Abstract
Specific and well-organized chromosome architecture in human sperm cells is supported by the prominent interactions between centromeres and between telomeres. The telomere-telomere interactions result in telomere dimers that are positioned at the nuclear periphery. It is unknown whether composition of sperm telomere dimers is random or specific. We now report that telomere dimers result from specific interactions between the two ends of each chromosome. FISH using pairs of subtelomeric DNA probes that correspond to the small and long arms of seven human chromosomes demonstrates that subtelomeres of one chromosome are brought together. Statistical analysis confirmed that telomere associations could not result from the random proximity of DNA sequences. Therefore, chromosomes in human sperm nuclei adopt a looped conformation. This higher-order chromosome structure is most likely required for chromosome withdrawal/decondensation during the early fertilization events leading to zygote formation.
Keywords: chromosome, fluorescence in-situ hybridization, human, spermatozoa, telomere
Introduction
Heterochromatic chromosomal domains such as centromeres and telomeres demonstrate an intrinsic tendency for association. Thus, centromere clusters and arrays were observed in mammalian interphase nuclei (Hadlaczky et al. 1986, Haaf & Schmidt 1989), while centromere pair-wise associations are characteristic to postmeiotic spermatogenic cells of different vertebrate species (Haaf et al. 1990). In budding yeast, telomeres cluster at the nuclear periphery in groups of four (Taddei & Gasser 2004). Terminal adhesions between non-homologous ends of polythene chromosomes seem to be an inherent feature of telomeres (Hinton 1945). Pronounced telomere-telomere interactions resulting in chromosome looping at the bouquet stage are prominent in meiotic cells at leptotene/zygotene stages (reviewed by Scherthan 2001, 2003). Walker and co-authors (Walker et al. 1991) described tight association between Xpter and Xqter segments in inactive X chromosomes (looped structure), while in active X chromosomes, such an association was not observed. In contrast, however, more recent publication presents evidence against such a looped structure of the inactive X chromosome (Dietzel et al. 1998).
In terminally differentiated spermatozoa, the extent of interactions between centromeres of non-homologous chromosomes and between telomeres is extremely high. These interactions result in the formation of a well-defined sperm chromocentre (Zalensky et al. 1993, 1995, Meyer-Ficca et al. 1998, Hazzouri et al. 2000) and obvious telomere-telomere complexes (Zalensky et al. 1997, Meyer-Ficca et al. 1998). Detailed quantitative analysis of telomere FISH performed in minimally decondensed human sperm cells, showed that, in 95% of nuclei, the observed number of telomere hybridization spots is 23, half of the telomere number expected for 23 chromosomes (Zalensky et al. 1997). Therefore, most, if not all telomeres in sperm are joined in dimers, and this feature is preserved in several mammalian species (Zalensky et al. 1997). Telomere dimers may result from contacts between two ends of one chromosome, from random adherence between ends of different chromosomes, or from specific interchromosomal interactions. The nature of these telomere dimers could not be established using in-situ hybridization with (TTAGGG)N DNA probe since this telomeric DNA is common for all chromosomes.
In this paper, we address the composition of telomere dimers in human sperm nuclei through in-situ hybridization with chromosome-specific subtelomeric DNA probes for seven human chromosomes. We demonstrate that the p and q subtelomeres of metacentric and submetacentric, large and small, chromosomes are localized in close proximity. Thus, telomere dimers in sperm correspond to interactions between the ends of each chromosome, and therefore, chromosomes in the nucleus of human spermatozoa are looped.
Materials and methods
Sperm cells were collected from semen obtained from donors with proven fertility. Cells were washed with PBS and fixed in 0.5% formaldehyde/PBS for 5 min. Limited nuclear decondensation was performed as described earlier (Zalensky et al. 1993, 1995). Briefly, cells were incubated in 10 mmol/L DTT, 0.04 mg/ml Heparin, PBS for 30 min, and then deposited onto microscope slides. In some experiments, sperm cells were first deposited onto slides, and then nuclear swelling was achieved by treatment with 10 mmol/L of lithium diiodosalicylate according to Celik-Ozenci (Celik-Ozenci et al. 2003). Such pretreatment resulted in more efficient hybridization and brighter signals. The final data obtained using these two procedures of nuclear swelling were identical within experimental error. Telomere FISH was performed essentially as described earlier (Zalensky et al. 1997) using biotinylated (TTAGGG)6 probe and signal detection with Avidin-FITC (Vector Inc.).
To localize subtelomeric DNA sequences, we used two-colour FISH with the directly labelled DNA probes from Vysis and/or from Aquarius. Hybridization and post-hybridization procedures were according to the manufacturer's instructions.
Chromosome 6 microdissected probes, kindly provided by Dr. M. Bittner (NIH), were PCR labelled with Biotin or Digoxigenin. Hybridization signals were detected using FITC-labelled anti-Dig antibodies (Boehringer Inc.) and Avidin-Texas Red (Vector Inc.).
FISH results were visualized using a Leitz Ortholux microscope, oil immersion objective 60 × 1.4NA, and a triple band-pass filter set. Images were collected using a digital colour camera (MagnaFire, Optronics Inc.) and processed with Adobe Photoshop 7.0 software. Measurements of distances in digitized images were performed in Sigma Scan Pro 5.0 software for individual sperm nuclei that had an unequivocal pattern of hybridization and a good morphology. In each experiment, intersubtelomere distance DNpNq and long nuclear axis L were measured in 200-300 cells. To correct for the slight variations in nuclear swelling between cells that may affect DNpNq we performed normalization and used DNpNq/L as a parameter describing distance between hybridization spots. Statistical analysis and graphical representation was performed using Microsoft Excel program supplemented with Analyse_It software.
Results and discussion
Recently, a complete set of human subtelomeric probes was developed (Knight et al. 1997, Kingsley et al. 1997, Knight & Flint 2000). Subtelomeric sequences are chromosome- and arm-specific; therefore, they were used here for localization of the physical ends of the individual chromosomes.
Extreme compactness of chromatin in sperm dictates the absolute necessity for artificial nuclear swelling to perform FISH. The degree of swelling depends on the type of DNA hybridization probe used. Subtelomere-specific probes need higher decondensation than a pantelomere probe. Following decondensation using these conditions native associations of telomeres into dimers (Zalensky et al. 1997) are partially destroyed (Figure 1A). Therefore, in the discussion of the data below, “extrapolation to the native state” of condensed sperm nuclei should be considered.
Figure 1.
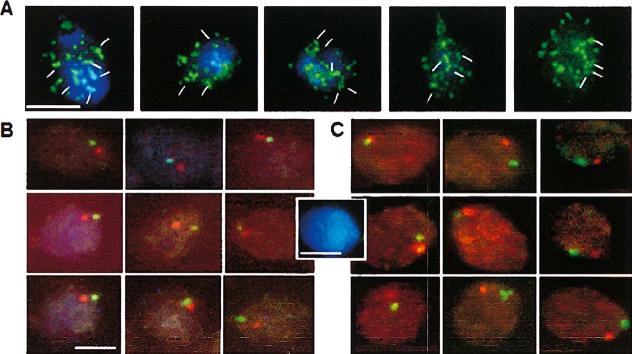
Telomere dimers and typical patterns of subtelomere localization in human sperm nuclei. (A) FISH with TTAGGG telomere probe (green), total nuclear DNA counterstained with DAPI (blue). White lines mark some telomere dimers. (B) FISH with subtelomere probes specific to chromosome 3: 3p (green) and 3q (red). (C) FISH with subtelomere probes specific to chromosome 18: 18p (green) and 18q (red). Scale bars correspond to 5 μm; DAPI-stained sperm cell in the centre shows the size of the untreated cell for comparison with sizes of mildly decondensed cells in (B) and (C).
Figure 1B provides an example of typical localization patterns of the p- and q-arm subtelomeric probes of chromosomes 3 and 18 in human sperm cells. The tips of chromosome arms often overlap; in nuclei that have been artificially pre-swollen before hybridization, their close proximity is evident. On average, the mean distance between subtelomeres is approximately 2 mm within an ellipsoid nuclear area of about 40 μm2. Since (1) subtelomeric probes are positioned within 400 kb of the telomere (Knight & Flint 2000) and (2) the nuclei have been decondensed resulting in an about three-fold increase in linear size (compare dimensions of swollen and native nuclei in Figure 1B), the actual distance between chromosome ends in native sperm cells is less than 0.6 mm.
To prove this observation and demonstrate that the p- and q-ends of one chromosome indeed join, we performed systematic measurements of distances between p and q subtelomere pairs of chromosomes 2, 3 and 4. Subtelomeres belonging to different chromosomes (3p-4q and 3p-2q) were localized as a control. We constructed frequency histograms (Figure 2A) to assess the scatter of data and to determine whether the data were normally distributed. Such descriptive presentation provides the mean value of the distance between chromosome ends and, more importantly, shows deviation of the distribution from random. Figure 2A shows that the distances between p- and q-subtelomeres of chromosome 2, 3 and 4 have a non-random dispersion and the plots are skewed, while the distances between subtelomeres belonging to different chromosomes D3p2q and D3p4q are distributed normally. The box-plot presentation in Figure 2B shows that D2p2q, D3p3q, D4p4q are noticeably less than the distances between pairs of subtelomeres belonging to different chromosomes. Therefore, in sperm nuclei, subtelomeric regions of each of these three chromosomes are closely located and this intranuclear position does not happen by chance.
Figure 2.
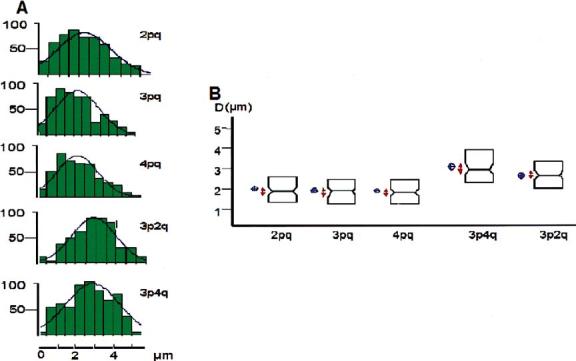
In human sperm nuclei, subtelomeric sequences located at the p- and q-arms of one chromosome are spatially adjacent. (A) Frequency distribution plots of distances between subtelomeres. Abscissa = DNpNg; ordinate = number of cells. (B) Box-plot presentation of distances between selected pairs of subtelomeres. Blue diamonds show the mean and the confidence interval around the mean; notched boxes show the median and red arrows the confidence intervals around the median.
In the following experiments, we expanded our data by determining distances between p and q subtelomeres for a set of six chromosomes that are different in sizes and some of which are submetacentric. To account for a slight variation in the sizes of the nuclei that influence the distance between a given pair of chromosome probes, we also performed normalization of each DNpNq to the length of the nuclei long axis (L) as described in the Methods. The normality of distributions was determined by the Kolmogorov-Smirnov test. Table 1 summarizes data for all seven chromosomes analysed. First, both the mean values of distances D and normalized distances D/L between the two ends of all chromosomes studied are notably smaller than the distance between subtelomeres that belong to different chromosomes. Second, distributions DNpNq and D/LNpNq for all chromosomes studied are non-random (p<0.02) while both distributions for 3p-4q and 3p-2q are random (p>0.15). The differences between the two groups are statistically significant.
Table 1.
Distances between subtelomeres in the partially decondensed nuclei of human sperm.
| D (μm)±SE | p | D/L±SE | p | |
|---|---|---|---|---|
| D = mean distance between centres of subtelomere hybridization signals; D/L = mean normalized distance where L is the length of the nuclei ellipsoid long axis; p = Kolmogorov-Smirnov test p-value (H0 normally distributed data). | ||||
| 1pq | 1.88±0.01 | <0.01 | 0.27±0.01 | <0.01 |
| 2pq | 2.17±0.01 | 0.02 | 0.28±0.01 | <0.01 |
| 3pq | 1.82±0.01 | <0.01 | 0.29±0.01 | <0.01 |
| 4pq | 2.02±0.01 | <0.01 | 0.28±0.01 | <0.01 |
| 7pq | 2.11±0.02 | 0.02 | 0.28±0.01 | <0.01 |
| 18pq | 1.39±0.01 | <0.01 | 0.21±0.01 | 0.02 |
| 20pq | 1.52±0.01 | <0.01 | 0.22±0.01 | <0.01 |
| 3p4q | 3.31±0.01 | >0.15 | 0.41±0.01 | >0.15 |
| 3p2q | 2.90±0.01 | >0.15 | 0.39±0.01 | >0.15 |
Quite interestingly, distances between subtelomeric signals of large chromosomes (1, 2, 3, 4 and 7) are larger than distances between subtelomeres of smaller chromosomes (18 and 20). This is statistically significant, as shown by comparison of the mean subtelomere distances for different chromosomes using t-test (Table 2). At the same time, differences between mean DNpNq of more metacentric (1 and 3) and less metacentric (2 and 4) chromosomes are statistically insignificant (Table 2). Pairwise comparison of the cumulative distributions using the Kolmogorov-Smirnov test supported the above conclusions (data not shown). The only considerable differences are between D of large and small chromosomes. This might be a consequence of the territorial chromosome organization (reviewed by Cremer & Cremer 2001) that is preserved in human sperm nuclei (Haaf & Ward 1995, Zalensky et al. 1995). Alternatively, larger chromosomes have a higher potential to relax during decondensation which will effectively result in greater intertelomere distances.
Table 2.
Statistical analysis of the differences between mean subtelomere distances.
| Δ(μm) | t | p | |
|---|---|---|---|
| t-test results, Δ=differences between mean pq-subtelomere distances for the indicated chromosome pairs. | |||
| 1pq/3pq | 0.025 | 1.68 | 0.1 |
| 1pq/2pq | 0.007 | 0.45 | 0.7 |
| 1pq/4pq | 0.004 | 0.31 | 0.7 |
| 2pq/4pq | 0.005 | 0.29 | 0.8 |
| 3pq/2pq | 0.017 | 1.06 | 0.3 |
| 3pq/4pq | 0.008 | 0.56 | 0.6 |
| 18pq/20pq | 0.011 | 1.4 | 0.3 |
| 1pq/18pq | 0.069 | 5.2 | <0.001 |
| 1pq/20pq | 0.054 | 3.7 | <0.001 |
In summary, data of Tables 1 and 2 demonstrate that, even in decondensed nuclei of human spermatozoa, p- and q-subtelomeres belonging to one chromosome are notably closer then subtelomeres from different chromosomes. This means that, in the ‘native’ sperm nucleus, chromosome ends are joined. Therefore, telomere dimers, the hallmark of mammalian sperm nuclear architecture (Zalensky et al. 1997, Meyer-Ficca et al. 1998), at least in humans, represent the associated ends of each chromosome.
The immediate consequence of such associations would be chromosome looping in sperm nuclei. We performed FISH using a combination of region-specific probes microdissected from chromosome 6 that included near telomere (6p-ter and 6q-ter) probes (Figure 3, upper drawing). FISH patterns in Figure 3 are typical, observed in the majority of cells (90%) at this degree of decondensation. Schemes of hybridization localization (Figure 3, right) illustrate trajectories of this chromosome in sperm nuclei corresponding to looped structures. A detailed study of chromosome higher-order structure using arm-specific probes (Mudrak & Zalensky, in preparation) also supports the looped configuration of chromosome in human spermatozoa. Overall, the chromosome hairpin protrudes from the nuclear interior, where centromeres of non-homologous chromosomes form a chromocentre (Zalensky et al. 1993, 1995; Haaf & Ward 1995, Hazzouri et al. 2000) to the nuclear surface where it is anchored by telomere-membrane interactions (Zalensky et al. 1997, Meyer-Ficca et al. 1998).
Figure 3.
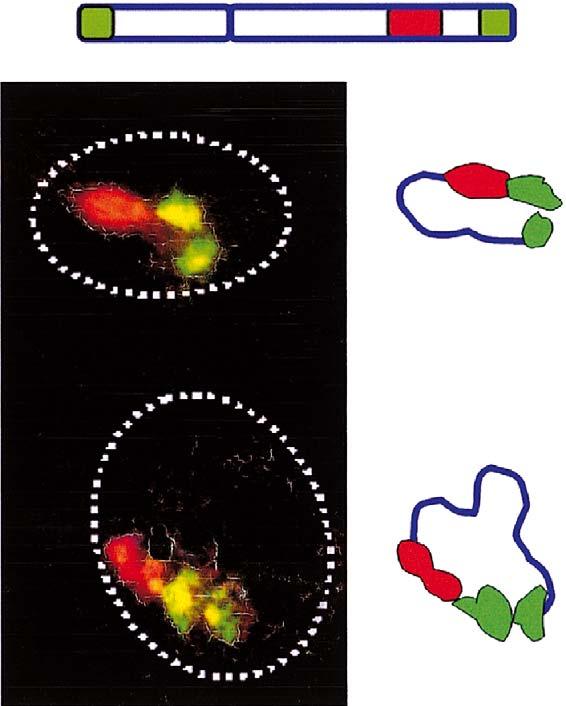
Looping of chromosome 6 as demonstrated by FISH with microdissected probes. Scheme on top shows the relative localization of the three probes used in this experiment and colour coding. Left panel - typical patterns of hybridization, stacked images obtained using laser scanning confocal microscopy; dotted lines show nuclei borders. Right panel - schematic diagrams of the chromosome trajectories that indicate their looped configurations.
Data presented here, demonstrate that chromosome looping is a consequence of the specific associations between DNA/chromatin structures located in terminal domains of chromosome arms. Such interactions could not be provided by the universal TTAGGG repeats. We propose that these interactions depend on chromosome arm-specific subtelomeric sequences and may involve protein complexes. Nuclear proteins specifically binding subtelomere sequences are yet to be established.
DNA (chromosome) looping is used in nature in a wide variety of contexts with the loop size ranging from hundreds of base pairs to whole chromosomes. Overall, looping plays an important role in the management and functional organization of genomes (Paul & Ferl l999). For example, actively transcribed gene loci (or loci poised for activation) have been found to loop away from their respective chromosome territories (Ragoczy et al. 2003, Chambeyron & Bickmore 2004). Walker and co-authors (Walker et al. 1991) observed looping of the inactive X chromosome. Similarly, X and Y chromosomes in the sex vesicle of humans, chimpanzees and mice are looped and their telomeres are clustered in a small area at the nuclear membrane (Metzler-Guillemain et al. 2000).
In sperm, chromosome territories (Haaf & Ward 1995, Zalensky et al. 1995) are non-randomly positioned within nuclei (Greaves et al. 2003, Zalenskaya & Zalensky 2004). Chromosome-size loops, described here in human spermatozoa, add one more level to the well-organized nuclear architecture characteristic of these cells. This architecture is most likely required for the early fertilization events leading to zygote formation.
Acknowledgements
This work was supported by the NIH grant R01 HD-42748 and, in part by the NIH grant HD-39830 to A. Zalensky.
References
- Celik-Ozenci C, Catalanotti J, Jakab A, et al. Human sperm maintain their shape following decondensation and denaturation for fluorescent in situ hybridization: shape analysis and objective morphometry. Biol Reprod. 2003;69:347–355. doi: 10.1095/biolreprod.103.019596. [DOI] [PubMed] [Google Scholar]
- Chambeyron S, Bickmore WA. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004;18:1119–1130. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- Dietzel S, Eils R, Satzler K, et al. Evidence against a looped structure of the inactive human X-chromosome territory. Exp Cell Res. 1998;240:187–196. doi: 10.1006/excr.1998.3934. [DOI] [PubMed] [Google Scholar]
- Greaves I, Rens KW, Ferguson-Smith MA, Griffin D, Marshall Graves JA. Conservation of chromosome arrangement and position of the X in mammalian sperm suggests functional significance. Chromosome Res. 2003;11:503–512. doi: 10.1023/a:1024982929452. [DOI] [PubMed] [Google Scholar]
- Haaf T, Schmid M. Centromeric association and non-random distribution of centromeres in human tumour cells. Hum Genet. 1989;81:137–143. doi: 10.1007/BF00293889. [DOI] [PubMed] [Google Scholar]
- Haaf T, Ward DC. Higher order nuclear structure in mammalian sperm revealed by in situ hybridization and extended chromatin fibers. Exp Cell Res. 1995;219:604–611. doi: 10.1006/excr.1995.1270. [DOI] [PubMed] [Google Scholar]
- Haaf T, Grunenberg H, Schmid M. Paired arrangement of nonhomologous centromeres during vertebrate spermiogenesis. Exp Cell Res. 1990;187:157–161. doi: 10.1016/0014-4827(90)90130-3. [DOI] [PubMed] [Google Scholar]
- Hadlaczky G, Went M, Ringertz NR. Direct evidence for the non-random localization of mammalian chromosomes in the interphase nucleus. Exp Cell Res. 1986;167:1–15. doi: 10.1016/0014-4827(86)90199-0. [DOI] [PubMed] [Google Scholar]
- Hazzouri M, Rousseaux S, Mongelard F, et al. Genome organization in the human sperm nucleus studied by FISH and confocal microscopy. Mol Reprod Dev. 2000;55:307–315. doi: 10.1002/(SICI)1098-2795(200003)55:3<307::AID-MRD9>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Hinton T. A study of chromosome ends in salivary gland nuclei of Drosophila. Biol Bull. 1945;88:144–165. [Google Scholar]
- Kingsley K, Wirth J, van der Maarel S, Freier S, Ropers HH, Haaf T. Complex FISH probes for the subtelomeric regions of all human chromosomes: comparative hybridization of CEPH YACs to chromosomes of the Old World monkey Presbytis cristata and great apes. Cytogenet Cell Genet. 1997;78:12–19. doi: 10.1159/000134616. [DOI] [PubMed] [Google Scholar]
- Knight SJ, Flint J. Perfect endings: a review of subtelomeric probes and their use in clinical diagnosis. J Med Genet. 2000;37:401–409. doi: 10.1136/jmg.37.6.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SJ, Horsley SW, Regan R, et al. Development and clinical application of an innovative fluorescence in situ hybridization technique which detects submicroscopic rearrangements involving telomeres. Eur J Hum Genet. 1997;5:1–8. [PubMed] [Google Scholar]
- Metzler-Guillemain C, Usson Y, Mignon C, et al. Organization of the X and Y chromosomes in human, chimpanzee and mouse pachytene nuclei using molecular cytogenetics and three-dimensional confocal analyses. Chromosome Res. 2000;8:571–584. doi: 10.1023/a:1009277722579. [DOI] [PubMed] [Google Scholar]
- Meyer-Ficca M, Muller-Navia J, Scherthan H. Clustering of pericentromeres initiates in step 9 of spermiogenesis of the rat (Rattus norvegicus) and contributes to a well defined genome architecture in the sperm nucleus. J Cell Sci. 1998;111:363–1370. doi: 10.1242/jcs.111.10.1363. [DOI] [PubMed] [Google Scholar]
- Paul AL, Ferl RJ. Higher-order chromatin structure: looping long molecules. Plant Mol Biol. 1999;41:713–720. doi: 10.1023/a:1006323222693. [DOI] [PubMed] [Google Scholar]
- Ragoczy T, Telling A, Sawado T, Groudine M, Kosak ST. A genetic analysis of chromosome territory looping: diverse roles for distal regulatory elements. Chromosome Res. 2003;11:513–525. doi: 10.1023/a:1024939130361. [DOI] [PubMed] [Google Scholar]
- Scherthan H. A bouquet makes ends meet. Nat Rev Mol Cell Biol. 2001;2:621–627. doi: 10.1038/35085086. [DOI] [PubMed] [Google Scholar]
- Scherthan H. Knockout mice provide novel insights into meiotic chromosome and telomere dynamics. Cytogenet Genome Res. 2003;103:235–244. doi: 10.1159/000076809. [DOI] [PubMed] [Google Scholar]
- Taddei A, Gasser SM. Multiple pathways for telomere tethering: functional implications of subnuclear position for heterochromatin formation. Biochim Biophys Acta. 2004;1677:l20–l28. doi: 10.1016/j.bbaexp.2003.11.014. [DOI] [PubMed] [Google Scholar]
- Walker CL, Cargile CB, Floy KM, Delannoy M, Migeon BR. The Barr body is a looped X chromosome formed by telomere association. Proc Natl Acad Sci USA. 1991;88:6191–6195. doi: 10.1073/pnas.88.14.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalenskaya IA, Zalensky AO. Non-random positioning of chromosomes in human sperm nuclei. Chromosome Res. 2004;12(2):163–173. doi: 10.1023/b:chro.0000013166.04629.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalensky AO, Breneman J, Zalenskaya IA, Brinkley BR, Bradbury EM. Organization of centromeres in the decondensed nuclei of mature human sperm. Chromosoma. 1993;102:509–518. doi: 10.1007/BF00368344. [DOI] [PubMed] [Google Scholar]
- Zalensky AO, Allen MJ, Kobayashi A, Zalenskaya IA, Balhorn R, Bradbury EM. Well-defined genome architecture in the human sperm nucleus. Chromosoma. 1995;103:577–590. doi: 10.1007/BF00357684. [DOI] [PubMed] [Google Scholar]
- Zalensky AO, Tomilin NV, Zalenskaya IA, Teplitz R, Bradbury EM. Telomere-telomere interactions and telomere binding proteins in mammalian sperm. Exp Cell Res. 1997;232:29–41. doi: 10.1006/excr.1997.3482. [DOI] [PubMed] [Google Scholar]