Abstract
By using DNA heteroduplexes that inhibit rewinding of the upstream part of the transcription bubble, we show that transcript release in termination by the enzymes Mfd and Rho is facilitated by reannealing of DNA in the upstream region of the transcription bubble, as is also true for termination by intrinsic terminators. We also show that, like Mfd, the Rho termination factor promotes forward translocation of RNA polymerase. These results support termination models in which external forces imposed on nucleic acids induce concerted rewinding of DNA and unwinding of the DNA/RNA hybrid, possibly accompanied by forward translocation of RNA polymerase, leading to transcription complex dissociation.
Keywords: RNA polymerase, Mfd protein, Rho termination factor
Termination of transcription and the release of RNA polymerase (RNAP) from its templating complex with DNA are essential for providing a boundary for gene expression and removing stalled enzymes that may obstruct gene expression and replication. Three mechanisms are known that cause the otherwise notably stable transcription complex of Escherichia coli RNAP to dissociate. These mechanisms are (i) the intrinsic terminator, consisting of nucleic acid structures that interact with RNAP (1), (ii) the termination factor Rho, an RNA-dependent ATPase and RNA “helicase” (or, more accurately, RNA translocase) that acts by binding the emerging transcript and (presumably) RNAP (2, 3), and (iii) Mfd, an ATP-dependent DNA translocase that acts on RNAP and DNA upstream of the transcription bubble (4–6) (Fig. 1). [The replication fork apparatus could contain a fourth mechanism that removes obstructing transcription complexes (7).] Understanding these termination pathways may reveal important aspects of transcription complex stability, as well as the nature of regulation that acts through antitermination.
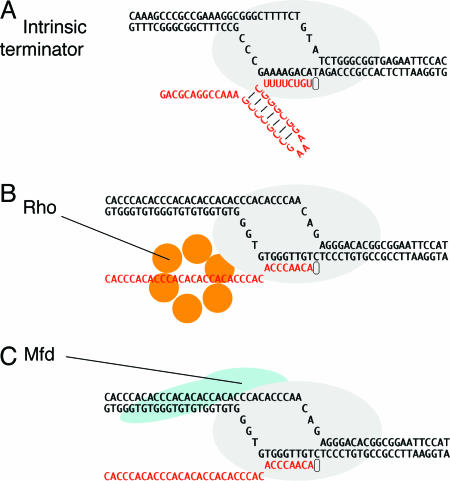
Fig. 1.
Three mechanisms of transcription termination. (A) Intrinsic termination is driven by formation of an RNA hairpin in the emerging transcript, the base of which occurs 8–9 nt from the site of release. Release also requires a uridine-rich segment downstream of the hairpin, particularly in the region immediately adjacent to the G/C-rich end of the stem. Although not illustrated, we suggest (as described in the text) that the DNA bubble is partly rewound and that the RNA/DNA hybrid is partly unwound when the hairpin is fully formed. (B) The termination factor Rho is a hexameric RNA translocase that binds ≈60 nt of emerging transcript, moving along it in a 5′–3′ direction in an ATP-dependent reaction. This movement is believed to extract the transcript. (C) Mfd is a DNA translocase that binds duplex DNA upstream of the transcription bubble and RNAP in a region near the site of DNA rewinding. The activity of the translocase causes dissociation of the complex in conditions that do not allow the RNA chain to advance through NTP polymerization.
Two classes of models of termination describe how nucleic acids could move relative to the enzyme such that the RNA becomes weakly held and can dissociate, leading to RNAP dissociation. The first class of models (“mechanical models”), illustrated in Fig. 2, proposes that rewinding of the upstream boundary of the transcription bubble is coupled with unwinding of the RNA/DNA hybrid within the enclosing structure of the enzyme (Fig. 2 B and C). In one mechanical model (Fig. 2B), the enzyme translocates forward without RNA synthesis, retaining protein–nucleic acid contacts of the elongation complex (8, 9); in another (Fig. 2C), the transcription bubble collapses within the channel as the hybrid unwinds, without enzyme translocation. The second class of models (“allosteric”) is less defined but proposes that long-range conformational changes in RNAP induced by some element of the terminator (e.g., the RNA hairpin or activities of the enzymatic terminators) destabilize the enzyme–nucleic acid contacts and lead to complex collapse (10).
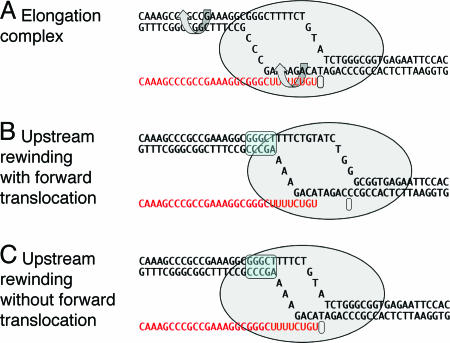
Fig. 2.
The elongation complex and models of RNA release in termination. (A) A model of an elongation complex showing helix rotation that would accompany branch migration in mechanical models of termination. (B) Termination by forward translocation. (C) Termination by bubble collapse. A shows a particular intrinsic transcription terminator poised at the site of release, but B and C are general to both intrinsic and enzymatic termination. The upstream rewound segment is indicated by the blue overscreen.
One apparent similarity among intrinsic and enzymatic terminators favoring mechanical models is that all three involve forces exerted on upstream nucleic acid elements at the site of termination.
The intrinsic terminator encodes an RNA hairpin that forms adjacent to a uridine-rich transcript segment at the site of RNA release. Formation of the hairpin is believed to initiate dissociation of the transcription complex by disrupting the upstream segment of the templating RNA/DNA hybrid, with the overall disruption process favored by the weak hybrid (8, 11). Formation of this hairpin also exerts forward force on RNAP in a transcription complex, because it can assist the enzyme in dislodging a blocking protein that is just downstream (9). Because a DNA oligonucleotide applied in trans can substitute for the upstream strand of the RNA hairpin, the critical event is the formation of a duplex structure that engages the emerging transcript and not any action of the RNA hairpin per se (8).
The enzyme Rho also binds the emerging transcript, possibly translocating along it to contact RNAP; Rho likely uses ATP energy to move 5′–3′ along the RNA and thereby to extract the transcript from the complex while it binds RNAP, although there is no evidence of a specific binding interaction between Rho and RNAP.
The activity of Mfd in exerting force on upstream DNA is particularly revealing of the mechanism of RNA release. Mfd is the transcription repair coupling protein of bacterial cells, serving to recognize RNAP stalled by damaged DNA, to remove it (and the transcript) from DNA in an ATP-dependent reaction (or, more conveniently for biochemical analysis, a dATP-dependent reaction), and to recruit DNA excision repair machinery to the site (4, 5). Two segments of the Mfd polypeptide are known to contribute to the RNAP release activity: an RNAP-binding domain (5, 12) and a DNA translocase region that interacts with ≈25 bp of duplex DNA upstream of the transcription bubble in the complex adjacent to the site of DNA rewinding (5). The structure and function of the translocase domain are well understood through comparison to the strongly homologous domain of the Holliday junction migration enzyme RecG (13) and through mutational analysis (14).
The role of the DNA translocase activity in RNAP release by Mfd is shown by its ability to translocate RNAP along DNA (or vice versa) in the direction of synthesis. In this way, Mfd can rescue a backtracked and arrested complex into productive elongation; however, if elongation fails because the NTP substrate is not provided in vitro, a condition presumed to model a site of template strand damage that blocks elongation, Mfd causes transcript release and dissociation of RNAP (5). Because essentially all arrested RNAP can be rescued into elongation in such an experiment, the enzyme must be translocated forward until the RNA 3′ end is in the active site before release occurs. This biochemical evidence, in addition to mutational confirmation that the translocase activity is required for complex release (14), strongly suggests that the force exerted by the Mfd–RNAP complex on DNA that causes forward translocation is also the direct cause of dissociation, which we suggest occurs as in Fig. 2B. If forward translocation is blocked [e.g., by a DNA-binding protein or possibly a DNA interstrand crosslink (9)], the rotational motion of the Mfd–RNAP complex tracking along DNA would be converted to a torque imposed on the upstream DNA; we propose that bubble collapse imposed by this torque is the prime element in Mfd-mediated transcription complex dissociation of a blocked complex, which we suggest occurs as in Fig. 2C.
Study of the intrinsic terminator has provided related information about the movements of nucleic acids that can provoke dissociation of the complex: Intrinsic termination is inhibited if rewinding of DNA in the region of the transcription bubble is prevented through nucleotide substitutions in the nontemplate DNA strand of the bubble (15). Because substituents of different sequences have similar effects, their effect is most likely to impair rewinding of the DNA strands in the region of the bubble. The stronger effect of heteroduplexes close to the upstream boundary of the bubble is consistent with rewinding that initiates from upstream, a process that we suggest is coordinated with unwinding of the hybrid. Finally, there is evidence that RNA release by an intrinsic terminator is facilitated by downstream translocation of the enzyme, in which downstream DNA is unwound in concert with upstream rewinding (9).
We provide evidence here that enzymatic mechanisms of termination have properties in common with the intrinsic mechanism. They are inhibited if rewinding of DNA in the upstream region of the transcription bubble is impaired (a result specifically consistent with the notion that Mfd and Rho act by imposing bubble rewinding), and they induce forward translocation of the enzyme. We suggest that all three mechanisms initiate transcript release through a concerted rewinding of upstream DNA and unwinding of the RNA/DNA hybrid, in effect a branch migration that also tends to promote forward translocation of the complex.
Results
Heteroduplex DNA in the Transcription Bubble Region Inhibits Mfd-Induced Transcript Release.
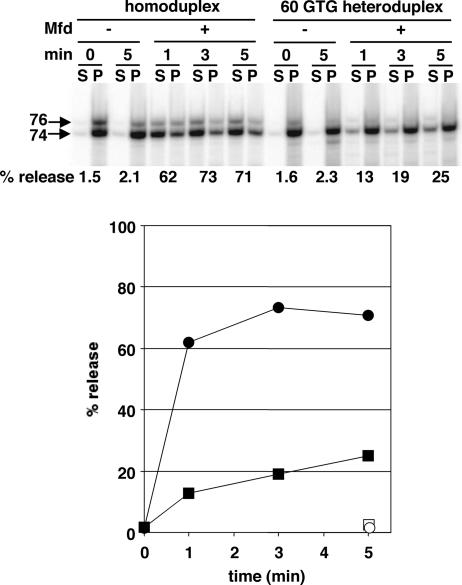
The transcript release activity of Mfd is conveniently measured by using defined, stalled transcription complexes that are affixed through a DNA end to paramagnetic beads; after incubation with or without Mfd, magnetic pelleting of complexes separates retained RNA from RNA released into the supernatant (5). Release can also be detected as loss of the DNA exonuclease III digestion boundary of RNAP (5). A consideration of forces maintaining the elongation complex implies that RNA and enzyme are removed simultaneously, because neither transcript nor core RNAP alone would be stable in complex with DNA. Fig. 3 shows the time course of RNA release by E. coli Mfd from transcription complexes stalled by NTP deprivation at position 74 of an experimental DNA template (as well as a derived heteroduplex template) that is described below. The reaction generally continues until ≥75% of the RNA is released; in these conditions, release from homoduplex DNA is nearly complete by 1 min, the fastest convenient time for the manual separation process. However, the reaction is roughly linear at shorter times, as shown for the heteroduplex DNA.
Fig. 3.
Mfd-mediated RNA release from homoduplex and 60 GTG heteroduplex DNA. Each pair of lanes shows a gel analysis of transcripts of stopped elongation complexes affixed through a biotinylated DNA end to magnetic beads; the released or supernatant (S) fraction and retained or pellet (P) fraction after magnetic partitioning is shown. The position of the 60 GTG nontemplate strand substitution is illustrated. The data illustrated in Upper were quantified and are plotted in Lower. Circles, homoduplex DNA; squares, heteroduplex DNA; filled symbols, +Mfd; open symbols, −Mfd.
To determine whether DNA strand rewinding affects transcript release by Mfd, we prepared a series of heteroduplex templates containing substitutions of three nucleotide segments that prevent base-pairing in the transcription bubble or in the duplex region upstream of the bubble where Mfd is believed to bind. Nontemplate strand substitutions were used because the base composition of the template strand is constrained by the requirement to stop transcription complexes at the same site by nucleotide starvation. Fig. 1C shows a portion of the template and the presumed nucleic acid structure of the transcription complex at this site; the entire transcript up to position +74 consists of A and C, so that RNA synthesis with only ATP and CTP produces a complex with a defined 74-nt transcript. The depicted size of the transcription bubble fits experimental determinations from crosslinking, as well as an independent detection of the site of rewinding at the rear of the bubble, albeit in a different sequence context (15).
We find that Mfd releases RNA more slowly from heteroduplex DNA than from homoduplex DNA, for substitutions both within the transcription bubble and just upstream of the bubble (Fig. 4). The rate of release of transcript from one such substitution (60 GTG, named for the position of the first substituted nucleotide) is compared with the rate of release from homoduplex DNA in Fig. 3. This rate is reduced at least 3-fold by the substitution, although the reduction at very early times could be considerably larger, because there is no information about the initial rate of release from homoduplex DNA. Because of some variability in the maximum transcript release and the difficulty of manipulating the release assay for times <1 min, we show in Fig. 4 RNA released at 1 min as a fraction of RNA released from homoduplex DNA.
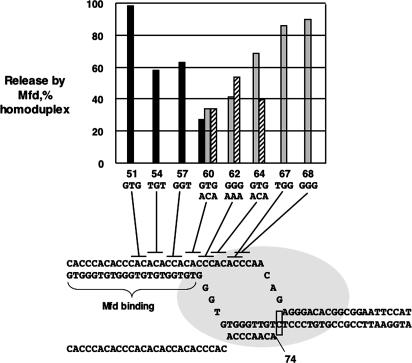
Fig. 4.
Effect of different nontemplate strand trinucleotide substitutions on Mfd-mediated RNA release. The indicated nontemplate strand DNA substitutions were used in release experiments in comparison with homoduplex wild-type DNA; the percentage of release relative to homoduplex DNA is shown. Black bars, heteroduplex series 1 (upper row of trinucleotide substitutions), experiment 1; gray bars, heteroduplex series 1 (upper row of trinucleotide substitutions), experiment 2; hatched bars, heteroduplex series 2 (lower row of trinucleotide substitutions). The incubation time was 1 min.
The important result of Fig. 4 is that heteroduplex substitutions mostly (substitution 62) or completely (substitution 64) within the region of single-stranded DNA of the transcription bubble inhibit release. Because substitutions of different base compositions are effective, we conclude that impairment of RNA release results from inhibition of DNA rewinding through loss of base-pairing and, therefore, that DNA rewinding in the transcription bubble occurs in the process of Mfd-mediated transcript release.
Heteroduplexes across ≈9 bp of the duplex region upstream of the transcription bubble (substitutions 54, 57, and 60) also inhibit transcript release. This effect could be attributed to lack of the natural duplex substrate for Mfd binding or possibly to a requirement for an upstream duplex DNA to bind an (unknown) site in RNAP. However, mismatches in duplex DNA upstream of the transcription bubble also are expected to stabilize a backtracked state of the elongation complex, which likely would inhibit release if Mfd must rewind DNA in the bubble to act.
Heteroduplex DNA in the Transcription Bubble Region also Inhibits Rho-Induced Transcript Release.
Rho factor is believed to have an entirely different primary substrate than Mfd, namely the emerging transcript, and there is no evidence that Rho interacts directly with DNA. Crystallographic analysis of a Rho-RNA structure shows that Rho binds ≈60 nt of transcript, which circles the RNA-binding domain of the Rho hexamer and penetrates into the translocase active center of Rho, oriented such that Rho can track in a 5′–3′ direction along the RNA (3). Although Rho is very unlikely to contact RNA in the region of the RNA/DNA hybrid in the transcription complex directly, a “helicase” activity could result from translocation that effectively pulls RNA from the complex, requiring that Rho be braced against RNAP at some undefined interaction site or possibly through an intermediary protein like NusG, which is known to bind both Rho and RNAP core (16).
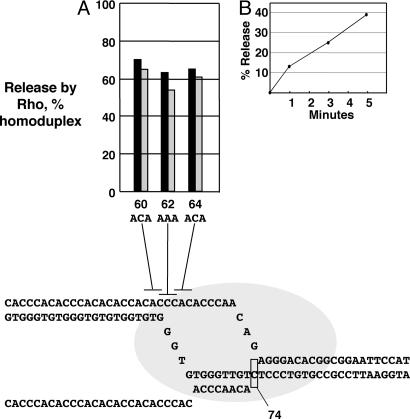
If the mechanisms of Rho and Mfd have in common that DNA rewinding provides energy to enable dissolution of the RNA/DNA hybrid and separation of the RNA, the DNA heteroduplexes in the nontemplate region of the transcription bubble also should inhibit Rho activity. We show in Fig. 5 that this is indeed the case. Rho activity can be measured by the magnetic bead-based RNA release assay described above; for 20 nM Rho (Fig. 5B), the rate of dATP-dependent release is approximately constant over 5 min. We assayed release with three nontemplate strand heteroduplexes in the same elongation complex described in Fig. 3, which was designed to contain the cytidine-rich transcript that optimally activates Rho.
Fig. 5.
Effect of nontemplate strand trinucleotide substitutions on Rho-mediated RNA release. (A) Rho-mediated release of RNA was measured from complexes stopped on nontemplate heteroduplex DNAs made with the substitutions shown. Black bars, release compared with wild-type homoduplex DNA; gray bars, release compared with mutant homoduplex DNA. (B) The time course of RNA release from complexes on wild-type homoduplex DNA.
As for Mfd, substitutions in the region of single-stranded DNA of the transcription bubble (substitutions 62 and 64) reduce the rate of RNA release by Rho by ≈2-fold. Thus, impairing DNA rewinding of the transcription bubble inhibits transcript release by Rho. Furthermore, a substitution in the upstream duplex region (substitution 60) also inhibits release. Because Rho does not contact DNA, we attribute the effect of this substitution to stabilizing a backtracked state of the stalled elongation complex and thus opposing upstream bubble rewinding that accompanies release.
Forward Translocation Is Promoted by Mfd and Rho During Transcript Release.
We have proposed that RNA release in intrinsic termination is facilitated by downstream translocation of the elongation complex, including unwinding of duplex DNA downstream of the bubble and movement of RNAP downstream along the DNA (8, 9), as illustrated in Fig. 2B. In this view, formation of the hairpin promotes a concerted rewinding of upstream DNA and unwinding of downstream DNA, effectively translocating the bubble without addition of nucleotides to the end of the RNA, thereby shortening the hybrid and favoring its dissolution. Some evidence in favor of this view is that a hairpin, or a DNA oligonucleotide that simulates the hairpin, provides enough force to increase transcription read-through of a DNA-binding protein (9).
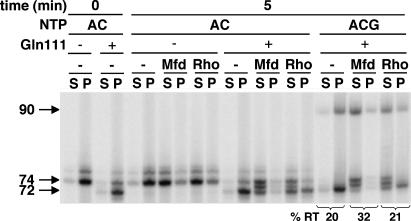
We show here that both Mfd and Rho also induce forward translocation as they release RNA from the complex. This result is not surprising for Mfd, which was shown to induce forward translocation of backtracked transcription complexes (5). For the experiment of Fig. 6, complexes were made and stalled by nucleotide deprivation on the (homoduplex) template illustrated in Fig. 4, with the additional condition that the downstream end was blocked by the DNA-binding protein EcoRI Gln-111 (17), an enzymatically inactive derivative of the EcoRI restriction enzyme, bound to the EcoRI sequence (GAATTC) that begins at position +87. Its effect is to stop most transcription at position +72 in the conditions used (50 μM ATP and CTP substrates), although the template sequence would allow elongation by ATP and CTP to position +74. (Higher concentrations of substrates allow synthesis to +74 against the roadblock, presumably by providing energy to force the enzyme forward against an elastic force provided by the EcoRI block.) When NTP substrates are removed and dATP is added as an energy source, both Mfd and Rho release complexes stopped by EcoRI Gln-111 at +72, confirming previous results (17, 18) (data not shown). However, when 7 μM ATP and CTP are included in the reaction to permit transcript elongation, the release is largely at positions +73 and +74 (Fig. 6); thus, both enzymes induce forward translocation of RNAP during the process of release. Presumably, Rho uses dATP energy to drive branch migration of the nucleic acids, against the force applied by EcoRI Gln-111, allowing further incorporation as forward translocation occurs. This experiment does not, of course, reveal whether translocation proceeds beyond the final site of incorporation as RNA release occurs.
Fig. 6.
Effect of Mfd and Rho on the translocation state of RNAP during RNA release. Transcription complexes were stopped on the JP-CA74-R1 template by EcoRI Gln-111 protein or by nucleotide deprivation at +74 through synthesis with only CTP and ATP. The EcoRI Gln-111 block stops RNAP at +72 in these conditions (50 μM ATP and CTP), although at higher NTP concentrations, the stop is at +74, in agreement with ref. 17. Released and retained RNA were measured after incubation with Mfd or Rho and the indicated NTP. In the last six lanes, 7 μM GTP was included, allowing continued synthesis if the EcoRI Gln-111 block could be removed during the reaction.
When 7 μM ATP, CTP, and GTP are provided to allow elongation beyond +74, Mfd causes a slight increase in transcription past the EcoRI Gln-111 block relative to the reaction in the absence of Mfd (Fig. 6). Presumably, the force exerted by Mfd is enough to displace EcoRI occasionally while the RNA 3′ end is still present in the RNAP active center, also a characteristic of the intrinsic terminator RNA hairpin (9).
Discussion
We have shown that disruption of the E. coli transcription complex by the two enzymes Mfd and Rho is inhibited if DNA strands of the open transcription bubble cannot pair, supporting mechanical models of termination. This result complements previous evidence that transcription termination by the RNA hairpin-based intrinsic terminator requires re-pairing of DNA strands in the transcription bubble and suggests a common underlying mechanism of RNA release. It is obvious that the end point of RNAP complex release must be reannealed DNA strands, because neither core RNAP nor RNA alone would engage DNA in a stable complex with unwound strands. However, because heteroduplexes inhibit release, the process also must involve intermediate stages in which DNA strands are partially rewound; in effect, the energy of DNA rewinding is used to drive the process of termination. For the intrinsic terminator, there are strong effects of heteroduplexes in DNA corresponding to the upstream half of the RNA/DNA hybrid and much smaller effects in the downstream half (15). Similarly, heteroduplex effects on Mfd activity are restricted to the upstream half of the hybrid region; the Rho results had less resolution.
We suggest that termination by all three mechanisms involves a concerted rewinding of DNA, initiating at the upstream edge of the transcription bubble, with unwinding of the RNA/DNA hybrid (essentially a branch migration, as in normal translocation) that proceeds until the RNA is sufficiently destabilized to dissociate from the complex. Furthermore, we suggest that the normal pathway involves forward translocation of RNAP with downstream DNA unwinding, as illustrated in Fig. 2B. Relative to normal translocation, the energy provided by advance of the RNA chain is lacking, but we suggest that this deficit is made up by the energy of hairpin formation or ATP or dATP utilization by Mfd or Rho. When a blocking agent prevents downstream translocation of RNAP, RNA release would occur as in Fig. 2C; the migrating branch invades the RNAP channel, collapsing the bubble and freeing the RNA, leading to dissociation of the complex. Previous results indicated that either a blocking agent (EcoRI Gln-111) or an intrastrand crosslink slows but does not prevent release of RNA at an intrinsic terminator, a result that is consistent with a more intrusive mechanism like that shown in Fig. 2C. If downstream unwinding were unfavorable, the pathway of Fig. 2C might be followed in the absence of a direct obstruction, or there might be partitioning between the two pathways. Our evidence argues against an allosteric model in which destabilization is induced only by conformational changes within the enzyme. However, conformational changes could accompany the models we propose, particularly that of Fig. 2C.
Our results allow a more complete description of the activity of Mfd on transcription complexes. We showed previously that Mfd uses ATP energy to drive forward translocation of the complex, visualized most directly with persistently backtracked (arrested) complexes that fail to elongate unless they are acted on by Mfd (5). There are two known important components of the Mfd polypeptide: a domain that binds the β-subunit of RNAP near the site where the DNA strands rewind at the upstream edge of the transcription bubble and a DNA translocase domain that binds upstream duplex DNA. Presumably, this assembly is oriented such that movement of the translocase along DNA forces the enzyme forward; the result is that RNAP tracks helically along the DNA, as long as forward movement allows the enzyme to continue melting downstream DNA. If the enzyme is blocked, the rotational motion becomes a torque imposed by the Mfd translocase activity on upstream DNA in such a direction as to collapse the transcription bubble. Although the release mechanism may well involve other interactions of the Mfd polypeptide with RNAP, we propose that complex dissociation results primarily from this collapse of the transcription bubble. This hypothesis is supported by mutational evidence that impairing translocase activity also inhibits the release function of Mfd (14).
An interesting, and contrasting, analogy can be drawn between models for the activity of Mfd and the eukaryotic polymerase II transcription initiation factor IIH. Whereas Mfd is proposed to use ATP energy to torque the bubble closed when forward translocation does not occur, transcription factor IIH is proposed to open the transcription bubble (and thus promote initiation) by exerting the opposite torque on downstream DNA (19).
Both Rho and Mfd induce some forward translocation against a transcription block, the EcoRI Gln-111 protein, allowing one or two more nucleotides to be added to the growing end as release occurs. We presume that the EcoRI Gln-111 protein provides an elastic barrier that can be compressed by a few nucleotides at high-substrate NTP or through the ATP (or dATP) energy transduced into nucleic acid movement by Mfd and Rho. Release then occurs when this translocation finally fails and bubble collapse ensues, coupled with unwinding of the RNA/DNA hybrid (Fig. 2C).
Because Rho acts only on the RNA, even though its activity still is influenced by the ability of DNA strands of the transcription bubble to rewind, it seems clear that DNA rewinding and RNA/DNA hybrid unwinding are coupled in some direct manner. This coupled movement presumably initiates within the elongation complex structure so that branch migration occurs while the RNA/DNA hybrid, upstream template strand, and emerging RNA are bound in the enzyme. (It is possible that upstream duplex DNA also is bound by RNAP, although no such contacts are known.) Either extraction of the RNA by Rho or forced translocation along DNA by Mfd would drive the coupled event that leads to release of RNA from the complex.
Materials and Methods
Proteins, Plasmids, and Templates for Transcription.
Mfd protein was purified from DH5α cells harboring pMFD19 as described (20). RNAP was purified as described (21). Rho protein was a gift from M. Kainz and R. Gourse (University of Wisconsin, Madison). EcoRI Gln-111 protein was a gift from P. Modrich (Duke University, Durham, NC) and I. Artsimovitch (Ohio State University, Columbus).
Templates and Plasmids.
Transcription templates were made by PCR of selected segments of plasmids. JP-CA74-R1 template contains the sequence: TTGCAAAACTGGATTAAAAAGCATATATTTCATATACCACCACACCCACACA CCCACACCCACACACCACACCCACACCCACACCCACACACCACACCCACACC. CAACAGAGGGACACGGCGGAATTC, where the −35 and −10 promoter sequences derived from the phage 82 late gene promoter are set in italics, the start site is the bold italicized A, and the last six nucleotides are an EcoRI site.
Terminally biotinylated templates were synthesized by PCR using biotinylated primers. They were purified with either the QIAquick PCR purification kit or the QIAquick gel extraction kit (Qiagen, Valencia, CA).
Heteroduplex Templates.
JP-CA74-RI and its derivatives are flanked by T7 and T3 primer sequences, which are upstream and downstream of the promoter, respectively. To produce heteroduplex DNA (22), wild-type JP-CA74-RI was amplified by PCR using a biotinylated T7 primer and a nonbiotinylated T3 primer, and mutant JP-CA74-RI carrying a 3-bp mutation was amplified by PCR using a nonbiotinylated T7 primer with four additional T residues at the 5′ end and a biotinylated T3 primer. After purification with the QIAquick PCR purification kit, PCR products were mixed, denatured, and reannealed. To isolate nonbiotinylated dsDNA, the reannealed DNAs were incubated with an excess amount of streptavidin for 30 min at room temperature and resolved on a Tris–acetate–EDTA agarose gel at 4°C. When biotin is bound to streptavidin, nonbiotinylated templates migrate faster than singly or doubly biotinylated templates on an agarose gel. Nonbiotinylated DNA was purified by using the MinElute gel purification kit (Qiagen), filled in with biotinylated dATP (Promega) using exonuclease-defective polymerase (VentR; NEB, Beverly, MA), and purified by using the QIAquick PCR purification kit.
In Vitro Transcription.
RNAP (50 nM) was added to 5 nM template bound to magnetic beads in transcription buffer [20 mM Tris·HCl, pH 8.0/0.1 mM EDTA/50 mM potassium glutamate/50 μg/ml acetylated BSA/50 μM ATP and CTP/0.2–1.0 μCi/μl [α-32P]CTP (1 Ci = 37 GBq)]. After 10 min of incubation at 37°C, 4 mM MgCl2 and 10 μg/ml rifampicin were added, and the reactions were incubated for 5 min at 37°C. The transcription buffer was replaced by the incubation buffer (20 mM Tris·HCl, pH 8.0/0.1 mM EDTA/50 mM potassium glutamate/50 μg/ml acetylated BSA/4 mM MgCl2/50 μM dATP). When indicated, 50 nM Mfd, 20 nM Rho, or the same volume of the storage buffer (10 mM Tris·HCl, pH 7.5/500 mM NaCl/1 mM DTT/1 mM EDTA/50% glycerol) was added. The reaction was incubated at 37°C for 1 min unless indicated otherwise. Each reaction was divided into supernatant and pellet fractions by magnetic partitioning and diluted with the precipitation buffer (500 mM Tris·HCl, pH 7.5/10 mM EDTA/100 μg/ml tRNA). After extraction with phenol/chloroform/isoamyl alcohol (50:50:1) and ethanol precipitation, samples were resolved on a polyacrylamide gel and bands were resolved and quantified with a PhosphorImager.
For the experiment shown in Fig. 6, the transcription buffer was supplemented with 50 nM Gln-111, and the incubation buffer contained 10 mM MgCl2 and 4 mM dATP plus 7 μM of the indicated NTP. The reaction was incubated at 37°C for 5 min before magnetic partitioning.
Acknowledgments
We thank members of our laboratory for discussions and reading the manuscript and Richard Ebright for helpful suggestions. This work was supported by National Institutes of Health Grant GM21941.
Abbreviation
- RNAP
RNA polymerase.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Nudler E., Gottesman M. E. Genes Cells. 2002;7:755–768. doi: 10.1046/j.1365-2443.2002.00563.x. [DOI] [PubMed] [Google Scholar]
- 2.Richardson J. P. Cell. 2003;114:157–159. doi: 10.1016/s0092-8674(03)00554-3. [DOI] [PubMed] [Google Scholar]
- 3.Skordalakes E., Berger J. M. Cell. 2003;114:135–146. doi: 10.1016/s0092-8674(03)00512-9. [DOI] [PubMed] [Google Scholar]
- 4.Selby C. P., Sancar A. Microbiol. Rev. 1994;58:317–329. doi: 10.1128/mr.58.3.317-329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park J. S., Marr M. T., Roberts J. W. Cell. 2002;109:757–767. doi: 10.1016/s0092-8674(02)00769-9. [DOI] [PubMed] [Google Scholar]
- 6.Roberts J., Park J. S. Curr. Opin. Microbiol. 2004;7:120–125. doi: 10.1016/j.mib.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 7.French S. Science. 1992;258:1362–1365. doi: 10.1126/science.1455232. [DOI] [PubMed] [Google Scholar]
- 8.Yarnell W. S., Roberts J. W. Science. 1999;284:611–615. doi: 10.1126/science.284.5414.611. [DOI] [PubMed] [Google Scholar]
- 9.Santangelo T. J., Roberts J. W. Mol. Cell. 2004;14:117–126. doi: 10.1016/s1097-2765(04)00154-6. [DOI] [PubMed] [Google Scholar]
- 10.Toulokhonov I., Artsimovitch I., Landick R. Science. 2001;292:730–733. doi: 10.1126/science.1057738. [DOI] [PubMed] [Google Scholar]
- 11.Komissarova N., Becker J., Solter S., Kireeva M., Kashlev M. Mol. Cell. 2002;10:1151–1162. doi: 10.1016/s1097-2765(02)00738-4. [DOI] [PubMed] [Google Scholar]
- 12.Selby C. P., Sancar A. J. Biol. Chem. 1995;270:4882–4889. doi: 10.1074/jbc.270.9.4882. [DOI] [PubMed] [Google Scholar]
- 13.Singleton M. R., Scaife S., Wigley D. B. Cell. 2001;107:79–89. doi: 10.1016/s0092-8674(01)00501-3. [DOI] [PubMed] [Google Scholar]
- 14.Chambers A. L., Smith A. J., Savery N. J. Nucleic Acids Res. 2003;31:6409–6418. doi: 10.1093/nar/gkg868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ryder A. M., Roberts J. W. J. Mol. Biol. 2003;334:205–213. doi: 10.1016/j.jmb.2003.09.039. [DOI] [PubMed] [Google Scholar]
- 16.Li J., Mason S. W., Greenblatt J. Genes Dev. 1993;7:161–172. doi: 10.1101/gad.7.1.161. [DOI] [PubMed] [Google Scholar]
- 17.Pavco P. A., Steege D. A. J. Biol. Chem. 1990;265:9960–9969. [PubMed] [Google Scholar]
- 18.Selby C. P., Sancar A. J. Biol. Chem. 1995;270:4890–4895. doi: 10.1074/jbc.270.9.4890. [DOI] [PubMed] [Google Scholar]
- 19.Kim T.-K., Ebright R. H., Reinberg D. Science. 2000;288:1418–1421. doi: 10.1126/science.288.5470.1418. [DOI] [PubMed] [Google Scholar]
- 20.Selby C. P., Sancar A. Science. 1993;260:53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- 21.Marr M. T., Roberts J. W. Mol. Cell. 2000;6:1275–1285. doi: 10.1016/s1097-2765(00)00126-x. [DOI] [PubMed] [Google Scholar]
- 22.Ring B. Z., Yarnell W. S., Roberts J. W. Cell. 1996;86:485–493. doi: 10.1016/s0092-8674(00)80121-x. [DOI] [PubMed] [Google Scholar]