Abstract
Rabies virus (RV) vaccine strain-based vectors show great promise as vaccines against other viral diseases such as human immunodeficiency virus type 1 (HIV-1) infection and hepatitis C, but a low residual pathogenicity remains a concern for their use. Here we describe several highly attenuated second-generation RV-based vaccine vehicles expressing HIV-1 Gag. For this approach, we modified the previously described RV vaccine vector SPBN by replacing the arginine at position 333 (R333) within the RV glycoprotein (G) with glutamic acid (E333), deleting 43 amino acids of the RV G cytoplasmic domain (CD), or combining the R333 exchange and the CD deletion. In addition, we constructed a new RV vector that expresses HIV-1 Gag from an RV transcription unit upstream of the RV phosphoprotein gene (BNSP-Gag) instead of upstream of the G gene. As expected and as demonstrated for SPBN-Gag, all vaccine vehicles were apathogenic after peripheral administration. However, the new, second-generation vaccine vectors containing modifications in the RV G were also apathogenic after intracranial infection with 105 infectious particles, and BNSP-Gag produced a 50%-reduced mortality in mice. Of note, the observed attenuation of pathogenicity did not result in either the attenuation of the humoral response against the RV G or the previously observed robust cellular response against HIV-1 Gag. These findings demonstrate that very safe and highly effective RV-based vaccines can be constructed and further emphasize their potential utility as efficacious antiviral vaccines.
Rabies virus (RV) is a nonsegmented negative-strand RNA virus of the Rhabdoviridae family. The 12-kb RV genome encodes five monocistronic RNAs encoding the nucleocapsid protein (N), phosphoprotein (P), matrix protein (M), the single transmembrane protein G, and the viral polymerase (L). The RV virion consists of an internal core or ribonucleoprotein (RNP) complex, which is composed of the viral RNA encased in the N protein and associated with P and L (32) and an external component, the viral envelope, which consists of the host-cell derived membrane with the M protein located at its inner surface and the membrane-spanning G protein (18, 19). The G protein is responsible for both the interaction with a cellular receptor(s) and pH-dependent membrane fusion (33).
RV pathogenicity has been studied for more than 100 years, with research results indicating that RV consists of a wide array of variants. These can range from highly pathogenic strains, such as silver-haired bat virus, to extremely attenuated RV vaccine strains, such as SAG-2, which are not pathogenic in severe combined immunodeficiency (SCID) mice after oral application (20, 23; C. Hanlon, M. Fiorello, C. L. Schumacher, V. Shankar, A. Hamir, and C. Rupprecht, Abstr. 4th Annu. Int. Meet. Adv. Rabies Control Americas, 1993). Two proteins have been associated with RV pathogenesis, namely, P and G. It has been suggested that a specific interaction of a conserved domain within RV P and the cytoplasmic dynein light chain (LC8) is responsible for retrograde transport to the central nervous system and, therefore, is partly responsible for RV pathogenesis (14, 24, 25). Recently, Mebatsion showed that the deletion of 11 amino acids (aa) within P abolishes the P-LC8 interaction and reduces the efficiency of the peripheral spread of RV (17). A large body of evidence, however, shows that RV G, the only target for neutralizing antibodies, is the major contributor to the pathogenicity of the virus. The RV G protein must interact effectively with cell surface molecules that can mediate rapid virus uptake (6, 22, 30). An important pathogenicity marker of RV is the arginine residue at position 333 (R333) of RV G, and a change to glutamic acid (E333) or aspartic acid (D333) not only abolishes RV pathogenesis but also hampers the virus ability to enter motor neurons (5). Certain partially attenuated vaccine strains can be completely attenuated, even after intracranial (i.c.) inoculation, by the E333 exchange; however some pathogenic RV strains cannot be fully attenuated with this mutation, indicating that other mutations in the RV G may be necessary (23).
It has been shown that the deletion of the RV G cytoplasmic domain (CD) reduces both the viral titer and spread of RV (18), but changes in the pathogenicity of such a modified RV were not analyzed. However, a recombinant vesicular stomatitis virus (VSV) expressing influenza A virus hemagglutinin (HA) that contains a truncation of the VSV G CD was completely attenuated in a small-animal model. Mice immunized intranasally with VSV carrying the CD-deleted G showed no weight loss and were protected against influenza A virus challenge. However, the parental virus expressing HA caused a transient 20% weight loss (26).
It has also been shown for VSV that certain rearrangements of the VSV gene order and expression of VSV G from a further-3′ location can eliminate the potential for the virus to cause disease and protect the natural host from VSV challenge (2, 10, 11). However, the usefulness of vectors with these changes as vaccine vehicles has not been analyzed.
Earlier studies from our laboratory have shown that RV is a highly effective vaccine vehicle. Immunization with RV vaccine strain-based vectors expressing human immunodeficiency virus type 1 (HIV-1) Gag or Env induces humoral and vigorous cellular responses against the expressed antigens. In addition, immunization with killed RV virions containing hepatitis C virus (HCV) E2 in their envelopes resulted in mouse seroconversion against HCV E2 after two inoculations without adjuvant. Whereas the use of killed RV is no concern in humans, the use of a viral vector always raises safety concerns. Our currently utilized RV-based vectors are very safe after peripheral application, but even a low vaccine vector-induced pathogenesis is undesirable.
Here we describe a second generation of RV-based vectors. To define the role of the R333 and the CD of RV G for vaccine vector-related pathogenesis, we constructed several recombinant RV vaccine strain-based vectors expressing HIV-1 Gag containing the mutated G proteins. In addition, we constructed a new RV vector that expresses HIV-1 Gag from an RV transcription unit upstream of the RV phosphoprotein gene (BNSP-Gag). As expected, all vaccine vehicles were apathogenic after peripheral administration. However, the new, second-generation vaccine vectors containing modifications in the RV G were also apathogenic after i.c. infection with 105 infectious particles, and BNSP-Gag produced a 50%-reduced mortality in mice. These data indicate that we are able to eliminate vector-associated pathogenesis and show that it is possible to create vaccine vectors for human use which may be safer than other recombinant vaccine vectors currently under investigation.
MATERIALS AND METHODS
Plasmid construction.
The plasmid encoding a recombinant RV vaccine vector (pSPBN) and pSPBN containing HIV-1 Gag (pSPBN-Gag) were described previously (17). To create a new RV vaccine vector expressing a foreign gene from a further-3′ position, pSPBN was digested with BsiWI and NheI and 5′ overhangs were filled in with DNA polymerase I large fragments (Klenow) and religated. The plasmid was designated pSP. pSP was the target to introduce a new transcription stop-start sequence, as well as a single BsiWI and NheI site by using a PCR strategy. First, two fragments were amplified by PCR from pSPBN using Vent polymerase (New England Biolabs) and the forward primer RP64 (5′-TGTGCTGCAAGGCGATTAAG-3′) or RP100 (5′-CCCGCTAGCCATGAAAAAAACTAACACCCCTCC-3′). The reverse primer was RP60 (5′-GACTTGGATCGTTGAAAG-3′) or RP99 (5′-CCCGCTAGCAAAACGTACGGGAGGGGTGTTAGTTTTTTTCATGTTATGAGTCACTCGAATATG-3′). The PCR products were digested with NheI and were ligated, and the 3.5-kb fragment was eluted from a gel. The fragment was reamplified with primers RP60 and RP64, gel purified, and digested with NcoI and PstI and was ligated to pSP previously digested with NcoI and PstI. The plasmid was designated pBNSP. To construct a recombinant BNSP expressing HIV-1 Gag, pSPBN-Gag was digested with BsiWI and NheI and the 1.5-kb fragment was eluted from an agarose gel and cloned into pBNSP previously digested with BsiWI and NheI.
The second-generation RV-based vaccine strain vehicles containing modified G proteins were constructed by using a PCR strategy and site-directed mutagenesis. First, we replaced the arginine at position 333 within the RV G with glutamic acid by using site-directed mutagenesis (GeneEditor; Promega Inc.) with RP10 (5′-CACTACAAGTCAGTCGAGACTTGGAATGAGATC-3′) and with pSBN (28) as a target. The resulting plasmid was designated pSBN-333. The coding sequence for RV G-333 was amplified by PCR from pSBN-333 and used to replace that for wild-type G in pSPBN-Gag by utilizing the XbaI/HpaI sites flanking G (Fig. 1). The plasmid was designated pSPBN-333-Gag. The recombinant RV pSPBN-ΔCD-Gag and pSPBN-ΔCD-333-Gag were constructed by replacing the RV G gene in SPBN-Gag with the PCR product from the template SPBN or SBN-333 by using the primers RP11 (5′-CCTCAAAAGACCCCGGGAAAGATGGTTCCTCAG-3′) and RP118 (5′-CCCTTAATTAATTATCTACAACATGTCATCAGG-3′). The PCR products were digested with XbaI and HpaI (the HpaI site is italicized) and cloned into pSPBN-Gag, previously digested with XbaI and HpaI.
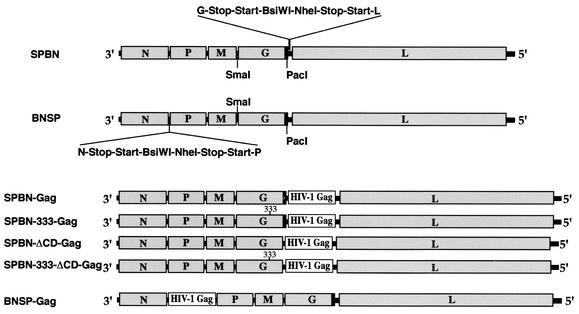
FIG. 1.
Construction of different recombinant vaccine vectors expressing HIV-1 Gag. At the top are two RV vaccine strain-based vectors containing an additional transcription stop-start signal flanked by two unique restriction sites between the G and L genes (SPBN) or the N and P genes (BNSP). These vectors were the targets to introduce the gene encoding HIV-1 Gag (SPBN-Gag, SPBN-333-Gag, SPBN-ΔCD-Gag, SPBN-333-ΔCD-Gag, and BNSP-Gag). By site-directed mutagenesis and a PCR strategy, an amino acid change at position 333 within the RV G from arginine (R) to glutamic acid (E) was introduced (SPBN-333-Gag and SPBN-333-ΔCD-Gag) and/or 43 of 44 aa of the RV G CD (black boxes) were deleted (SPBN-ΔCD-Gag and SPBN-333-ΔCD-Gag).
Generation of recombinant viruses.
For experiments involving recovery of the recombinant RVs, the previously described RV recovery system was used (9, 28). Briefly, BSR T7 cells (3), which stably express T7 RNA polymerase, were transfected with 5 μg of full-length RV cDNA (SPBN-333-Gag, SPBN-ΔCD-Gag, SPBN-333-ΔCD-Gag, BNSP-Gag, and BNSP) in addition to plasmids encoding the RV N, P, L, and G proteins by using a Ca2PO4 transfection kit (Stratagene), as instructed by the vendor. Three days posttransfection, supernatants were transferred onto fresh BSR cells and infectious RV was detected 3 days later by immunostaining with fluorescein isothiocyanate for the RV N protein (Centacor Inc.).
Western blotting.
BSR cells were infected with SPBN, SPBN-Gag, SPBN-333-Gag, SPBN-ΔCD-Gag, SPBN-333-ΔCD-Gag, BNSP, or BNSP-Gag at a multiplicity of infection (MOI) of 2 for 48 h and resuspended in lysis buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 1% NP-40, 0.1% sodium dodecyl sulfate [SDS], 1× protease inhibitor cocktail [Sigma]) on ice for 5 min. The suspension was transferred to a microcentrifuge tube and spun for 1 min at 14,000 rpm to remove cell debris. Proteins were separated by SDS-10% polyacrylamide gel electrophoresis (PAGE) and transferred to a PVDF-Plus membrane (Osmonics, Minnetonka, Minn.). Blots were blocked for 1 h (5% dry milk powder in phosphate-buffered saline [PBS; pH 7.4]) and then washed three times with a 0.1% PBS-Tween 20 solution and incubated with a human anti-p24 antibody (1:1,000), a polyclonal rabbit anti-RV G antibody (1:10,000), or a polyclonal rabbit anti-RV RNP antibody (1:2,000) overnight at 4°C (12). Blots were then washed three times with 0.1% PBS-Tween. Secondary Alexa Fluor 546 goat anti-human immunoglobulin G (IgG) (1:500) or Alexa Fluor 532 goat anti-rabbit IgG (1:1,000) (Molecular Probes, Inc.) was added, and blots were incubated for 2 h at room temperature. Blots were washed three times with 0.1% PBS-Tween and washed once with PBS (pH 7.4). Fluorescence analysis was performed as instructed by the vendor.
HIV-1 p24 antigen capture ELISA.
HeLa cells were infected at a MOI of 5, and, 48 h later, supernatants were collected and cells were resuspended in lysing buffer (Triton X-100 in PBS-2-chloroacetamide). The supernatants and cell suspension were transferred to microcentrifuge tubes and spun for 2 min at 14,000 rpm to remove cell debris. The quantification of Gag p24 protein in cell supernatants and lysates was performed by using the p24 antigen enzyme-linked immunosorbent assay (ELISA), as described by the manufacturer (ZeptoMetrix, Inc.).
Multicycle growth and one-step growth curves.
BSR cells (a BHK-21 clone) were plated in 60-mm-diameter dishes and 16 h later were infected at a MOI of 0.01 (multicycle growth) or 5 (one-step growth) with SPBN, SPBN-Gag, SPBN-333-Gag, SPBN-ΔCD-Gag, SPBN-333-ΔCD-Gag, BNSP, or BNSP-Gag. After incubation at 37°C for 1 h, inocula were removed and cells were washed four times with PBS to remove any unabsorbed virus. Three milliliters of complete medium was added back, and 100 μl of tissue culture supernatants was removed at various time points after infection (see Fig. 3). Virus aliquots were titrated in duplicate on BSR cells.
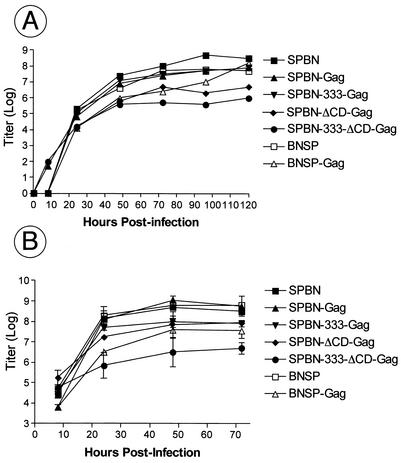
FIG. 3.
Multicycle replication and one-step growth curve of recombinant vaccine vectors. BSR cells were infected with SPBN, SPBN-Gag, SPBN-333-Gag, SPBN-ΔCD-Gag, SPBN-333-ΔCD-5, BNSP, or BNSP-Gag at a MOI of 0.01 (multicycle growth; A) or 5 (one-step growth curve; B). Aliquots of tissue culture supernatants were collected, and viral titers were determined in duplicate.
Studies of pathogenicity in mice.
Groups of 10 6- to 8-week-old female Swiss-Webster mice (Taconic Farms) were inoculated i.c. with 10 μl containing 105 infectious particles of the different recombinant vaccines. One group (10 mice) of uninfected mice served as a control. After inoculation, the mice were observed daily for the first 2 weeks and every other day for another 2 weeks for any signs of disease, i.e., weight loss. Mice were euthanized at the onset of neurological symptoms.
RV G protein ELISA.
RV G protein was purified from sucrose-purified RV virions as described previously. RV G was resuspended in coating buffer (50 mM Na2CO3, pH 9.6) at a concentration of 200 μg/ml and plated in 96-well ELISA MaxiSorp plates (Nunc) with 100 μl in each well. After overnight incubation at 4°C, plates were washed three times (PBS [pH 7.4], 0.1% Tween 20), blocked with blocking buffer (PBS [pH 7.4], 5% dry milk powder) for 30 min at room temperature, and incubated with serial dilutions of sera for 1 h. Plates were washed three times, followed by the addition of horseradish peroxidase-conjugated goat anti-mouse IgG (heavy plus light chains) secondary antibody (1:5,000; Jackson ImmunoResearch Laboratories). After a 30-min incubation at 37°C, plates were washed three times and 200 μl of OPD (o-phenylenediamine dihydrochloride; Sigma) substrate was added to each well. The reaction was stopped by the addition of 50 μl of 3 M H2SO4 per well. Optical density was determined at 490 nm.
Immunization and enzyme-linked immunospot (ELISPOT) assays.
Groups of five 6- to 8-week-old female BALB/c mice (Harlan) were inoculated intraperitoneally with 3.4 × 106 focus-forming units of SPBN, SPBN-Gag, SPBN-333-Gag, SPBN-ΔCD-Gag, SPBN-333-ΔCD-Gag, BNSP, BNSP-Gag, or VSV-Gag. Four to 5 weeks postimmunization, mice were challenged with 107 PFU of vaccinia virus expressing HIV-1 Gag. Five days after the challenge, two mice per group were sacrificed, spleens were removed and pooled, and single-cell suspensions were prepared. Red blood cells were removed with ACK lysing buffer (BioSource), and cells were washed twice in RPMI 10 supplemented with 10% fetal bovine serum (FBS). Ninety-six-well filtration plates (Millipore, Bedford, Mass.) were coated overnight at 4°C with 10 μg of anti-mouse gamma interferon (IFN-γ; Pharmingen)/ml in sterile PBS. The plates were blocked for 1 h with 5% bovine serum albumin (BSA) in PBS at 37°C. The plates were prepared for the splenocytes by the addition of 100 μl of RPMI 1640 supplemented with 10% FBS and incubated an additional 2 h. Dilutions of splenocytes were added to the plates and incubated with or without a major histocompatibility complex class I-restricted p24 peptide (AMQMLKETI) for 16 h at 37°C. Plates were washed 10 times with PBS containing 0.25% Tween 20 and then once with sterile distilled water. Wells were incubated with 5 μg of biotinylated rat anti-mouse antibody/ml for 2 h and then washed five times in 0.25% Tween 20 in PBS. The wells were treated with 1 mg of horseradish peroxidase-conjugated streptavidin/ml in PBS containing 1% BSA, incubated for 2 h at room temperature, and washed four times. IFN-γ-secreting spot-forming cells were detected by the addition of diaminobenzidine-4-chloro-1-naphthol in cold methanol.
RESULTS
Construction of attenuated RVs expressing HIV-1 Gag protein.
Recent studies show that foreign proteins such as HIV-1 Env and Gag are stably expressed by RV-based vaccine vectors and induce long-lasting and vigorous immune responses in vaccinated mice (15, 16, 28). However, some pathogenesis associated with the vector itself remains a concern for its use in humans. A large body of evidence supports the suggestion that a single surface glycoprotein, G, of RV is responsible for RV pathogenesis (6, 8, 20, 23, 30). We therefore constructed several new, second-generation vaccine vectors by genetically modifying the RV G. Using site-directed mutagenesis and PCR, we introduced an amino acid change at position 333 from arginine (R) to glutamic acid (E), deleted 43 of 44 aa of the RV G cytoplasmic tail, or did both. The position 333 mutation has previously been shown to result in slower uptake of the virus by cells (6), to interfere with the possibility of certain RV strains infecting motor neurons, and to diminish the efficient spread of the virus in neuroblastoma cells (5). One study by Mebatsion and colleagues showed that the RVs with CD-deleted Gs form smaller foci and have reduced viral titers (18), which also should result in a reduction of the vector-associated pathogenesis. However, the deletion of the G CD itself is not likely to affect the neurotropism of the recombinant RV, which is most likely determined by the RV G ectodomain (12, 23, 34). The SmaI/PacI site flanking the RV G gene was used to introduce the coding regions of the modified G genes into the previously described RV vaccine vector expressing HIV-1 Gag (SPBN-Gag; Fig. 1). The resulting plasmids were designated pSPBN-333-Gag, pSPBN-ΔCD-Gag, and pSPBN-333-ΔCD-Gag (Fig. 1). In addition, a new RV vector containing a new RV transcription unit between the RV N and P genes was constructed. The goal of this construct was twofold. First, a location further upstream of the RV 3′ end is expected to result in a higher expression level of the foreign protein; second, a gene at this location should lower the expression levels of the downstream proteins, which may result in slower replication and therefore reduced pathogenicity. The new vaccine vector was designated BNSP and was the target used to introduce the HIV-1 Gag gene, resulting in BNSP-Gag (Fig. 1).
The cDNAs encoding the plus-strand RNA of the respective recombinant RVs were cotransfected with plasmids encoding the RV N, P, G, and L proteins into cells stably expressing T7 RNA polymerase (3) by standard methods (15, 16), and infectious viruses for all six recombinant RVs were recovered. The sequences of the modified G genes were verified by sequencing reverse transcription-PCR products, and the results indicated the alteration within the RV genome.
Expression of HIV-1 Gag by recombinant RVs.
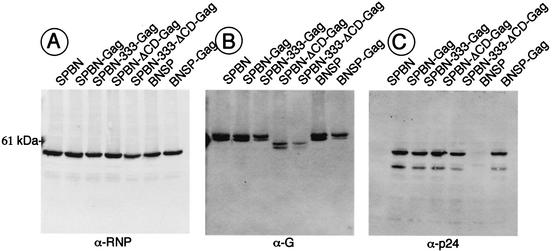
To determine whether the recombinant RVs express HIV-1 Gag and the modified G proteins correctly, BSR cells were infected at a MOI of 5 with SPBN-Gag, SPBN-333-Gag, SPBN-ΔCD-Gag, SPBN-333-ΔCD-Gag, and BNSP-Gag, with the parental vectors SPBN and BNSP as controls. Forty-eight hours later, cell extracts were prepared and proteins were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and analyzed by Western blotting with antibodies directed against RV RNP, RV G, or HIV-1 p24 (Fig. 2). Quantification with a “quantifier” indicated similar amounts of RV N expressed by all recombinant RVs (Fig. 2A). A polyclonal antibody directed against RV G detected the G protein of the expected size of RV G, and the slightly faster migrating G proteins with 43 aa of the CD deleted, expressed from SPBN-ΔCD-Gag and SPBN-333-ΔCD-Gag, were also detected. The weaker signals detected for the two CD-deleted G proteins may be due to their release from the cell membrane into the supernatants of the infected cells or to a weaker reaction of the polyclonal antibody with the CD-deleted G proteins. Blotting with an antibody directed against HIV-1 p24 detected a protein of ∼55 kDa, which is the size expected for unprocessed HIV-1 Gag p55. Quantification with the quantifier indicated similar amounts of p55 in cell extracts infected with all five recombinant RVs expressing HIV-1 Gag. These results were not surprising for cell extracts infected with SPBN-Gag, SPBN-333-Gag, SPBN-ΔCD-Gag, or SPBN-333-ΔCD-Gag, but larger amounts of p55 were expected for BNSP-Gag. We therefore decided to analyze the amount of HIV-1 Gag released into the supernatants of SPBN-Gag- or BNSP-Gag-infected cells 48 h after infection. In both cases, approximately 2 ng of Gag protein/ml was detected by a p24 ELISA. These data indicate that neither the production of HIV-1 Gag within the BNSP-Gag-infected cells nor the amount of secreted p55 is enhanced after expression from a further-upstream location.
FIG. 2.
Western blot analysis of recombinant vaccine vectors. BSR cells were infected with SPBN, SPBN-Gag, SPBN-333-Gag, SPBN-ΔCD-Gag, SPBN-333-ΔCD-Gag, BNSP, or BNSP-Gag (MOI of 2) and lysed 48 h later. Proteins were separated by SDS-PAGE and subjected to Western blotting with antibodies specific for RV RNP (A), RV G (B), or HIV-1 p24 (C). A protein of the expected size for RV N was detected for all recombinant RVs (A), whereas an RV G-specific antibody detected a 62-kDa protein for SPBN, SPBN-Gag, SPBN-333-Gag, BNSP, and BNSP-Gag and a slightly faster migrating protein in cell extracts from SPBN-ΔCD-Gag- and SPBN-333-ΔCD-Gag-infected cells (B). Expression of HIV-1 p55 from SPBN-Gag, SPBN-333-Gag, SPBN-ΔCD-Gag, SPBN-333-ΔCD-Gag, and BNSP-Gag was confirmed with an HIV-1 Gag-specific antibody.
Multicycle and one-step growth curve of recombinant RVs.
During the production of our initial virus stocks, we realized that several of the recombinant viruses grew to lower titers or reached their final titers only after a longer incubation time. To study the replication of our recombinant RVs in greater detail, a multicycle growth curve was produced by infecting BSR cells at a MOI of 0.01. As shown in Fig. 3A, both RVs with CD-deleted Gs and BNSP-Gag had lower titers than the other four recombinant viruses at 20 h after infection. However, the titer of BNSP-Gag reached wild-type-like levels over 5 days, whereas the final titers of the viruses with CD-deleted Gs remained approximately 10-fold lower. These results are similar to previous findings by Mebatsion et al., which indicated a 10-fold titer reduction for an RV with CD-deleted G (18). The results noted above were also observed in a one-step growth curve. The results shown in Fig. 3B indicate very similar titers for SPBN, BNSP, SPBN-Gag, and SPBN-333-Gag but reduced titers for SPBN-ΔCD-Gag and SPBN-333-ΔCD-Gag. However, the one-step growth curve indicates that the growth impairment of BNSP-Gag is not caused by a slower spread of BNSP-Gag but rather a slower production of virus. This may be due to changes in the ratio of the RV proteins and is confirmed with our observation that BNSP-Gag forms foci similar in size to those formed by SPBN-Gag, whereas both RVs with CD-deleted Gs had greatly reduced focus sizes (data not shown).
It is interesting that the vector BNSP grows similarly to SPBN; this indicates that a new transcription unit without a foreign gene is ignored by the polymerase complex, an observation we previously made for transcription units in VSV- based vectors (M. J. Schnell and J. K. Rose, unpublished data).
Attenuated vaccine vectors are safe after i.c. inoculation in mice.
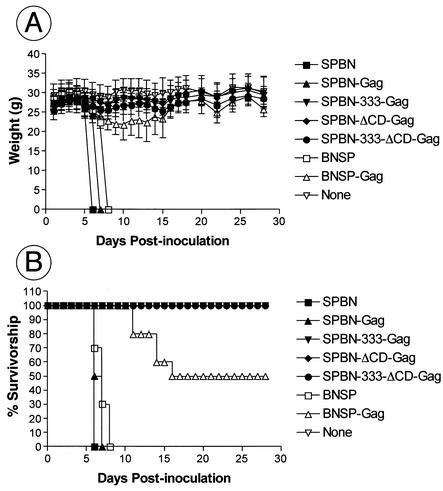
Our previous studies indicate that the vaccine vector SPBN is apathogenic after peripheral inoculation in mice but lethal after i.c. inoculation. It has also been shown by our group and others that a similar vector containing an amino acid change from arginine (R) to glutamic acid (E) or aspartic acid (D) in the RV G protein was completely apathogenic, even after i.c. injection (17, 20, 23). However, it was unknown how the deletion of the G CD or the combination of the CD deletion with the 333 mutation would affect the pathogenicity of these vaccine vehicles. In addition, no data are available for RV-based vectors expressing a foreign gene from a position further upstream than the G gene. Therefore, seven groups of 10 8-week-old Swiss-Webster mice were infected i.c. with 2 × 105 infectious particles. The mice were observed daily for any sign of rabies, and their weights were recorded. As expected and as shown in Fig. 4B, all mice infected with SPBN, BNSP, and SPBN-Gag died 6 to 8 days after i.c. inoculation. We observed a 100% survival rate of mice inoculated i.c. with SPBN-333-Gag, SPBN-ΔCD-Gag, or SPBN-333-ΔCD-Gag. Of note, no weight loss was observed for the animals in the SPBN-333-Gag, SPBN-ΔCD-Gag, and SPBN-333-ΔCD-Gag groups, which reemphasizes the extensive attenuation of the RV-based live vaccine vehicles.
FIG. 4.
New RV-based vaccine vectors are safe even after i.c. inoculation in mice. Six- to 8-week-old Swiss-Webster mice were inoculated i.c. with 105 infectious particles of SPBN, SPBN-Gag, SPBN-333-Gag, SPBN-ΔCD-Gag, SPBN-333-ΔCD-Gag, BNSP, or BNSP-Gag. Mice were observed for 4 weeks, and average weights (A) and mortalities (B) were recorded. Error bars (A), standard deviations.
Notably, only a 50% mortality occurred in the BNSP-Gag group, and the first sign of rabies was observed several days later than for SPBN or BNSP. However, these data reflect the observation made on the basis of the multicycle growth curve, which showed that growth of BNSP-Gag, but not BNSP, was slower than that of SPBN. The slower replication of SPBN-ΔCD-Gag, the other nonpathogenic virus not containing the 333 mutation in the RV G, probably results in the attenuation of this virus as well.
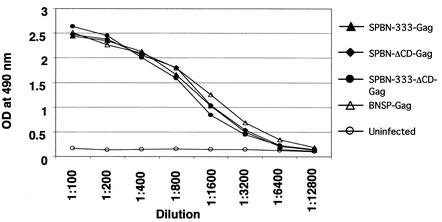
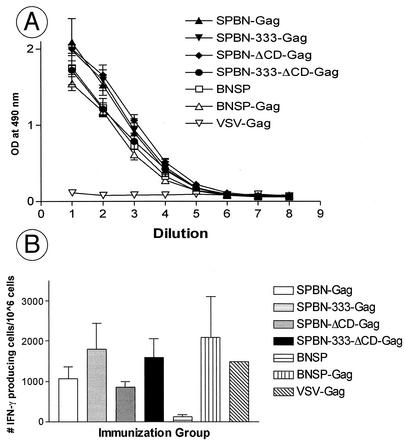
Last, we wanted to ensure that all surviving mice had been infected with the respective RV vector and therefore collected blood from each mouse of each group. Sera were screened for antibodies against RV G by ELISA (Fig. 5), and similar antibody titers were observed for mice injected with SPBN-333-Gag, SPBN-ΔCD-Gag, or SPBN-333-ΔCD-Gag and for the five surviving mice from the BNSP-Gag group.
FIG. 5.
Surviving mice develop a strong immune response against RV G. Surviving mice were bled 28 days after i.c. injection, and pooled sera from each group were analyzed for ELISA reactivity against RV G in serial dilutions. OD, optical density.
Attenuation does not interfere with immunogenicity.
Our results identified at least three extensively attenuated RV-based vaccine vectors, which may be able to replace our previously utilized vaccine vehicle, SPBN, expressing HIV-1 and HCV genes. In the next step, we wanted to analyze whether the attenuation factor(s) introduced into the RV genome changed the immunogenicity of the vaccine vector itself, the expressed foreign antigen, or both. We chose to analyze RV G as the only target for neutralizing antibodies against the vaccine vector and HIV-1 Gag as a well-characterized foreign antigen. Eight groups of five mice were immunized with 3.4 × 106 infectious particles of the respective recombinant RVs or a recombinant VSV expressing HIV-1 Gag as a control. Three weeks later, mice were bled and pooled serum of each group was analyzed by ELISA using RV G as an antigen. The results in Fig. 6A indicate that all RV-based vaccine vehicles developed similar humoral responses against RV G. These results were promising, and our next experiments were designed to analyze if cellular immune responses against HIV-1 Gag similar to those described previously for SPBN-Gag could be achieved with the highly attenuated RV vaccine vectors. BALB/c mice were immunized with the respective RV vector expressing HIV-1 Gag and 4 to 5 weeks later were challenged with a recombinant vaccinia virus expressing the same antigen. Two mice from each group were scarified 5 days after the vaccinia virus challenge, and Gag-specific CD8+ effector cells were determined after restimulation with an HIV-1 Gag p24 peptide (AMQMLKETI) by three different, independent ELISPOT assays for cells secreting IFN-γ. The results in Fig. 6B show a low background level of IFN-γ-secreting splenocytes from mice primed the vaccine vector BNSP, whereas a high number of IFN-γ-secreting cells were detected for SPBN-Gag. Of note, we observed approximately the same number of IFN-γ-secreting cells for all the new extensively attenuated RV vaccine vectors. In addition, a recombinant VSV expressing HIV-1 Gag, which was included in the last round of experiments, induced similar numbers of IFN-γ-secreting cells in a similar experimental setting, indicating the potency of the rhabdovirus group as vaccine vehicles.
FIG. 6.
Recombinant vaccine vehicles induce similar humoral and cellular immune responses in immunized mice. (A) RV G ELISA. Groups of five BALB/c mice were immunized with SPBN, SPBN-Gag, SPBN-333-Gag, SPBN-ΔCD-Gag, SPBN-333-ΔCD-Gag, BNSP,BNSP-Gag, or VSV-Gag and bled 3 weeks after immunization. Serial dilutions from pooled sera from each immunization group were used to determine the antibody titers against RV G by ELISA. 1 to 9 on the x axis indicates twofold dilutions from 1:100 to 1:25,600. (B) ELISPOT assays. Groups of two BALB/c mice were immunized with SPBN, SPBN-Gag, SPBN-333-Gag, SPBN-ΔCD-Gag, SPBN-333-ΔCD-Gag, BNSP, and BNSP-Gag and challenged 4 to 5 weeks after immunization with a recombinant vaccinia virus expressing HIV-1 Gag. Pooled splenocytes from two mice were analyzed at different dilutions for cells secreting IFN-γ. Error bars, standard deviations from three independent experiments. The recombinant VSV expressing HIV-1 Gag (VSV-Gag) was included in the last round of experiments (no error bar). OD, optical density.
DISCUSSION
Recombinant vaccines based on the RV vaccine strain SAD B19 (4, 29) expressing foreign genes such as the HIV-1 env and gag genes or the HCV glycoproteins have been proven to be highly effective in inducing both humoral and vigorous cellular immune responses in mice (15, 16, 28, 31). These currently used RV-based vectors are very safe after peripheral application, but even a low vaccine vector-induced pathogenesis is undesirable. Previous results clearly indicate that RV G is largely responsible for RV-associated pathogenicity (8, 20-22). It has been shown that a G-deleted RV can neither invade neurons nor cause disease even after i.c. inoculation (7). In addition, it is well established that an arginine-to-glutamic acid exchange at position 333 within the RV G dramatically reduces RV pathogenicity for some RV strains (6, 17, 30). We therefore introduced this mutation in our previously described vaccine vehicle SPBN-Gag. SPBN-333-Gag was completely apathogenic after i.c. inoculation into mouse brains but was still as potent as SPBN-Gag in inducing cellular and humoral responses. The results of these experiments indicate that the neurotropic infection caused by RV can be abolished without affecting the immunogenicity, indicating that tropism and immunogenicity can be uncoupled. These results are consistent with the finding by Yan et al., who used stereotaxic inoculation of RV infectious particles into the hippocampi of rats. Their results showed that an RV containing the RV G with an R-to-E exchange at position 333 lost its ability to spread to other areas of the brain and infected neurons only at the site of inoculation (34). However, the replacement of the RV G with VSV G in the same vector supported spread in the rat brain, but in a pattern similar to that observed for VSV rather than for RV. It is interesting that the VSV G-containing RV still produces 40% mortality in mice after i.c. inoculation (J. P. McGettigan, M. J. Schnell, and B. Dietzschold, unpublished data).
The second method we used to attenuate SPBN-Gag was the deletion of the RV G CD domain. We hypothesized that a slower spread of RV might enable the immune system to clear the virus before it causes disease. Previous work by Mebatsion et al. indicates that the deletion of RV G CD reduces both the viral titers and the RV focus size (18). In addition, a similar method has been successfully used to attenuate a VSV-based vaccine vector expressing influenza A virus HA. The results indicated that the deletion of the VSV G CD in the vaccine vector resulted in attenuation in immunized mice compared to the parental VSV expressing HA (26). However, neutralizing antibodies against influenza A virus were also approximately 10-fold reduced for the attenuated version of the VSV-based vaccine. For RV, the detected immune responses against the HIV-1 Gag and RV G were not affected by the deletion of the RV G CD.
We next combined both attenuation methods listed above in one vector expressing HIV-1 Gag (SPBN-333-ΔCD-Gag). These experiments were performed because of concerns that reversion of the attenuating mutation(s) may occur in vivo. It has been shown that a CD-deleted VSV acquired a nonspecific G CD sequence after serial passage and was able to drive efficient budding of the recombinant VSV (27). In addition, the natural mutation rate of the rhabdovirus polymerase creates a mixed virus population and therefore some RVs may contain an amino acid other than E at position 333 of the RV G. It is, however, highly unlikely that reversions would occur in two different attenuating mutations, as is the case for SPBN-333-ΔCD-Gag. In addition, for SPBN-333-ΔCD-Gag there were no differences between the induced immune responses against the vector and those against HIV-1 Gag compared to SPBN-Gag, and therefore this approach is feasible.
Last, we constructed a new RV vector that expresses HIV-1 Gag from an RV transcription unit downstream of the RV N gene instead of downstream of the G gene. This construct was also attenuated after i.c. inoculation in mice. The observed attenuation of BNSP-Gag is probably due to the altered growth of the virus, as previously demonstrated for SPBN-ΔCD-Gag. As shown in Fig. 3, production of BNSP-Gag is delayed compared with production of BNSP or SPBN-Gag, resulting in about 50%-reduced mortality. Of note, these results fit well with the more-extensive attenuation of SPBN-ΔCD-Gag and SPBN-ΔCD-333-Gag, RVs with even more impaired growth patterns.
It is important that the observed attenuation of the different RVs is probably due to two different factors: (i) the neurotropism of the respective virus and (ii) the viral spread, which determines if the host immune responses can detain the virus and therefore prevent vector-induced disease or death. The current RV vector work in our laboratory is focused on introducing a second mutation, which interferes with the RV neurotropism, such as the recently described deletion of the LC8 domain that binds with P (17). Such a combination of three or four attenuation markers in one RV vector may prove to be safe even in immunocompromised hosts.
In summary, the results presented herein indicate that an extremely safe RV-based vaccine vector can be constructed and multiple mutations can be introduced without reducing the immune responses against both the vector and the expressed foreign gene. We feel that the use of RV-based vectors is advantageous because the parameters of their pathogenicity are better understood than those of other vectors. We are currently in the process of analyzing these second-generation RV-based vaccines expressing HIV-1 Env and simian immunodeficiency virus Gag in the rhesus macaque model system, and these experiments will further explore the safety and efficiency of this new vaccine approach.
Acknowledgments
The recombinant VSV encoding HIV-1 Gag containing VSV was a generous gift from J. K. Rose and K. Haglund, Yale University, New Haven, Conn. The human monoclonal antibody directed against p24 (1) and the plasmid pNL4-3 (13), encoding an infectious clone of HIV-1NL4-3, were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
This study was supported by NIH grants AI49153 to M.J.S and AI45097 to B.D.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ball, L. A., C. R. Pringle, B. Flanagan, V. P. Perepelitsa, and G. W. Wertz. 1999. Phenotypic consequences of rearranging the P, M, and G genes of vesicular stomatitis virus. J. Virol. 73:4705-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchholz, U. J., S. Finke, and K. K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conzelmann, K. K., J. H. Cox, L. G. Schneider, and H. J. Thiel. 1990. Molecular cloning and complete nucleotide sequence of the attenuated rabies virus SAD B19. Virology 175:485-499. [DOI] [PubMed] [Google Scholar]
- 5.Coulon, P., J. P. Ternaux, A. Flamand, and C. Tuffereau. 1998. An avirulent mutant of rabies virus is unable to infect motoneurons in vivo and in vitro. J. Virol. 72:273-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dietzschold, B., W. H. Wunner, T. J. Wiktor, A. D. Lopes, M. Lafon, C. L. Smith, and H. Koprowski. 1983. Characterization of an antigenic determinant of the glycoprotein that correlates with pathogenicity of rabies virus. Proc. Natl. Acad. Sci. USA 80:70-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Etessami, R., K. K. Conzelmann, B. Fadai-Ghotbi, B. Natelson, H. Tsiang, and P. E. Ceccaldi. 2000. Spread and pathogenic characteristics of a G-deficient rabies virus recombinant: an in vitro and in vivo study. J. Gen. Virol. 81:2147-2153. [DOI] [PubMed] [Google Scholar]
- 8.Faber, M., R. Pulmanausahakul, S. S. Hodawadekar, S. Spitsin, J. P. McGettigan, M. J. Schnell, and B. Dietzschold. 2002. Overexpression of the rabies virus glycoprotein results in enhancement of apoptosis and antiviral immune response. J. Virol. 76:3374-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finke, S., and K. K. Conzelmann. 1999. Virus promoters determine interference by defective RNAs: selective amplification of mini-RNA vectors and rescue from cDNA by a 3′ copy-back ambisense rabies virus. J. Virol. 73:3818-3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flanagan, E. B., L. A. Ball, and G. W. Wertz. 2000. Moving the glycoprotein gene of vesicular stomatitis virus to promoter-proximal positions accelerates and enhances the protective immune response. J. Virol. 74:7895-7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flanagan, E. B., J. M. Zamparo, L. A. Ball, L. L. Rodriguez, and G. W. Wertz. 2001. Rearrangement of the genes of vesicular stomatitis virus eliminates clinical disease in the natural host: new strategy for vaccine development. J. Virol. 75:6107-6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foley, H. D., J. P. McGettigan, C. A. Siler, B. Dietzschold, and M. J. Schnell. 2000. A recombinant rabies virus expressing vesicular stomatitis virus glycoprotein fails to protect against rabies virus infection. Proc. Natl. Acad. Sci. USA 97:14680-14685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorny, M. K., V. Gianakakos, S. Sharpe, and S. Zolla-Pazner. 1989. Generation of human monoclonal antibodies to human immunodeficiency virus. Proc. Natl. Acad. Sci. USA 86:1624-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacob, Y., H. Badrane, P. E. Ceccaldi, and N. Tordo. 2000. Cytoplasmic dynein LC8 interacts with lyssavirus phosphoprotein. J. Virol. 74:10217-10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGettigan, J. P., H. D. Foley, I. M. Belyakov, J. A. Berzofsky, R. J. Pomerantz, and M. J. Schnell. 2001. Rabies virus-based vectors expressing human immunodeficiency virus type 1 (HIV-1) envelope protein induce a strong, cross-reactive cytotoxic T-lymphocyte response against envelope proteins from different HIV-1 isolates. J. Virol. 75:4430-4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGettigan, J. P., S. Sarma, J. M. Orenstein, R. J. Pomerantz, and M. J. Schnell. 2001. Expression and immunogenicity of human immunodeficiency virus type 1 Gag expressed by a replication-competent rhabdovirus-based vaccine vector. J. Virol. 75:8724-8732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mebatsion, T. 2001. Extensive attenuation of rabies virus by simultaneously modifying the dynein light chain binding site in the P protein and replacing Arg333 in the G protein. J. Virol. 75:11496-11502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mebatsion, T., M. Konig, and K. K. Conzelmann. 1996. Budding of rabies virus particles in the absence of the spike glycoprotein. Cell 84:941-951. [DOI] [PubMed] [Google Scholar]
- 19.Mebatsion, T., F. Weiland, and K. K. Conzelmann. 1999. Matrix protein of rabies virus is responsible for the assembly and budding of bullet-shaped particles and interacts with the transmembrane spike glycoprotein G. J. Virol. 73:242-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morimoto, K., H. D. Foley, J. P. McGettigan, M. J. Schnell, and B. Dietzschold. 2000. Reinvestigation of the role of the rabies virus glycoprotein in viral pathogenesis using a reverse genetics approach. J. Neurovirol. 6:373-381. [DOI] [PubMed] [Google Scholar]
- 21.Morimoto, K., D. C. Hooper, H. Carbaugh, Z. F. Fu, H. Koprowski, and B. Dietzschold. 1998. Rabies virus quasispecies: implications for pathogenesis. Proc. Natl. Acad. Sci. USA 95:3152-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morimoto, K., D. C. Hooper, S. Spitsin, H. Koprowski, and B. Dietzschold. 1999. Pathogenicity of different rabies virus variants inversely correlates with apoptosis and rabies virus glycoprotein expression in infected primary neuron cultures. J. Virol. 73:510-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morimoto, K., J. P. McGettigan, H. D. Foley, D. C. Hooper, B. Dietzschold, and M. J. Schnell. 2001. Genetic engineering of live rabies vaccines. Vaccine 19:3543-3551. [DOI] [PubMed] [Google Scholar]
- 24.Poisson, N., E. Real, Y. Gaudin, M. C. Vaney, S. King, Y. Jacob, N. Tordo, and D. Blondel. 2001. Molecular basis for the interaction between rabies virus phosphoprotein P and the dynein light chain LC8: dissociation of dynein-binding properties and transcriptional functionality of P. J. Gen. Virol. 82:2691-2696. [DOI] [PubMed] [Google Scholar]
- 25.Raux, H., A. Flamand, and D. Blondel. 2000. Interaction of the rabies virus P protein with the LC8 dynein light chain. J. Virol. 74:10212-10216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roberts, A., L. Buonocore, R. Price, J. Forman, and J. K. Rose. 1999. Attenuated vesicular stomatitis viruses as vaccine vectors. J. Virol. 73:3723-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schnell, M. J., L. Buonocore, E. Boritz, H. P. Ghosh, R. Chernish, and J. K. Rose. 1998. Requirement for a non-specific glycoprotein cytoplasmic domain sequence to drive efficient budding of vesicular stomatitis virus. EMBO J. 17:1289-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schnell, M. J., H. D. Foley, C. A. Siler, J. P. McGettigan, B. Dietzschold, and R. J. Pomerantz. 2000. Recombinant rabies virus as potential live-viral vaccines for HIV-1. Proc. Natl. Acad. Sci. 97:3544-3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schnell, M. J., T. Mebatsion, and K. K. Conzelmann. 1994. Infectious rabies viruses from cloned cDNA. EMBO J. 13:4195-4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seif, I., P. Coulon, P. E. Rollin, and A. Flamand. 1985. Rabies virulence: effect on pathogenicity and sequence characterization of rabies virus mutations affecting antigenic site III of the glycoprotein. J. Virol. 53:926-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siler, C. A., J. P. McGettigan, B. Dietzschold, S. K. Herrine, J. Dubuisson, R. J. Pomerantz, and M. J. Schnell. 2002. Live and killed rhabdovirus-based vectors as potential hepatitis C vaccines. Virology 292:24-34. [DOI] [PubMed] [Google Scholar]
- 32.Wagner, R. R., and J. K. Rose. 1996. Rhabdoviridae: the viruses and their replication, p. 1121-1135. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 1. Lippincott-Raven Publishers, Philadelphia, Pa.
- 33.Whitt, M., L. Buonocore, C. Prehaud, and J. K. Rose. 1991. Membrane fusion activity, oligomerization, and assembly of the rabies virus glycoprotein. Virology 185:681-688. [DOI] [PubMed] [Google Scholar]
- 34.Yan, X. Y., P. S. Mohankumar, B. Dietzschold, M. J. Schnell, and Z. F. Fu. 2002. The glycoprotein determines the spreading of different rabies virus strains in the CNS. J. Neurovirol. 8:345-352. [DOI] [PubMed] [Google Scholar]