) | NO | Nuclear import/export |
| STE22/AXL1 (YPR122W) | NA | a-factor processing |
| STE23 (YLR389C) | NA | a-factor processing |
| STE24 (YJR117W) | YES | a-factor processing |
| STE50 (YCL032W) | YES | Ste11 regulator |
| SIR2 (YDL042C) | YES | Transcription silencing |
List of genes known to be required for pheromone signaling were identified (“ID”) by α-factor screening (“YES”) or unidentified because they are required for a-factor responsiveness or for pheromone processing and are not applicable (NA) or unidentified because they were absent from the collection and not tested (NT) or strains containing them failed to grow and so they were not done (ND). Deletion of STE21 results in partial sterility (http://www.yeastgenome.org).
TABLE 4.
Group VI mutants are constitutively activea
| Gene | ORFΔ | α-MF | ORF+ STE+ | orfΔ STE+ | orfΔ ste2Δ | orfΔ ste4Δ | orfΔ ste7Δ | orfΔ ste12Δ |
|---|---|---|---|---|---|---|---|---|
| ylr452c | sst2Δ | − | 1.0 | 12.3 | ||||
| + | 143 | 189 | ||||||
| ygl024w | − | 1.0 | 11.4 | 10.6 | 1.7 | 1.4 | 0.8 | |
| + | 161 | 188 | 8.2 | 1.6 | 1.2 | 0.76 | ||
| yjl128c | pbs2Δ | − | 1.0 | 6.7 | 26.2 | 0.3 | 0.3 | 0.3 |
| + | 269 | 280 | 23.3 | 0.51 | 0.35 | 0.57 | ||
| ymr307w | gas1Δ | − | 1.0 | 14.1 | 26.8 | 0.15 | 0.18 | 0.14 |
| + | 116 | 146 | 29.2 | 0.19 | 0.20 | 0.23 |
Group VI gene (“ORF”) or gene deletion mutants (“ORFΔ”) in mating-competent (“STE+”) or the indicated sterile mutant strain background. Values represent reporter transcription activities expressed as ratios, where activity of the unstimulated WT equals 1.0. Complete data for sst2Δ have been published elsewhere (105).
The proportion of all strains tested was approximately 89%. This value was calculated as the number of mutants for which full dose-response profiles were obtained (4,349) divided by the total number of mutant strains available (4,847). Of the strains not analyzed, 54 failed to transform and the remainder grew poorly and failed to reach a suitable cell density even after two or more attempts. The identity of strains not tested is provided in Table S2 in the supplemental material.
We focused our initial efforts on mutants that alter the EC50 for pheromone, since this is the parameter most affected by deletion of the prototypical G protein regulator SST2. To obtain more accurate EC50 values, we retested the best-performing strains from group I (eight mutants originally) and group II (three mutants originally) over a wider range of α-factor concentrations (0.001 to 100 μM). Examples of these higher-resolution dose-response curves are presented in Fig. 5. In these experiments, an increase in agonist potency was evident for group I mutants, in order of magnitude as follows: sst2Δ, bar1Δ, ygl024wΔ, asc1Δ, and ubc4Δ (Table 2). Note that, upon retesting, the activity of the ubc4Δ strain fell just below the twofold threshold. The group II mutant cla4Δ exhibited an ∼50% reduction in agonist potency (Table 2). Additional mutants from groups I and II produced a reproducible phenotype but could not be validated using a genetic complementation test (see below); these putative false positives are listed in Table S1 in the supplemental material but are not listed in Table 2 and were dropped from further consideration.
FIG. 5.
Pheromone response after deletion of group I or group II genes. WT cells and the indicated mutant cells were transformed with the FUS1-lacZ reporter plasmid and treated with the indicated concentration of α-factor for 90 min. (A) Wild-type strain and group I control mutants sst2Δ and bar1Δ. (B) Wild-type strain and representative group I mutant asc1Δ. (C) Wild-type strain and the group II mutant cla4Δ. Data shown are typical of three independent experiments performed in triplicate. Error bars indicate SEM.
To further validate the mutants we performed a growth arrest assay on the 90 strains identified in our initial screen (see Table S1 in the supplemental material). As expected for group I mutants, substantially larger-than-normal halos were observed for, in order of magnitude, sst2Δ, bar1Δ, asc1Δ, and ubc4Δ. The group II mutant cla4Δ responded much like the wild-type strain (Table 2).
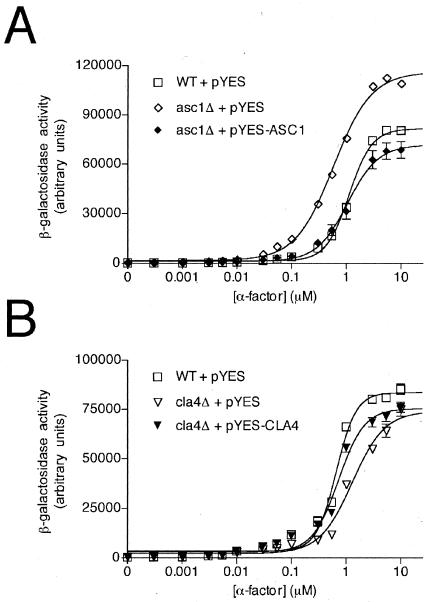
We then investigated whether the phenotype of each mutant from group I and group II could be reversed by transformation with a plasmid-borne copy of the wild-type gene. This control was necessary to ensure that each deletion was properly constructed and catalogued and that the observed phenotype was due to the original mutation and not due to a suppressor mutation or another secondary mutation. Each gene (including the native promoter and transcription termination sequence) was PCR amplified and cloned into a yeast expression vector. These expression plasmids were then transformed together with the FUS1-lacZ reporter plasmid into the corresponding deletion strain and tested using the growth arrest and reporter transcription assays. Plasmid-borne expression of the wild-type gene conferred wild-type growth arrest and transcription induction responses to the asc1Δ, ygl024wΔ, and cla4Δ mutants as well as the control mutants bar1Δ and sst2Δ (Table 2). Figure 6 shows rescue data for a representative group I mutant (asc1Δ) and the sole group II mutant cla4Δ. Rescue was not observed for yjl123cΔ, eap1Δ, ylr068wΔ, or trf5Δ (see Table S1 in the supplemental material). Considering that each plasmid-borne gene was fully sequenced and each gene deletion was authenticated by PCR, it appears that a second genomic mutation was responsible for the phenotype observed in these strains. For trf5Δ, the signaling defect appears to result from alterations in CLA4. Within the yeast genome, TRF5 and CLA4 are juxtaposed, suggesting that deletion of TRF5 interferes with the expression of CLA4. For yjl123cΔ, the additional mutation appears to be in SST2, since in this case full rescue was observed with transformation of the SST2 expression plasmid (data not shown).
FIG. 6.
Plasmid rescue of group I or group II mutants. WT cells and the indicated mutant cells were transformed with the FUS1-lacZ reporter plasmid and either an empty vector (pYES) or the same vector containing the absent gene, as indicated. (A) Wild-type strain and the group I mutant asc1Δ. (B) Wild-type strain and the group II mutant cla4Δ. Data for the remaining mutants are summarized in Table 2.
We then examined the function of the group VI mutants sst2Δ, pbs2Δ, gas1Δ, and ygl024wΔ, all of which exhibited pheromone-independent signaling activity. Since sst2Δ and ygl024wΔ exhibited an elevated basal signal as well as increased pheromone sensitivity, these two mutants met the criteria of both group I and group VI mutants. We first attempted to establish where in the pathway this activation occurs. For example, high basal transcriptional activity could result from hyperactivation of the receptor, G protein, effector kinase, or transcription factor. A similar analysis indicated previously that deletion of SST2 promotes a moderate level of constitutive activity and that this activity was dependent on components downstream of the receptor (105). To determine whether the novel mutants functioned similarly, the activities of pbs2Δ, gas1Δ, and ygl024wΔ strains were examined in cells lacking either the pheromone receptor (ste2Δ), G protein β subunit (ste4Δ), effector MAP kinase kinase (ste7Δ), or the transcription factor (ste12Δ). Elevated basal activity was detected for all three mutants in an otherwise mating-competent (STE+) strain, as well as in the absence of the receptor gene STE2 (Table 4). However, basal activity was not elevated in cells lacking STE4, STE7, or STE12. As expected, all of the ste− mutants also lost the ability to respond to α-factor pheromone. We conclude from this analysis that Ygl024w, Pbs2, and Gas1 regulate the pheromone pathway downstream of the receptor and most likely at the level of the G protein.
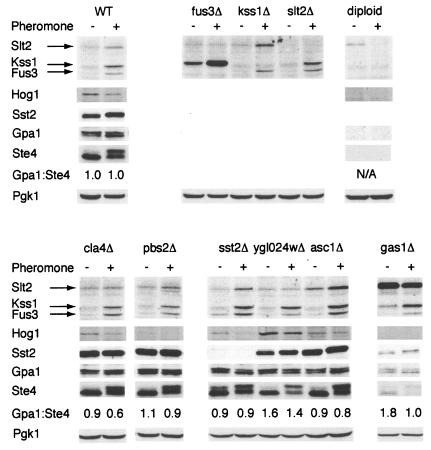
We then considered possible mechanisms by which the group VI mutants might increase basal activity, all of which have some precedent in the literature. Initially, we considered whether any mutant altered the expression of Sst2, Gpa1, or Ste4. Deletion of SST2 was shown previously to result in pheromone-independent activation of the mating pathway, as described above (105). An even more dramatic response occurs upon deletion of GPA1 or overexpression of STE4 (24, 34, 83, 92, 130). Thus, a reduction of Sst2 or a change in the relative abundance of Gpa1 versus Ste4 could account for the constitutive signaling phenotype of the group VI mutants. To test these possibilities, lysates from wild-type and constitutively active mutant cells (group VI), as well as from all validated group I and II mutants, were resolved by gel electrophoresis and immunoblotting using antibodies against Gpa1, Ste4, and Sst2. Equal loading of each lane was confirmed using anti-Pgk1 antibodies. In most of the mutants, all three proteins were expressed at normal levels, making it unlikely that the constitutive signaling phenotype resulted from altered expression of the RGS or G protein (Fig. 7), but there were two exceptions. First, in the gas1Δ strain the apparent expression levels of Gpa1, Ste4, and Sst2 were reduced to similar degrees (Pgk1 expression was only slightly diminished). Perhaps this is a result of the upregulation of the cell integrity pathway with the deletion of GAS1 as a competing signaling pathway (97). Second, in the ygl024wΔ strain, we observed a substantial decrease in Sst2 abundance, with no alteration in Gpa1, Ste4, or Pgk1. Indeed, ygl024wΔ resembles sst2Δ in having elevated basal signaling as well as a leftward shift in the pheromone dose-response profile. These data, together with the genetic epistasis data presented above, suggest that deletion of the YGL024W open reading frame regulates the pheromone pathway by promoting full expression of Sst2. However, since Ygl024w does not resemble any other known protein, the mechanism of action is not obvious. Unfortunately, our repeated attempts to epitope tag the protein were unsuccessful, making it difficult to characterize potential interactions with the G protein or Sst2.
FIG. 7.
Expression of G protein components and activated MAP kinases. For immunoblot detection of phosphorylated MAP kinases, WT cells and the indicated mutant cells were grown to mid-log phase, treated for 1 h with (+) or without (−) 2.5 μM α-factor pheromone as indicated, lysed, and resolved by SDS-PAGE and immunoblotting. Membranes were probed with antibodies to phospho-p44/p42 (to detect phosphorylated Fus3, Kss1, and Slt2), phospho-Tyr (to detect phosphorylated Hog1), Sst2, Gpa1, Ste4, or Pgk1 (loading control), as indicated. Specificity of antibody detection was established using the corresponding gene deletion mutant and/or diploid cells (which do not normally express Sst2, Gpa1, Ste4, or Fus3) (data not shown). Gpa1:Ste4, ratio of Gpa1 and Ste4 expression, as calculated by scanning densitometry; N/A, not analyzed.
We then considered whether any of the group I, II, or VI mutants could alter signaling by MAP kinases other than Fus3. To this end, we examined the phosphorylation state of Kss1 (which promotes invasive growth and, under some circumstances, can substitute for Fus3), Hog1 (which functions in the high-osmolarity glycerol pathway), and Slt2 (which functions in the cell integrity pathway). It has been shown previously that deletion of Hog1 or its upstream activator, the MAP kinase kinase Pbs2, blocks the high-osmolarity response but also allows spurious activation of the mating pathway (52, 94, 118). Similarly, deletion of GAS1 was previously shown to result in activation of the cell integrity pathway, including hyperphosphorylation of the MAP kinase Slt2 (14, 31, 67, 96), as well as spurious activation of the mating response pathway (25). Thus, whole-cell extracts were probed by immunoblotting using phospho-p42/p44 antibodies, which recognize the dually phosphorylated and activated forms of Fus3, Kss1, and Slt2, as well as with antibodies against phospho-p38, which detect the phosphorylated form of Hog1. As shown in Fig. 7, pheromone treatment of wild-type cells resulted in increased phosphorylation of Fus3, Kss1, and Slt2 but decreased phosphorylation of Hog1. In the sst2Δ strain it appears the MAP kinases Fus3, Kss1, and Slt2 were activated to a greater degree than in the wild-type strain; however, it was interesting to note that in the absence of pheromone, Fus3 alone appeared to be upregulated. In the gas1Δ strain there was a striking increase in basal phosphorylation of Slt2 (Fig. 7); this phenomenon has been noted before, and suggests that cell wall defects activate the cell integrity MAP kinase signaling pathway (14, 67) and perhaps indirectly activate the pheromone response pathway. In addition, the GAS1 null mutant showed activation of Kss1 in the absence of pheromone, suggesting that the constitutive upregulation of the yeast pheromone pathway could be mediated through Kss1. However, fus3Δ strains demonstrate even more robust activation of Kss1, both with and without the addition of pheromone, mitigating this possibility and supporting the model that pheromone pathway activation is mediated through, or occurs in concert with, Slt2 activation. Slt2 phosphorylation was also elevated slightly in the asc1Δ strain, and Hog1 phosphorylation was enhanced in the ygl024wΔ strain. Thus, it is possible that asc1Δ and ygl024wΔ mutants promote spurious activation of the mating pathway via dysregulation of Slt2 or Hog1.
DISCUSSION
Previous genetic studies in yeast have revealed a large number of components required for signal propagation. Most were identified through the isolation of mating-deficient, or sterile (ste), gene mutations. Examples include genes that encode the α-factor pheromone receptor (STE2), G protein βγ subunits (STE4 and STE18), downstream protein kinases (STE20, STE11, and STE7), kinase-binding proteins (STE5 and STE50), and a transcription factor (STE12) (40).
Yeast genetic analysis has also led to the discovery of several components required for signal modulation, most notably BAR1 (SST1) and SST2. The BAR1 gene encodes a secreted pepsin-like protease that cleaves and inactivates α-factor (78). The SST2 gene product is the founding member of the RGS protein family and is known to accelerate Gpa1 GTPase activity (2). Cells deficient in either BAR1 or SST2 exhibit sustained activation of the Gpa1 signaling pathway and an inability to recover from prolonged pheromone stimulation (15, 16).
A second RGS protein in yeast, Rgs2, was identified as a multicopy suppressor of Gpa2-dependent loss of heat shock resistance in stationary-phase cells and was shown also to accelerate GTP binding and hydrolysis by Gpa2 (123). The role of the remaining two RGS protein family members, Rax1 and Mdm1, has not been characterized previously with respect to G protein signaling.
Acceleration of GTPase activity is likely the predominant mechanism by which RGS proteins regulate Gα signaling in vivo. However, there is growing evidence that RGS proteins can regulate signaling even in the absence of GAP activity (62, 89). Regardless of the mechanism, it is important to know whether a given RGS protein regulates a specific Gα subtype and, if so, under what circumstances and, ultimately, by what mechanism. A related question is whether other proteins also contribute to G protein-mediated signaling. Even in the genetically tractable yeast system, few such regulatory factors have been identified or characterized.
Our objective here was to identify new pheromone response regulators through systematic analysis of all RGS proteins and nearly all other nonessential genes in yeast. Our approach was to construct cells that lack the four RGS-encoding genes, individually or in combination, and compare their abilities to regulate the well-characterized Gpa1 signaling pathway. We found that Sst2 alone binds preferentially to the transition state conformation of Gpa1, while the other RGS proteins bind equally to the transition state and inactive-state conformations. Sst2 functions primarily to reduce basal signaling and to reduce pheromone potency, while Rgs2 and Rax1 regulate pheromone efficacy.
Based on our detailed understanding of pathway regulation by Sst2, we devised a strategy to also assess regulation by other nonessential, non-RGS genes in yeast. We found that asc1Δ and ygl024wΔ mutants reduce the EC50 for pheromone stimulation, in the manner of the known regulatory mutants sst2Δ and bar1Δ. Conversely, we found that deletion of CLA4 results in an unusual increase in the EC50 for pheromone. Additional mutants exhibit some degree of pheromone-independent signaling; these include those with deletions of PBS2 and YGL024W as well as the previously documented GAS1 and SST2 (25, 105). These findings suggest that although Sst2 is the principal regulator of Gpa1 signaling, many other proteins also modulate pathway activity.
Our analysis benefited from powerful genomic tools, many of which are unique to the yeast system. With the sequencing of the yeast genome, all RGS domain-containing proteins could be readily identified through sequence similarity analysis. With the deletion of most open reading frames, new regulatory factors could be screened in a systematic manner (10, 49, 131). Since each mutant has been arrayed and annotated, new pathway components could be identified immediately and with few false negatives. By using a highly specific reporter, only pathway components would be identified and with few false positives. By using a full range of pheromone concentrations, even small differences in signal responsiveness could be detected and quantified.
A systematic approach has also been used to study the function of all RGS proteins in Caenorhabditis elegans (41, 51, 56). It has long been known that the RGS protein EGL-10 inhibits signaling by GOA-1 (Goα) (65). Activation of GOA-1 leads to diminished egg-laying and locomotor functions, and these activities are amplified in egl-10-deficient mutants. To identify other RGS regulators of GOA-1, Koelle and colleagues overexpressed each of the 13 RGS-homologous proteins in C. elegans and examined their effects on Goα-dependent signaling. By this approach they found two, the products of rgs-1 and rgs-2, that mimic the effects of egl-10 overexpression. The ability of RGS-1 and RGS-2 to regulate GOA-1 was confirmed by isolating loss-of-function mutants for the RGS genes, as well as by demonstrating that they too are needed for proper egg-laying behavior; however, the effects of rgs-1 and rgs-2 mutants differ from those for egl-10, in that they are only manifest following a period of starvation and refeeding (41, 56). These findings were interpreted to mean that EGL-10 controls “baseline” signaling while RGS-1 and RGS-2 modulate signaling under specific physiological circumstances. We observed a similar pattern of activity in yeast; whereas Sst2 controls basal signaling as well as signaling at low doses of pheromone, the other RGS proteins modulate signaling exclusively at high doses of pheromone.
An unresolved question is how RGS proteins that fail to bind preferentially to the transition state form of Gpa1 can nevertheless regulate Gpa1 signaling. One possibility is that these proteins have a function similar to GRK2. GRK2 was originally identified as a kinase that phosphorylates and desensitizes G protein-coupled receptors but was later shown to also have an N-terminal RGS homology domain that binds specifically to Gqα (13, 101, 104). In contrast to most other RGS proteins, however, GRK2 binds with high affinity to the active conformation as well as the transition state conformation of the Gqα protein and has a negligible effect on GTP hydrolysis (13, 101). GRK2 appears to function primarily by blocking Gqα coupling to its effector enzyme, phospholipase Cβ. The structural basis for this activity was first deduced from the available crystal structure of the RGS4-Giα complex. Whereas Giα binds to the so-called “A-site” of the RGS core domain (and in so doing stabilizes the transition state and reduces the activation energy for GTP hydrolysis) (115, 119), mutagenesis studies revealed that Gqα binds instead to a distinct site within the RGS core domain of GRK2 designated the “C-site” (116). Binding in this case would not be expected to influence the conformation of the G protein or to alter GTP hydrolysis but could easily account for the ability of GRK2 to diminish G protein-mediated signaling by effector antagonism.
Competition for Gα binding to effectors has likewise been reported for one of the more typical RGS family members, RGS2 (13, 55). Moreover, there is at least one report of a mammalian RGS protein that can directly bind and inhibit an effector enzyme, adenylyl cyclase (108). RGS proteins have also been reported to bind to receptors (8, 109), possibly in competition with Gα (8), as well as to Gβγ, possibly in competition with Gα or effectors (42, 103, 119, 125). Regardless of mechanism, inhibition of G protein subunit association will indirectly prevent receptor activation, since receptors recognize only the assembled G protein heterotrimer (127). Currently we are attempting to determine which, if any, of these mechanisms account for the ability of Rgs2, Rax1, and Mdm1 to modulate the Gpa1 signal.
Our analysis also revealed a number of non-RGS domain-containing regulators of the Gpa1 signaling pathway. Of these, Bar1, Ubc4, and Gas1 have been described before. Bar1 is a secreted pepsin-like protease that cleaves and inactivates α-factor pheromone (78). Like SST2, deletion of BAR1 dramatically amplifies the α-factor pheromone response. UBC4 encodes an E2 ubiquitin-conjugating enzyme that promotes pheromone-stimulated ubiquitination and down-regulation of the α-factor receptor Ste2 (57). GAS1 encodes a glycosylphosphatidylinositol-anchored β-1,3-glucanosyltransferase localized to the external face of the plasma membrane (85, 86, 93). A gas1 mutant exhibits anomalous cell wall composition, including reduced β-glucan and increased chitin and mannan content, suggesting a role in proper cell wall assembly (12, 68, 97, 122). In agreement with this idea, gas1 mutants exhibit chronic hyperactivation of Slt2, which is the MAP kinase that regulates the cell integrity pathway (14, 31, 67, 96).
We also identified genes that had not been previously associated with the yeast pheromone signaling pathway. YGL024W is a novel gene with no known function or obvious homologues. It encodes a 111-amino-acid protein with two putative transmembrane domains. However, there is uncertainty whether YGL024W is an authentic gene, and it has been designated as “dubious” by the Saccharomyces Genome Database (http://www.yeastgenome.org/). Neither the gene nor its deduced translation product is conserved in any other species. Further, the YGL024W open reading frame overlaps the promoter and 5′-coding regions of the PGD1 gene present on the complementary Crick strand. PGD1 encodes a protein reported to mediate RNA polymerase II activity (87). Thus, it is possible that loss of YGL024W disrupts mRNA synthesis and, in this manner, modulates expression of some protein involved in pheromone signaling. On the other hand, deletion of the PGD1 gene did not result in a signaling phenotype in our high-throughput screen. Moreover, we observed full rescue of the ygl024wΔ-mutant phenotype following plasmid-borne expression of the YGL024W gene, as demonstrated by both the reporter transcription and halo assays (data not shown). There are several possibilities that might explain these results. First, there may be a library annotation error or a secondary suppressor mutation within the pgd1 mutant strain. Indeed, secondary mutations were detected in multiple strains, as evidenced by the numerous false-positive sterile mutants identified. Another possibility is that the N-terminal and C-terminal domains of Pgd1 affect pheromone signaling by independent and functionally antagonistic mechanisms. According to this model, the Pgd1 N-terminal domain is a negative regulator and the C-terminal domain is a positive regulator of the pathway. Deletion of YGL024W removes the N-terminal portion of the Pgd1 protein and enhances signaling activity, while deletion of the entire PGD1 gene removes both the N-terminal and C-terminal portions, and signaling is unaffected. Despite numerous attempts, we have been unable to epitope tag Ygl024w and therefore unable to demonstrate expression or characterize the protein in situ. Thus, we have been unable to establish how YGL024W affects signaling, and considerable further investigation will be needed to fully resolve all of these complex issues.
Another novel regulator gene, ASC1, encodes a protein with seven WD40 repeats (17), a domain structure found in all G protein β subunits (110). Asc1 is functionally and structurally similar (54% sequence identity) to mammalian RACK1 (receptor of activated C kinase 1) (17, 59), a protein that is best known to serve as an adapter for protein kinase C (102). RACK1 has also been shown to interact with a variety of other proteins, including dynamin-1, Src, and β-integrins, all of which have been suggested to bind directly or indirectly to G proteins (102, 121). More recently, reports from the laboratory of Hamm and colleagues have reported that RACK1 binds directly with Gβγ subunits, either alone or in complex with Gα (21, 22, 30). We have obtained evidence that Asc1 functions similarly, associating preferentially with the inactive form of yeast G protein heterotrimers (C. Zeller and H. G. Dohlman, unpublished data). While the functional significance of these interactions is not known, it raises the intriguing possibilities that Gpa1 can regulate protein kinase C or some related protein kinase and that these interactions are in turn regulated by Asc1.
One model currently being considered is that Asc1 regulates Cla4. Cla4 and Ste20 are both members of the p21-activated protein kinase family implicated in cell signaling and in the establishment of cell polarity (27, 43, 70, 98, 128). Cla4 and Ste20 are at least partially functionally redundant, since neither is essential for viability but a double deletion is inviable (27, 82). Both kinases are activated by the GTP-bound form of Cdc42 (63, 95, 107, 136), and Ste20 is activated as well by binding to the Gβγ subunits Ste4/Ste18 (72, 136). Ste20 in turn phosphorylates Ste11 and ultimately promotes activation of the MAP kinases Fus3 and Kss1 (27, 43, 70-72, 84, 95, 98, 107, 136).
Another mechanism by which Asc1 could modulate signaling is by altering protein translation, possibly in conjunction with the candidate effector protein Scp160 (50). Asc1 has been identified as an integral component of the 40S ribosomal subunit (74) and is required for efficient association of Scp160 with ribosomes (5, 48). Although the precise function of Asc1 and Scp160 is not established, one possibility is that these proteins coordinately regulate translation efficiency or ribosome biosynthesis (5, 17, 69).
Finally, we found that deletion of YGL024W and PBS2 results in elevated basal pathway activation. A similar constitutively active phenotype was reported previously for sst2 and gas1 mutants (25, 105). In all four cases, the constitutively active phenotype was evident in the absence of the receptor gene STE2 but not in the absence of the G protein β subunit STE4 or other downstream signaling components (Table 4) (105). Based on the genetic epistasis data, we postulate that all of these proteins act at the level of the G protein. These proteins might prevent coupling of the receptor to its G protein, thereby leading to diminished G protein activation. Alternatively, they could indirectly diminish G protein activation or otherwise promote G protein subunit inactivation. Our data suggest that Ygl024w (or Pgd1) acts indirectly, by promoting full expression of Sst2 (Fig. 7). Yet another possibility is that these novel regulators bind to an effector kinase such as Ste20 or Cla4, thereby leading to diminished signaling. Currently we are attempting to establish which, if any, of these mechanisms account for the ability of YGL024W, GAS1, and ASC1 to modulate the pheromone signal.
The yeast system is now well recognized as a powerful resource for molecular pharmacology research. The discovery of Sst2 in yeast, and of RGS proteins generally, has established a new paradigm in signaling (19, 40, 89). The results presented here reveal several additional genes that also modulate agonist sensitivity in vivo. Most of the newly identified components have human homologues, which are thus likely to have functions similar to their yeast counterparts. The identification of new pathway modulators in yeast, whether affecting basal activity or agonist sensitivity, could eventually reveal potential new drug targets in humans. Drugs that modulate hormone or neurotransmitter signaling may be useful in situations where the receptor exhibits diminished activity or is chronically desensitized (89). A number of drugs that indirectly modulate G protein responsiveness have already been developed. For instance, inhibitors of serotonin reuptake have proven very useful in the treatment of depression. Competitive inhibitors of cGMP phosphodiesterase, an enzyme that degrades second messengers, have been developed for treatment of erectile dysfunction. Finally, if mutations or polymorphisms exist in agonist sensitivity genes such as those described here, that information could be used to reveal genetic predispositions to neurological or cardiovascular disorders or to dictate treatments for the most drug-sensitive individuals (100).
In conclusion, we have used a genome-scale approach to identify several new components of the pheromone signaling pathway. The identification of these agonist sensitivity genes provides a far more complete view of G protein signaling in yeast. Significantly, we found no gene as important to pathway regulation as SST2. Nevertheless we believe a similar analysis of agonist sensitivity in mammals would likewise reveal additional pathway components and may serve to validate potential drug targets within these fundamentally important signaling pathways. This goal may soon be realistic, given recent advances in homologous gene replacement and RNA interference technologies (77, 135).
Supplementary Material
Acknowledgments
This work was supported by American Heart Association fellowships 0020239T (to P.F.) and 0415413U (to N.H.) as well as by National Institutes of Health grant P01-GM65533 (to H.G.D. and D.P.S.).
We thank Ken Harden and Beverly Errede for their valuable comments and guidance.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Alani, E., L. Cao, and N. Kleckner. 1987. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics 116:541-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apanovitch, D. M., K. C. Slep, P. B. Sigler, and H. G. Dohlman. 1998. Sst2 is a GTPase-activating protein for Gpa1: purification and characterization of a cognate RGS-Gα protein pair in yeast. Biochemistry 37:4815-4822. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1987. Current protocols in molecular biology. Wiley Interscience, New York, N.Y.
- 4.Batlle, M., A. Lu, D. A. Green, Y. Xue, and J. P. Hirsch. 2003. Krh1p and Krh2p act downstream of the Gpa2p G(alpha) subunit to negatively regulate haploid invasive growth. J. Cell Sci. 116:701-710. [DOI] [PubMed] [Google Scholar]
- 5.Baum, S., M. Bittins, S. Frey, and M. Seedorf. 2004. Asc1p, a WD40-domain containing adaptor protein, is required for the interaction of the RNA-binding protein Scp160p with polysomes. Biochem. J. 380:823-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berman, D. M., T. Kozasa, and A. G. Gilman. 1996. The GTPase-activating protein RGS4 stabilizes the transition state for nucleotide hydrolysis. J. Biol. Chem. 271:27209-27212. [DOI] [PubMed] [Google Scholar]
- 7.Berman, D. M., T. M. Wilkie, and A. G. Gilman. 1996. GAIP and RGS4 are GTPase-activating proteins for the Gi subfamily of G protein α subunits. Cell 86:445-452. [DOI] [PubMed] [Google Scholar]
- 8.Bernstein, L. S., S. Ramineni, C. Hague, W. Cladman, P. Chidiac, A. I. Levey, and J. R. Hepler. 2004. RGS2 binds directly and selectively to the M1 muscarinic acetylcholine receptor third intracellular loop to modulate Gq/11alpha signaling. J. Biol. Chem. 279:21248-21256. [DOI] [PubMed] [Google Scholar]
- 9.Berstein, G., J. L. Blank, D. Y. Jhon, J. H. Exton, S. G. Rhee, and E. M. Ross. 1992. Phospholipase C-beta 1 is a GTPase-activating protein for Gq/11, its physiologic regulator. Cell 70:411-418. [DOI] [PubMed] [Google Scholar]
- 10.Birrell, G. W., G. Giaever, A. M. Chu, R. W. Davis, and J. M. Brown. 2001. A genome-wide screen in Saccharomyces cerevisiae for genes affecting UV radiation sensitivity. Proc. Natl. Acad. Sci. USA 98:12608-12613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brewster, J. L., T. de Valoir, N. D. Dwyer, E. Winter, and M. C. Gustin. 1993. An osmosensing signal transduction pathway in yeast. Science 259:1760-1763. [DOI] [PubMed] [Google Scholar]
- 12.Bulik, D. A., M. Olczak, H. A. Lucero, B. C. Osmond, P. W. Robbins, and C. A. Specht. 2003. Chitin synthesis in Saccharomyces cerevisiae in response to supplementation of growth medium with glucosamine and cell wall stress. Eukaryot. Cell 2:886-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carman, C. V., J. L. Parent, P. W. Day, A. N. Pronin, P. M. Sternweis, P. B. Wedegaertner, A. G. Gilman, J. L. Benovic, and T. Kozasa. 1999. Selective regulation of Gα(q/11) by an RGS domain in the G protein-coupled receptor kinase, GRK2. J. Biol. Chem. 274:34483-34492. [DOI] [PubMed] [Google Scholar]
- 14.Carotti, C., L. Ferrario, C. Roncero, M. H. Valdivieso, A. Duran, and L. Popolo. 2002. Maintenance of cell integrity in the gas1 mutant of Saccharomyces cerevisiae requires the Chs3p-targeting and activation pathway and involves an unusual Chs3p localization. Yeast 19:1113-1124. [DOI] [PubMed] [Google Scholar]
- 15.Chan, R. K., and C. A. Otte. 1982. Isolation and genetic analysis of Saccharomyces cerevisiae mutants supersensitive to G1 arrest by a factor and α factor pheromones. Mol. Cell. Biol. 2:11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan, R. K., and C. A. Otte. 1982. Physiological characterization of Saccharomyces cerevisiae mutants supersensitive to G1 arrest by a factor and α factor pheromones. Mol. Cell. Biol. 2:21-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chantrel, Y., M. Gaisne, C. Lions, and J. Verdiere. 1998. The transcriptional regulator Hap1p (Cyp1p) is essential for anaerobic or heme-deficient growth of Saccharomyces cerevisiae: genetic and molecular characterization of an extragenic suppressor that encodes a WD repeat protein. Genetics 148:559-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chasse, S. A., and H. G. Dohlman. 2004. Identification of yeast pheromone pathway modulators by high-throughput agonist response profiling of a yeast gene knockout strain collection. Methods Enzymol. 389:399-409. [DOI] [PubMed] [Google Scholar]
- 19.Chasse, S. A., and H. G. Dohlman. 2003. RGS proteins: G protein-coupled receptors meet their match. ASSAY Drug Dev. Technol. 1:357-364. [DOI] [PubMed] [Google Scholar]
- 20.Cheever, M. L., T. K. Sato, T. de Beer, T. G. Kutateladze, S. D. Emr, and M. Overduin. 2001. Phox domain interaction with PtdIns(3)P targets the Vam7 t-SNARE to vacuole membranes. Nat. Cell Biol. 3:613-618. [DOI] [PubMed] [Google Scholar]
- 21.Chen, S., E. J. Dell, F. Lin, J. Sai, and H. E. Hamm. 2004. RACK1 regulates specific functions of Gβγ. J. Biol. Chem. 279:17861-17868. [DOI] [PubMed] [Google Scholar]
- 22.Chen, S., B. D. Spiegelberg, F. Lin, E. J. Dell, and H. E. Hamm. 2004. Interaction of Gβγ with RACK1 and other WD40 repeat proteins. J. Mol. Cell. Cardiol. 37:399-406. [DOI] [PubMed] [Google Scholar]
- 23.Cid, V. J., A. Durán, F. del Rey, M. P. Snyder, C. Nombela, and M. Sánchez. 1995. Molecular basis of cell integrity and morphogenesis in Saccharomyces cerevisiae. Microbiol. Rev. 59:345-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cole, G. M., D. E. Stone, and S. I. Reed. 1990. Stoichiometry of G protein subunits affects the Saccharomyces cerevisiae mating pheromone signal transduction pathway. Mol. Cell. Biol. 10:510-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cullen, P. J., J. Schultz, J. Horecka, B. J. Stevenson, Y. Jigami, and G. F. Sprague, Jr. 2000. Defects in protein glycosylation cause SHO1-dependent activation of a STE12 signaling pathway in yeast. Genetics 155:1005-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cullen, P. J., and G. F. Sprague, Jr. 2002. The roles of bud-site-selection proteins during haploid invasive growth in yeast. Mol. Biol. Cell 13:2990-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cvrckova, F., C. De Virgilio, E. Manser, J. R. Pringle, and K. Nasmyth. 1995. Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes Dev. 9:1817-1830. [DOI] [PubMed] [Google Scholar]
- 28.Davis, J. L., R. Kunisawa, and J. Thorner. 1992. A presumptive helicase (MOT1 gene product) affects gene expression and is required for viability in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 12:1879-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Alba, E., L. De Vries, M. G. Farquhar, and N. Tjandra. 1999. Solution structure of human GAIP (Gα interacting protein): a regulator of G protein signaling. J. Mol. Biol. 291:927-939. [DOI] [PubMed] [Google Scholar]
- 30.Dell, E. J., J. Connor, S. Chen, E. G. Stebbins, N. P. Skiba, D. Mochly-Rosen, and H. E. Hamm. 2002. The βγ subunit of heterotrimeric G proteins interacts with RACK1 and two other WD repeat proteins. J. Biol. Chem. 277:49888-49895. [DOI] [PubMed] [Google Scholar]
- 31.de Nobel, H., C. Ruiz, H. Martin, W. Morris, S. Brul, M. Molina, and F. M. Klis. 2000. Cell wall perturbation in yeast results in dual phosphorylation of the Slt2/Mpk1 MAP kinase and in an Slt2-mediated increase in FKS2-lacZ expression, glucanase resistance and thermotolerance. Microbiology 146:2121-2132. [DOI] [PubMed] [Google Scholar]
- 32.Dhami, G. K., L. B. Dale, P. H. Anborgh, K. E. O'Connor-Halligan, R. Sterne-Marr, and S. S. Ferguson. 2004. G protein-coupled receptor kinase 2 regulator of G protein signaling homology domain binds to both metabotropic glutamate receptor 1a and Gαq to attenuate signaling. J. Biol. Chem. 279:16614-16620. [DOI] [PubMed] [Google Scholar]
- 33.DiBello, P. R., T. R. Garrison, D. M. Apanovitch, G. Hoffman, D. J. Shuey, K. Mason, M. I. Cockett, and H. G. Dohlman. 1998. Selective uncoupling of RGS action by a single point mutation in the G protein alpha-subunit. J. Biol. Chem. 273:5780-5784. [DOI] [PubMed] [Google Scholar]
- 34.Dietzel, C., and J. Kurjan. 1987. The yeast SCG1 gene: a G α-like protein implicated in the a- and α-factor response pathway. Cell 50:1001-1010. [DOI] [PubMed] [Google Scholar]
- 35.Diverse-Pierluissi, M. A., T. Fischer, J. D. Jordan, M. Schiff, D. F. Ortiz, M. G. Farquhar, and L. De Vries. 1999. Regulators of G protein signaling proteins as determinants of the rate of desensitization of presynaptic calcium channels. J. Biol. Chem. 274:14490-14494. [DOI] [PubMed] [Google Scholar]
- 36.Dohlman, H. G. 2002. G proteins and pheromone signaling. Annu. Rev. Physiol. 64:129-152. [DOI] [PubMed] [Google Scholar]
- 37.Dohlman, H. G., D. Apaniesk, Y. Chen, J. Song, and D. Nusskern. 1995. Inhibition of G-protein signaling by dominant gain-of-function mutations in Sst2p, a pheromone desensitization factor in Saccharomyces cerevisiae. Mol. Cell. Biol. 15:3635-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dohlman, H. G., P. Goldsmith, A. M. Spiegel, and J. Thorner. 1993. Pheromone action regulates G-protein α-subunit myristoylation in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 90:9688-9692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dohlman, H. G., J. Song, D. Ma, W. E. Courchesne, and J. Thorner. 1996. Sst2, a negative regulator of pheromone signaling in the yeast Saccharomyces cerevisiae: expression, localization, and genetic interaction and physical association with Gpa1 (the G-protein α subunit). Mol. Cell. Biol. 16:5194-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dohlman, H. G., and J. W. Thorner. 2001. Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu. Rev. Biochem. 70:703-754. [DOI] [PubMed] [Google Scholar]
- 41.Dong, M. Q., D. Chase, G. A. Patikoglou, and M. R. Koelle. 2000. Multiple RGS proteins alter neural G protein signaling to allow C. elegans to rapidly change behavior when fed. Genes Dev. 14:2003-2014. [PMC free article] [PubMed] [Google Scholar]
- 42.Dowal, L., J. Elliott, S. Popov, T. M. Wilkie, and S. Scarlata. 2001. Determination of the contact energies between a regulator of G protein signaling and G protein subunits and phospholipase Cβ1. Biochemistry 40:414-421. [DOI] [PubMed] [Google Scholar]
- 43.Eby, J. J., S. P. Holly, F. van Drogen, A. V. Grishin, M. Peter, D. G. Drubin, and K. J. Blumer. 1998. Actin cytoskeleton organization regulated by the PAK family of protein kinases. Curr. Biol. 8:967-970. [DOI] [PubMed] [Google Scholar]
- 44.Evan, G. I., G. K. Lewis, G. Ramsay, and J. M. Bishop. 1985. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol. Cell. Biol. 5:3610-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fisk, H. A., and M. P. Yaffe. 1997. Mutational analysis of Mdm1p function in nuclear and mitochondrial inheritance. J. Cell Biol. 138:485-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fujita, A., M. Lord, T. Hiroko, F. Hiroko, T. Chen, C. Oka, Y. Misumi, and J. Chant. 2004. Rax1, a protein required for the establishment of the bipolar budding pattern in yeast. Gene 327:161-169. [DOI] [PubMed] [Google Scholar]
- 47.Garrison, T. R., Y. Zhang, M. Pausch, D. Apanovitch, R. Aebersold, and H. G. Dohlman. 1999. Feedback phosphorylation of an RGS protein by MAP kinase in yeast. J. Biol. Chem. 274:36387-36391. [DOI] [PubMed] [Google Scholar]
- 48.Gerbasi, V. R., C. M. Weaver, S. Hill, D. B. Friedman, and A. J. Link. 2004. Yeast Asc1p and mammalian RACK1 are functionally orthologous core 40S ribosomal proteins that repress gene expression. Mol. Cell. Biol. 24:8276-8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. Arkin, A. Astromoff, M. El-Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 50.Guo, M., C. Aston, S. A. Burchett, C. Dyke, S. Fields, S. J. Rajarao, P. Uetz, Y. Wang, K. Young, and H. G. Dohlman. 2003. The yeast G protein α subunit Gpa1 transmits a signal through an RNA binding effector protein Scp160. Mol. Cell 12:517-524. [DOI] [PubMed] [Google Scholar]
- 51.Hajdu-Cronin, Y. M., W. J. Chen, G. Patikoglou, M. R. Koelle, and P. W. Sternberg. 1999. Antagonism between Goα and Gqα in Caenorhabditis elegans: the RGS protein EAT-16 is necessary for Goα signaling and regulates Gqα activity. Genes Dev. 13:1780-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hall, J. P., V. Cherkasova, E. Elion, M. C. Gustin, and E. Winter. 1996. The osmoregulatory pathway represses mating pathway activity in Saccharomyces cerevisiae: isolation of a FUS3 mutant that is insensitive to the repression mechanism. Mol. Cell. Biol. 16:6715-6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hao, N., N. Yildirim, Y. Wang, T. C. Elston, and H. G. Dohlman. 2003. Regulators of G protein signaling and transient activation of signaling: experimental and computational analysis reveals negative and positive feedback controls on G protein activity. J. Biol. Chem. 278:46506-46515. [DOI] [PubMed] [Google Scholar]
- 54.Harashima, T., and J. Heitman. 2002. The Gα protein Gpa2 controls yeast differentiation by interacting with kelch repeat proteins that mimic Gβ subunits. Mol. Cell 10:163-173. [DOI] [PubMed] [Google Scholar]
- 55.Hepler, J. R., D. M. Berman, A. G. Gilman, and T. Kozasa. 1997. RGS4 and GAIP are GTPase-activating proteins for Gqα and block activation of phospholipase Cβ by γ-thio-GTP-Gq α. Proc. Natl. Acad. Sci. USA 94:428-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hess, H. A., J. C. Roper, S. W. Grill, and M. R. Koelle. 2004. RGS-7 completes a receptor-independent heterotrimeric G protein cycle to asymmetrically regulate mitotic spindle positioning in C. elegans. Cell 119:209-218. [DOI] [PubMed] [Google Scholar]
- 57.Hicke, L., and H. Riezman. 1996. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell 84:277-287. [DOI] [PubMed] [Google Scholar]
- 58.Hiroaki, H., T. Ago, T. Ito, H. Sumimoto, and D. Kohda. 2001. Solution structure of the PX domain, a target of the SH3 domain. Nat. Struct. Biol. 8:526-530. [DOI] [PubMed] [Google Scholar]
- 59.Hoffmann, B., H. U. Mosch, E. Sattlegger, I. B. Barthelmess, A. Hinnebusch, and G. H. Braus. 1999. The WD protein Cpc2p is required for repression of Gcn4 protein activity in yeast in the absence of amino-acid starvation. Mol. Microbiol. 31:807-822. [DOI] [PubMed] [Google Scholar]
- 60.Hoffman, G., T. R. Garrison, and H. G. Dohlman. 2002. Analysis of RGS proteins in Saccharomyces cerevisiae. Methods Enzymol. 344:617-631. [DOI] [PubMed] [Google Scholar]
- 61.Hoffman, G. A., T. R. Garrison, and H. G. Dohlman. 2000. Endoproteolytic processing of Sst2, a multidomain regulator of G protein signaling in yeast. J. Biol. Chem. 275:37533-37541. [DOI] [PubMed] [Google Scholar]
- 62.Hollinger, S., and J. R. Hepler. 2002. Cellular regulation of RGS proteins: modulators and integrators of G protein signaling. Pharmacol. Rev. 54:527-559. [DOI] [PubMed] [Google Scholar]
- 63.Holly, S. P., and K. J. Blumer. 1999. PAK-family kinases regulate cell and actin polarization throughout the cell cycle of Saccharomyces cerevisiae. J. Cell Biol. 147:845-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hopkins, A. L., and C. R. Groom. 2002. The druggable genome. Nat. Rev. Drug Discov. 1:727-730. [DOI] [PubMed] [Google Scholar]
- 65.Koelle, M. R., and H. R. Horvitz. 1996. EGL-10 regulates G protein signaling in the C. elegans nervous system and shares a conserved domain with many mammalian proteins. Cell 84:115-125. [DOI] [PubMed] [Google Scholar]
- 66.Kraakman, L., K. Lemaire, P. Ma, A. W. Teunissen, M. C. Donaton, P. Van Dijck, J. Winderickx, J. H. de Winde, and J. M. Thevelein. 1999. A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol. Microbiol. 32:1002-1012. [DOI] [PubMed] [Google Scholar]
- 67.Lagorce, A., N. C. Hauser, D. Labourdette, C. Rodriguez, H. Martin-Yken, J. Arroyo, J. D. Hoheisel, and J. Francois. 2003. Genome-wide analysis of the response to cell wall mutations in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 278:20345-20357. [DOI] [PubMed] [Google Scholar]
- 68.Lagorce, A., V. Le Berre-Anton, B. Aguilar-Uscanga, H. Martin-Yken, A. Dagkessamanskaia, and J. Francois. 2002. Involvement of GFA1, which encodes glutamine-fructose-6-phosphate amidotransferase, in the activation of the chitin synthesis pathway in response to cell-wall defects in Saccharomyces cerevisiae. Eur. J. Biochem. 269:1697-1707. [DOI] [PubMed] [Google Scholar]
- 69.Lang, B. D., and J. L. Fridovich-Keil. 2000. Scp160p, a multiple KH-domain protein, is a component of mRNP complexes in yeast. Nucleic Acids Res. 28:1576-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Leberer, E., D. Dignard, D. Harcus, D. Y. Thomas, and M. Whiteway. 1992. The protein kinase homologue Ste20p is required to link the yeast pheromone response G-protein βγ subunits to downstream signalling components. EMBO J. 11:4815-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Leberer, E., D. Y. Thomas, and M. Whiteway. 1997. Pheromone signalling and polarized morphogenesis in yeast. Curr. Opin. Genet. Dev. 7:59-66. [DOI] [PubMed] [Google Scholar]
- 72.Leeuw, T., C. Wu, J. D. Schrag, M. Whiteway, D. Y. Thomas, and E. Leberer. 1998. Interaction of a G-protein β-subunit with a conserved sequence in Ste20/PAK family protein kinases. Nature 391:191-195. [DOI] [PubMed] [Google Scholar]
- 73.Lemaire, K., S. Van de Velde, P. Van Dijck, and J. M. Thevelein. 2004. Glucose and sucrose act as agonist and mannose as antagonist ligands of the G protein-coupled receptor Gpr1 in the yeast Saccharomyces cerevisiae. Mol. Cell 16:293-299. [DOI] [PubMed] [Google Scholar]
- 74.Link, A. J., J. Eng, D. M. Schieltz, E. Carmack, G. J. Mize, D. R. Morris, B. M. Garvik, and J. R. Yates III. 1999. Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17:676-682. [DOI] [PubMed] [Google Scholar]
- 75.Lord, M., F. Inose, T. Hiroko, T. Hata, A. Fujita, and J. Chant. 2002. Subcellular localization of Axl1, the cell type-specific regulator of polarity. Curr. Biol. 12:1347-1352. [DOI] [PubMed] [Google Scholar]
- 76.Lorenz, M. C., X. Pan, T. Harashima, M. E. Cardenas, Y. Xue, J. P. Hirsch, and J. Heitman. 2000. The G protein-coupled receptor gpr1 is a nutrient sensor that regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Genetics 154:609-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lum, L., S. Yao, B. Mozer, A. Rovescalli, D. Von Kessler, M. Nirenberg, and P. A. Beachy. 2003. Identification of Hedgehog pathway components by RNAi in Drosophila cultured cells. Science 299:2039-2045. [DOI] [PubMed] [Google Scholar]
- 78.MacKay, V. L., J. Armstrong, C. Yip, S. Welch, K. Walker, S. Osborn, P. Sheppard, and J. Forstrom. 1991. Characterization of the Bar proteinase, an extracellular enzyme from the yeast Saccharomyces cerevisiae. Adv. Exp. Med. Biol. 306:161-172. [DOI] [PubMed] [Google Scholar]
- 79.Marsh, L., and I. Herskowitz. 1988. From membrane to nucleus: the pathway of signal transduction in yeast and its genetic control. Cold Spring Harbor Symp. Quant. Biol. 53:557-565. [DOI] [PubMed] [Google Scholar]
- 80.McConnell, S. J., L. C. Stewart, A. Talin, and M. P. Yaffe. 1990. Temperature-sensitive yeast mutants defective in mitochondrial inheritance. J. Cell Biol. 111:967-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McGuffin, L. J., K. Bryson, and D. T. Jones. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404-405. [DOI] [PubMed] [Google Scholar]
- 82.Mitchell, D. A., and G. F. Sprague, Jr. 2001. The phosphotyrosyl phosphatase activator, Ncs1p (Rrd1p), functions with Cla4p to regulate the G2/M transition in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:488-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Miyajima, I., M. Nakafuku, N. Nakayama, C. Brenner, A. Miyajima, K. Kaibuchi, K. Arai, Y. Kaziro, and K. Matsumoto. 1987. GPA1, a haploid-specific essential gene, encodes a yeast homolog of mammalian G protein which may be involved in mating factor signal transduction. Cell 50:1011-1019. [DOI] [PubMed] [Google Scholar]
- 84.Moskow, J. J., A. S. Gladfelter, R. E. Lamson, P. M. Pryciak, and D. J. Lew. 2000. Role of Cdc42p in pheromone-stimulated signal transduction in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:7559-7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mouyna, I., T. Fontaine, M. Vai, M. Monod, W. A. Fonzi, M. Diaquin, L. Popolo, R. P. Hartland, and J. P. Latge. 2000. Glycosylphosphatidylinositol-anchored glucanosyltransferases play an active role in the biosynthesis of the fungal cell wall. J. Biol. Chem. 275:14882-14889. [DOI] [PubMed] [Google Scholar]
- 86.Muller, G., and W. Bandlow. 1991. A cAMP-binding ectoprotein in the yeast Saccharomyces cerevisiae. Biochemistry 30:10181-10190. [DOI] [PubMed] [Google Scholar]
- 87.Myers, L. C., C. M. Gustafsson, K. C. Hayashibara, P. O. Brown, and R. D. Kornberg. 1999. Mediator protein mutations that selectively abolish activated transcription. Proc. Natl. Acad. Sci. USA 96:67-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nakafuku, M., H. Itoh, S. Nakamura, and Y. Kaziro. 1987. Occurrence in Saccharomyces cerevisiae of a gene homologous to the cDNA coding for the α subunit of mammalian G proteins. Proc. Natl. Acad. Sci. USA 84:2140-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Neubig, R. R., and D. P. Siderovski. 2002. Regulators of G-protein signalling as new central nervous system drug targets. Nat. Rev. Drug Discov. 1:187-197. [DOI] [PubMed] [Google Scholar]
- 90.Neves, S. R., P. T. Ram, and R. Iyengar. 2002. G protein pathways. Science 296:1636-1639. [DOI] [PubMed] [Google Scholar]
- 91.Ni, L., and M. Snyder. 2001. A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol. Biol. Cell 12:2147-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nomoto, S., N. Nakayama, K. Arai, and K. Matsumoto. 1990. Regulation of the yeast pheromone response pathway by G protein subunits. EMBO J. 9:691-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nuoffer, C., P. Jenö, A. Conzelmann, and H. Riezman. 1991. Determinants for glycophospholipid anchoring of the Saccharomyces cerevisiae GAS1 protein to the plasma membrane. Mol. Cell. Biol. 11:27-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.O'Rourke, S. M., and I. Herskowitz. 1998. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 12:2874-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Peter, M., A. M. Neiman, H. O. Park, M. van Lohuizen, and I. Herskowitz. 1996. Functional analysis of the interaction between the small GTP binding protein Cdc42 and the Ste20 protein kinase in yeast. EMBO J. 15:7046-7059. [PMC free article] [PubMed] [Google Scholar]
- 96.Popolo, L., D. Gilardelli, P. Bonfante, and M. Vai. 1997. Increase in chitin as an essential response to defects in assembly of cell wall polymers in the ggp1Δ mutant of Saccharomyces cerevisiae. J. Bacteriol. 179:463-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ram, A. F. J., J. C. Kapteyn, R. C. Montijn, L. H. P. Caro, J. E. Douwes, W. Baginsky, P. Mazur, H. Van Den Ende, and F. M. Klis. 1998. Loss of the plasma membrane-bound protein Gas1p in Saccharomyces cerevisiae results in the release of β1,3-glucan into the medium and induces a compensation mechanism to ensure cell wall integrity. J. Bacteriol. 180:1418-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ramer, S. W., and R. W. Davis. 1993. A dominant truncation allele identifies a gene, STE20, that encodes a putative protein kinase necessary for mating in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 90:452-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rolland, F., J. H. De Winde, K. Lemaire, E. Boles, J. M. Thevelein, and J. Winderickx. 2000. Glucose-induced cAMP signalling in yeast requires both a G-protein coupled receptor system for extracellular glucose detection and a separable hexose kinase-dependent sensing process. Mol. Microbiol. 38:348-358. [DOI] [PubMed] [Google Scholar]
- 100.Roses, A. D. 2002. Genome-based pharmacogenetics and the pharmaceutical industry. Nat. Rev. Drug Discov. 1:541-549. [DOI] [PubMed] [Google Scholar]
- 101.Sallese, M., S. Mariggio, E. D'Urbano, L. Iacovelli, and A. De Blasi. 2000. Selective regulation of Gq signaling by G protein-coupled receptor kinase 2: direct interaction of kinase N terminus with activated Gαq. Mol. Pharmacol. 57:826-831. [PubMed] [Google Scholar]
- 102.Schechtman, D., and D. Mochly-Rosen. 2001. Adaptor proteins in protein kinase C-mediated signal transduction. Oncogene 20:6339-6347. [DOI] [PubMed] [Google Scholar]
- 103.Shi, C. S., S. B. Lee, S. Sinnarajah, C. W. Dessauer, S. G. Rhee, and J. H. Kehrl. 2001. Regulator of G-protein signaling 3 (RGS3) inhibits Gβ1γ2-induced inositol phosphate production, mitogen-activated protein kinase activation, and Akt activation. J. Biol. Chem. 276:24293-24300. [DOI] [PubMed] [Google Scholar]
- 104.Siderovski, D. P., A. Hessel, S. Chung, T. W. Mak, and M. Tyers. 1996. A new family of regulators of G-protein-coupled receptors? Curr. Biol. 6:211-212. [DOI] [PubMed] [Google Scholar]
- 105.Siekhaus, D. E., and D. G. Drubin. 2003. Spontaneous receptor-independent heterotrimeric G-protein signalling in an RGS mutant. Nat. Cell Biol. 5:231-235. [DOI] [PubMed] [Google Scholar]
- 106.Sierra, D. A., D. J. Gilbert, D. Householder, N. V. Grishin, K. Yu, P. Ukidwe, S. A. Barker, W. He, T. G. Wensel, G. Otero, G. Brown, N. G. Copeland, N. A. Jenkins, and T. M. Wilkie. 2002. Evolution of the regulators of G-protein signaling multigene family in mouse and human. Genomics 79:177-185. [DOI] [PubMed] [Google Scholar]
- 107.Simon, M. N., C. De Virgilio, B. Souza, J. R. Pringle, A. Abo, and S. I. Reed. 1995. Role for the Rho-family GTPase Cdc42 in yeast mating-pheromone signal pathway. Nature 376:702-705. [DOI] [PubMed] [Google Scholar]
- 108.Sinnarajah, S., C. W. Dessauer, D. Srikumar, J. Chen, J. Yuen, S. Yilma, J. C. Dennis, E. E. Morrison, V. Vodyanoy, and J. H. Kehrl. 2001. RGS2 regulates signal transduction in olfactory neurons by attenuating activation of adenylyl cyclase III. Nature 409:1051-1055. [DOI] [PubMed] [Google Scholar]
- 109.Snow, B. E., R. A. Hall, A. M. Krumins, G. M. Brothers, D. Bouchard, C. A. Brothers, S. Chung, J. Mangion, A. G. Gilman, R. J. Lefkowitz, and D. P. Siderovski. 1998. GTPase activating specificity of RGS12 and binding specificity of an alternatively spliced PDZ (PSD-95/Dlg/ZO-1) domain. J. Biol. Chem. 273:17749-17755. [DOI] [PubMed] [Google Scholar]
- 110.Sondek, J., A. Bohm, D. G. Lambright, H. E. Hamm, and P. B. Sigler. 1996. Crystal structure of a G-protein beta gamma dimer at 2.1A resolution. Nature 379:369-374. [DOI] [PubMed] [Google Scholar]
- 111.Sondek, J., D. G. Lambright, J. P. Noel, H. E. Hamm, and P. B. Sigler. 1994. GTPase mechanism of G proteins from the 1.7-A crystal structure of transducin α-GDP-AIF-4. Nature 372:276-279. [DOI] [PubMed] [Google Scholar]
- 112.Song, J., J. Hirschman, K. Gunn, and H. G. Dohlman. 1996. Regulation of membrane and subunit interactions by N-myristoylation of a G protein α subunit in yeast. J. Biol. Chem. 271:20273-20283. [DOI] [PubMed] [Google Scholar]
- 113.Spink, K. E., P. Polakis, and W. I. Weis. 2000. Structural basis of the Axin-adenomatous polyposis coli interaction. EMBO J. 19:2270-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sprague, G. F., Jr. 1991. Assay of yeast mating reaction. Methods Enzymol. 194:77-93. [DOI] [PubMed] [Google Scholar]
- 115.Srinivasa, S. P., N. Watson, M. C. Overton, and K. J. Blumer. 1998. Mechanism of RGS4, a GTPase-activating protein for G protein alpha subunits. J. Biol. Chem. 273:1529-1533. [DOI] [PubMed] [Google Scholar]
- 116.Sterne-Marr, R., J. J. Tesmer, P. W. Day, R. P. Stracquatanio, J. A. Cilente, K. E. O'Connor, A. N. Pronin, J. L. Benovic, and P. B. Wedegaertner. 2003. G protein-coupled receptor kinase 2/G alpha q/11 interaction. A novel surface on a regulator of G protein signaling homology domain for binding G alpha subunits. J. Biol. Chem. 278:6050-6058. [DOI] [PubMed] [Google Scholar]
- 117.Sternweis, P. C., and A. G. Gilman. 1982. Aluminum: a requirement for activation of the regulatory component of adenylate cyclase by fluoride. Proc. Natl. Acad. Sci. USA 79:4888-4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Stevenson, B. J., B. Ferguson, C. De Virgilio, E. Bi, J. R. Pringle, G. Ammerer, and G. F. Sprague, Jr. 1995. Mutation of RGA1, which encodes a putative GTPase-activating protein for the polarity-establishment protein Cdc42p, activates the pheromone-response pathway in the yeast Saccharomyces cerevisiae. Genes Dev. 9:2949-2963. [DOI] [PubMed] [Google Scholar]
- 119.Tesmer, J. J., D. M. Berman, A. G. Gilman, and S. R. Sprang. 1997. Structure of RGS4 bound to AlF4-activated G(iα1): stabilization of the transition state for GTP hydrolysis. Cell 89:251-261. [DOI] [PubMed] [Google Scholar]
- 120.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Thornton, C., K. C. Tang, K. Phamluong, K. Luong, A. Vagts, D. Nikanjam, R. Yaka, and D. Ron. 2004. Spatial and temporal regulation of RACK1 function and N-methyl-D-aspartate receptor activity through WD40 motif-mediated dimerization. J. Biol. Chem. 279:31357-31364. [DOI] [PubMed] [Google Scholar]
- 122.Valdivieso, M.-H., L. Ferrario, M. Vai, A. Duran, and L. Popolo. 2000. Chitin synthesis in a gas1 mutant of Saccharomyces cerevisiae. J. Bacteriol. 182:4752-4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Versele, M., J. H. de Winde, and J. M. Thevelein. 1999. A novel regulator of G protein signalling in yeast, Rgs2, downregulates glucose-activation of the cAMP pathway through direct inhibition of Gpa2. EMBO J. 18:5577-5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Versele, M., K. Lemaire, and J. M. Thevelein. 2001. Sex and sugar in yeast: two distinct GPCR systems. EMBO Rep. 2:574-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wang, J., A. Ducret, Y. Tu, T. Kozasa, R. Aebersold, and E. M. Ross. 1998. RGSZ1, a Gz-selective RGS protein in brain. Structure, membrane association, regulation by Gαz phosphorylation, and relationship to a Gz GTPase-activating protein subfamily. J. Biol. Chem. 273:26014-26025. [DOI] [PubMed] [Google Scholar]
- 126.Watson, N., M. E. Linder, K. M. Druey, J. H. Kehrl, and K. J. Blumer. 1996. RGS family members: GTPase-activating proteins for heterotrimeric G-protein α-subunits. Nature 383:172-175. [DOI] [PubMed] [Google Scholar]
- 127.Webb, C. K., C. R. McCudden, F. S. Willard, R. J. Kimple, D. P. Siderovski, and G. S. Oxford. 2005. D2 dopamine receptor activation of potassium channels is selectively decoupled by Gαi-specific GoLoco motif peptides. J. Neurochem. 92:1408-1418. [DOI] [PubMed] [Google Scholar]
- 128.Weiss, E. L., A. C. Bishop, K. M. Shokat, and D. G. Drubin. 2000. Chemical genetic analysis of the budding-yeast p21-activated kinase Cla4p. Nat. Cell Biol. 2:677-685. [DOI] [PubMed] [Google Scholar]
- 129.Whiteway, M., L. Hougan, D. Dignard, D. Y. Thomas, L. Bell, G. C. Saari, F. J. Grant, P. O'Hara, and V. L. MacKay. 1989. The STE4 and STE18 genes of yeast encode potential β and γ subunits of the mating factor receptor-coupled G protein. Cell 56:467-477. [DOI] [PubMed] [Google Scholar]
- 130.Whiteway, M., L. Hougan, and D. Y. Thomas. 1990. Overexpression of the STE4 gene leads to mating response in haploid Saccharomyces cerevisiae. Mol. Cell. Biol. 10:217-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, R. W. Davis, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]
- 132.Wittinghofer, A. 1997. Signaling mechanistics: aluminum fluoride for molecule of the year. Curr. Biol. 7:R682-R685. [DOI] [PubMed] [Google Scholar]
- 133.Yi, T. M., H. Kitano, and M. I. Simon. 2003. A quantitative characterization of the yeast heterotrimeric G protein cycle. Proc. Natl. Acad. Sci. USA 100:10764-10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yu, J. W., and M. A. Lemmon. 2001. All phox homology (PX) domains from Saccharomyces cerevisiae specifically recognize phosphatidylinositol 3-phosphate. J. Biol. Chem. 276:44179-44184. [DOI] [PubMed] [Google Scholar]
- 135.Zambrowicz, B. P., and A. T. Sands. 2003. Knockouts model the 100 best-selling drugs—will they model the next 100? Nat. Rev. Drug Discov. 2:38-51. [DOI] [PubMed] [Google Scholar]
- 136.Zhao, Z.-S., T. Leung, E. Manser, and L. Lim. 1995. Pheromone signalling in Saccharomyces cerevisiae requires the small GTP-binding protein Cdc42p and its activator CDC24. Mol. Cell. Biol. 15:5246-5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zheng, B., Y. C. Ma, R. S. Ostrom, C. Lavoie, G. N. Gill, P. A. Insel, X. Y. Huang, and M. G. Farquhar. 2001. RGS-PX1, a GAP for Gαs and sorting nexin in vesicular trafficking. Science 294:1939-1942. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.