Abstract
Nucleocapsid (NC) proteins in most retroviruses have a well-conserved Cys-His box(es) as well as more divergent flanking regions that are rich in basic residues. Mutations in the flanking regions can affect RNA packaging, virus assembly, and reverse transcription of the viral RNA. To gain a further understanding of the roles of NC flanking regions in the retroviral replication cycle, we generated and characterized chimeric gag-pol expression constructs derived from murine leukemia virus and spleen necrosis virus by replacing an NC flanking region from one virus with the counterpart from the other virus. We found that all four chimeras were able to generate virions, package viral RNA, and complete the viral replication cycle. Two chimeras had mild defects in virus assembly that correlated with a decrease in the isoelectric points of NCs, suggesting that the basic nature of NC is important in virus assembly. This finding indicates that, although the primary sequences of these flanking regions have little homology, the heterologous sequences are functional both as part of the Gag polyprotein and as processed NC protein.
The gag gene in retroviruses encodes the structural proteins of the virions (60). The proteolytic products of Gag polyprotein include the matrix, capsid (CA), nucleocapsid (NC), and other cleavage products, which vary in number and size among different retroviruses (57, 60).
NC is thought to play multiple roles during the viral replication cycle. As part of Gag, NC is important in directing virus assembly, specific encapsidation of the viral genomic RNA and primer tRNA, and the placement of tRNA onto viral RNA (2, 3, 8, 10, 31, 35, 36, 41, 46, 48, 51, 57, 60, 63, 64). Once the Gag polyprotein is processed, the NC protein is thought to affect viral RNA dimerization, reverse transcription of the viral genome, and integration of the newly synthesized viral DNA (4, 6, 7, 12, 20, 37, 51, 59, 60).
With the exception of spumaviruses, all retroviral NC proteins contain one or two Cys-His boxes (39, 57, 60), which have Cys-X2-Cys-X4-His-X4-Cys sequences that resemble the zinc finger motif. Mutations that destroy the Cys-His boxes render the virus noninfectious; these mutants lose the selectivity to package viral RNA (26, 28, 31, 32, 41, 42). Mutations that change the conserved sequences in the Cys-His boxes to other zinc finger-like motifs also affect NC function and could create notable defects in viral RNA packaging, reverse transcription, or integration in murine leukemia virus (MLV), human immunodeficiency virus type 1 (HIV-1), and simian immunodeficiency virus (27, 29, 30, 62).
The flanking regions are also important for NC function. Mutations in the flanking regions of NC have been shown to hamper or abolish virus assembly, specific viral RNA packaging, and reverse transcription (4, 5, 10, 13, 24, 34, 49, 50). The flanking regions contain multiple basic residues that are important to NC function; mutation of multiple basic residues in MLV NC results in a severe replication defect (34). In addition to the basic residues, noncharged residues also play important roles in NC function. Mutation of a leucine to an alanine in the NC flanking region causes a significant replication defect in MLV (25).
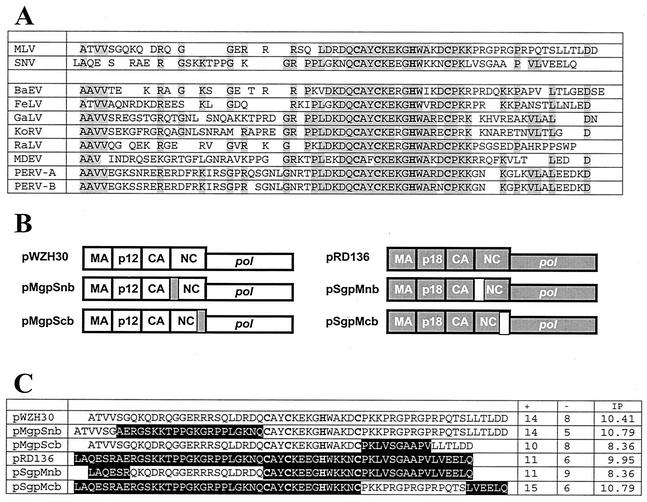
Comparison of NC sequences from multiple gammaretroviruses reveals that the amino acid sequences of the Cys-His boxes are highly conserved, whereas the flanking regions are quite divergent (Fig. 1A). We hypothesized that, despite the divergence in the primary sequences, the flanking regions of these NCs are functionally conserved and it is likely that a flanking region from one virus could replace the counterpart of another virus. To test our hypothesis, we generated viruses containing chimeric NC and examined their ability to replicate. As our model system, we used MLV and spleen necrosis virus (SNV), an avian pathogen distantly related to MLV (47). SNV proteins can support the replication of MLV vectors efficiently (8, 9, 19), thus allowing us to use the same MLV vector to characterize both MLV- and SNV-based chimeric proteins and their effects in the multiple steps of the replication cycle. In addition, the amino acid sequences of the NC flanking regions in MLV and SNV were very diverse (Fig. 1A). In the N-terminal flanking regions, only 7 of the 25 (MLV) and 27 (SNV) amino acids were identical. Similarly, only 6 of the 21 (MLV) and 16 (SNV) amino acids were identical in the C-terminal flanking regions. In this report, we generated four gag-pol expression constructs that have chimeric NCs containing one flanking region from the counterpart of the other virus and characterized their effects during multiple steps of the viral replication cycle.
FIG. 1.
Amino acid sequences of NCs and structures of the vectors. (A) Comparison of NC amino acid sequences among different gammaretroviruses. The conserved C and H residues that constitute the Cys-His boxes are shown in bold. Residues that are identical in more than half of the viruses are shown in gray. BaEV, baboon endogenous virus; FeLV, feline leukemia virus subgroup A; GaLV, gibbon ape leukemia virus; KoRV, Phascolarctos cinereus (koala) retrovirus; RaLV, rat leukemia virus; MDEV, Mus dunni endogenous virus; PERV-A, porcine endogenous virus class A; PERV-B, porcine endogenous virus class B. (B) General structures of the wild-type and chimeric gag-pol expression constructs. Sequences derived from MLV and SNV are shown in white and gray boxes, respectively. MA, matrix. (C) Amino acid sequences of the wild-type and chimeric NCs. MLV- and SNV-derived sequences are shown in white and black text, respectively. +, number of positively charged amino acids; −, number of negatively charged amino acids; IP, isoelectric point.
Experimental system used to study the roles of NC flanking regions in viral replication.
Using PCR and standard cloning techniques (54), we generated NC chimeras based on the MLV and SNV gag-pol expression constructs pWZH30 and pRD136, respectively (40, 43, 44) (Fig. 1B). In each chimera, one of the flanking regions was replaced with the heterologous counterpart, with the exception that the 6 amino acids immediately adjacent to the NC junction remained unchanged to preserve the protease cleavage site (Fig. 1C). The names of these chimeras reflect their genotypes; for example, pMgpSnb denotes the MLV gag-pol expression construct containing the SNV N-terminal flanking region.
MLV and SNV NCs both contain multiple basic residues, which are distributed differently in the two NCs. Along with the alteration in primary sequences, these chimeras also have different numbers of charged residues (Fig. 1C). For example, the NC in pMgpScb has only 10 basic residues, 4 basic residues fewer than the wild-type MLV NC.
To test the abilities of these gag-pol expression constructs to form virus particles and support viral vector replication, we introduced these constructs into E1 cells (8) by cotransfection with pBSpac (15), which conferred puromycin resistance. E1 is a D17-based cell line that expresses amphotropic MLV Env, a copy of an MLV vector that encodes the hygromycin phosphotransferase B gene (hygro), and a copy of an SNV vector that encodes the neomycin phosphotransferase gene (8, 17, 43, 61). D17 is a dog osteosarcoma cell line permissive for MLV and SNV replication (52); both MLV and SNV Gag can be pseudotyped by amphotropic MLV Env to form infectious particles. Transfected puromycin-resistant cells were pooled, and viruses were harvested from these cells. These viruses were serially diluted and used to infect target D17 cells; the viral titers generated were calculated from the numbers of drug-resistant colonies. Transfection, infection, and drug selection were performed as previously described (8). In addition, the biochemical nature of these viruses was also examined by measuring reverse transcriptase (RT) activity, performing Western analyses, and quantifying the amounts of packaged viral RNA. Although E1 cells contained both the MLV and SNV vectors, the analyses in this report focus on the characterization of the MLV vector because both the MLV and SNV proteins can support its replication efficiently (8, 19).
Biochemical analyses of cell-free virions released by Gag and Gag-Pol polyproteins containing chimeric NC.
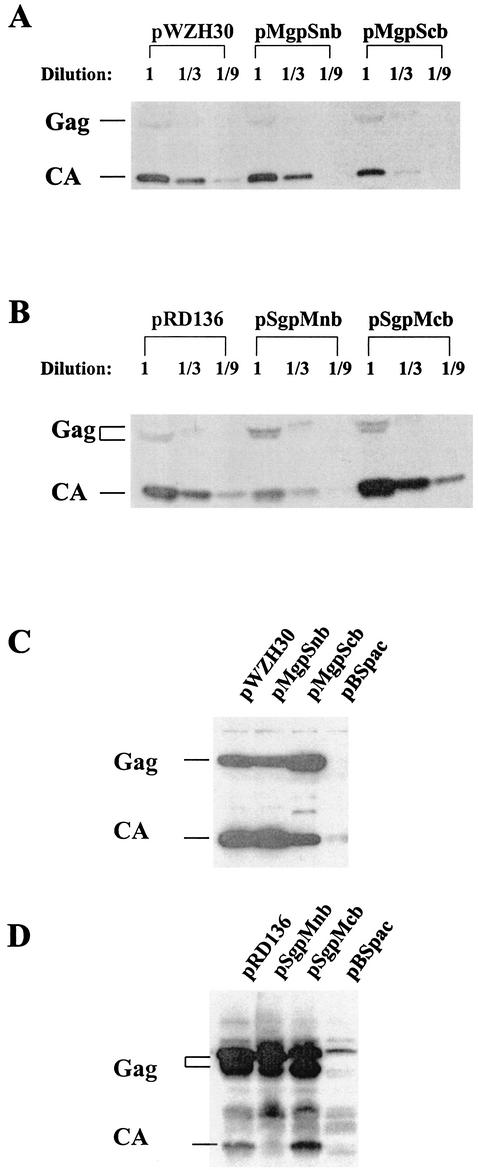
Altering the flanking regions of the NC could have a detrimental effect on viral assembly (10, 13, 34, 49). Therefore, we first examined whether these chimeras were able to generate cell-free virus particles and analyzed the properties of the virions. Western analyses of proteins generated from various gag-pol expression constructs are shown in Fig. 2. Western analyses of cell-free virions revealed that both MLV-based NC chimeras were able to produce virus particles (Fig. 2A). Compared with pWZH30-transfected cells, pMgpSnb-transfected cells generated similar amounts of cell-free virions whereas pMgpScb-transfected cells generated fewer virions (approximately one-half to one-third of that for the wild type). Western analyses indicated that both SNV-based NC chimeras were also able to produce virions (Fig. 2B). Compared with pRD136-transfected cells, pSgpMnb-transfected cells generated fewer cell-free virions (approximately one-third to one-quarter of that for the wild type) whereas pSgpMcb-transfected cells generated similar amounts of virions. Western analyses demonstrated that cell lysates from E1 transfected with pWZH30, pMgpSnb, or pMgpScb had similar amounts of viral proteins (Fig. 2C). Similarly, cells transfected with pRD136, pSgpMnb, or pSgpMcb also expressed similar amounts of viral proteins (Fig. 2D).
FIG. 2.
Western analyses of the proteins expressed by various gag-pol expression constructs in transfected cells and cell-free virions. (A and B) Western analyses of viral proteins in cell-free virions. Results from virions produced by cells transfected with MLV- and SNV-based constructs are shown in panels A and B, respectively. Equal numbers of stably transfected cells were plated; cell-free supernatants were harvested 48 h later. An equal portion of each sample was loaded in the lanes marked 1, whereas one-third and one-ninth of the portions were loaded in lanes marked 1/3 and 1/9, respectively. (C and D) Western analyses of cell lysates generated from transfected E1 cells.Results from MLV- and SNV-based constructs are shown in panels C and D, respectively. Cell lysates from E1 cells transfected with only pBSpac were used as controls. A rabbit anti-MLV CA antibody was used to probe the Western blots shown in panels A and C, whereas a rabbit anti-SNV CA antibody was used to probe the Western blots shown in panels B and D. Anti-MLV and -SNV CA antibodies were kind gifts from the AIDS Vaccine Program, Science Applications International Corporation, and Nancy Rice, NCI-Frederick, respectively.
The RT activities of these cell-free viral samples were also quantified as previously described (11, 58); these results paralleled the data from the Western analyses. The RT activities produced by cell-free virions generated by pMgpSnb and pSgpMcb were equivalent to those of pWZH30 and pRD136, respectively (data not shown). However, the RT activities generated by samples from pMgpScb were on average threefold lower than those from pWZH30, whereas samples from pSgpMnb showed RT activities an average of sixfold lower than those from pRD136 (data not shown).
Taken together, these data indicate that, despite the dramatic change in primary sequences, all four NC chimeras were able to express viral proteins in transfected cells; these viral proteins were able to assemble into viral particles and were released from the cells. Two of the chimeras, pMgpSnb and pSgpMcb, generated virions with efficiencies similar to those of the wild-type viruses, whereas the other two chimeras, pMgpScb and pSgpMnb, had mild defects in virion assembly and release.
Alteration of the NC sequences could also abolish specific viral RNA packaging. To examine whether Gag proteins with chimeric NC have the capacity to package viral RNA, we used quantitative real-time RT-PCR to characterize the amounts of MLV vector RNA in transfected cells as well as RNA encapsidated in the virions produced by the chimeras. MLV vector RNA was examined because it can be efficiently packaged by both the MLV and SNV proteins. RNA samples were reverse transcribed using random hexamers as primers and quantified using real-time PCR with primers and probes located in the hygro gene of the MLV vector. Data generated from three sets of independent experiments are summarized in Table 1. As expected, MLV vector RNA was expressed in all transfected cells at similar levels. Cell-free virion RNA analyses demonstrated that all of the Gag polyproteins with chimeric NC were able to package vector RNA with efficiencies that were within a twofold range of that for the wild-type Gag.
TABLE 1.
Comparisons of MLV vector RNA in cells transfected with MLV- and SNV-based gag-pol expression constructs and in cell-free virionsa
| Expt | % MLV vector RNA
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MLV basedb
|
SNV basedc
|
|||||||||||
| Cellular
|
Cell-free viriond
|
Cellular
|
Cell-free virione
|
|||||||||
| pWZH30 | pMgpSnb | pMgpScb | pWZH30 | pMgpSnb | pMgpScb | pRD136 | pSgpMnb | pSgpMcb | pRD136 | pSgpMnb | pSgpMcb | |
| 1 | 100 | 160 | 167 | 100 | 43 | 54 | 100 | 77 | 62 | 100 | 154 | 186 |
| 2 | 100 | 107 | 127 | 100 | 36 | 28 | 100 | 54 | 119 | 100 | 101 | 138 |
| 3 | 100 | 66 | 180 | 100 | 111 | 87 | 100 | 78 | 105 | 100 | 84 | 143 |
| Mean ± SE | 100 | 111 ± 27 | 158 ± 16 | 100 | 63 ± 24 | 56 ± 17 | 100 | 70 ± 8 | 95 ± 17 | 100 | 113 ± 21 | 155 ± 15 |
The primers (5′ ACGAGGTCGCCAACATCTTC 3′, 5′ AGCGCGTCTGCTGCTCC 3′) and probe (5′ FAMTCTGGAGGCCGTGGTTGGCTTGTATAMRA 3′) used in the analyses are located in hygro.
All RNAs were standardized to the amount of RNA in the pWZH30 samples.
All RNAs were standardized to the amount of RNA in the pRD136 samples.
RNAs from the pWZH30-, pMgpSnb-, and pMgpScb-derived samples with similar amounts of RT activities were used in these analyses.
RNAs from the pRD136-, pSgpMnb-, and pSgpMcb-derived samples with similar amounts of RT activities were used in these analyses.
Infectivity of virions containing chimeric NC.
To examine the relative infectivity of the virions generated by these chimeras, we determined viral titers and standardized them to the RT activities of their respective wild-type gag-pol expression constructs (Table 2). Compared with the viruses produced by pWZH30-transfected cells, those from pMgpSnb- and pMgpScb-transfected cells generated 61 to 25% and 56 to 18% of the wild-type titers, respectively. The viral titers from the wild type and the two chimeric viruses were not significantly different (one-way analysis of variance, P = 0.289). For SNV-based expression constructs, viruses produced by pSgpMnb- and pSgpMcb-transfected cells generated 31 to 18% and 13 to 3% of the titers generated by viruses from pRD136-transfected cells, respectively. In these three viruses, wild-type titers were significantly higher than those from the two chimeras (one-way analysis of variance and Dunnett's test, P < 0.05). Therefore, all four chimeras generated infectious virions that were capable of supporting vector replication in a single-cycle assay. Compared with the wild-type infectivity, virions from the chimeras varied from similar to a 10-fold reduction in the wild type.
TABLE 2.
Comparison of adjusted viral titers generated by various gag-pol expression constructs
| Expt |
gag-pol expression construct titer
|
|||||
|---|---|---|---|---|---|---|
| MLV derived (104 CFU/ml)a,b
|
SNV derived (103 CFU/ml)a
|
|||||
| pWZH30 | pMgpSnb | pMgpScb | pRD136 | pSgpMnb | pSgpMcb | |
| 1 | 4.2 | 1.3 | 0.84 | 0.45 | 0.08 | 0.06 |
| 2 | 7.2 | 4.4 | 4.0 | 1.4 | 0.36 | 0.04 |
| 3 | 2.6 | 0.65 | 0.48 | 2.1 | 0.65 | 0.25 |
| 4 | ND | ND | ND | 0.83 | 0.18 | 0.08 |
| 5 | ND | ND | ND | 2.0 | 0.43 | 0.18 |
| Mean ± SE | 4.7 ± 1.3 | 2.1 ± 1.2 | 1.8 ± 1.1 | 1.4 ± 0.3 | 0.34 ± 0.1 | 0.12 ± 0.04 |
Viral titers were standardized to the RT activities of the virions containing the appropriate wild-type viral proteins.
ND, not determined.
Characterization of reduction in viral replication from viruses generated by pSgpMcb: studies on RNA dimerization and reverse transcription.
Among the four chimeras, the largest reduction in infectivity was a 10-fold reduction in pSgpMcb-derived viruses. The virions generated by pSgpMcb were able to package viral RNA at an efficiency similar to that of the virions from wild-type SNV; therefore, this reduced infectivity is likely to be caused by blocks in other steps of viral replication. To investigate the possible blocks, we examined the dimerization of the viral RNA and reverse transcription of the viral genome.
Viral RNAs in the virions are present as immature dimers immediately after budding; these dimers are unstable and fragile (23). After processing of the viral proteins and maturation of the virion, viral RNAs also undergo maturation to form stable dimers (21, 23, 56). The chaperone activity of NC was proposed to play an important role in the maturation of the viral dimer (20, 51). Therefore, we questioned whether the chimeric NC encoded by pSgpMcb was able to promote the maturation of the viral RNA dimer and whether this impacted the infectivity of the virion. To address this possibility, we compared the physical state of the MLV vector RNA in the virions with that of the wild-type or chimeric proteins. We found that the RNAs isolated from virions with wild-type or chimeric proteins were indistinguishable in their mobility and thermostability; approximately half of the RNA dimers and all of the RNA dimers dissociated into monomers when RNA samples were heated to 65 and 70°C, respectively (data not shown). This result indicates that the proteins expressed by pSgpMcb, similar to the wild-type proteins, were able to induce the conformational change from the immature RNA dimers to mature RNA dimers.
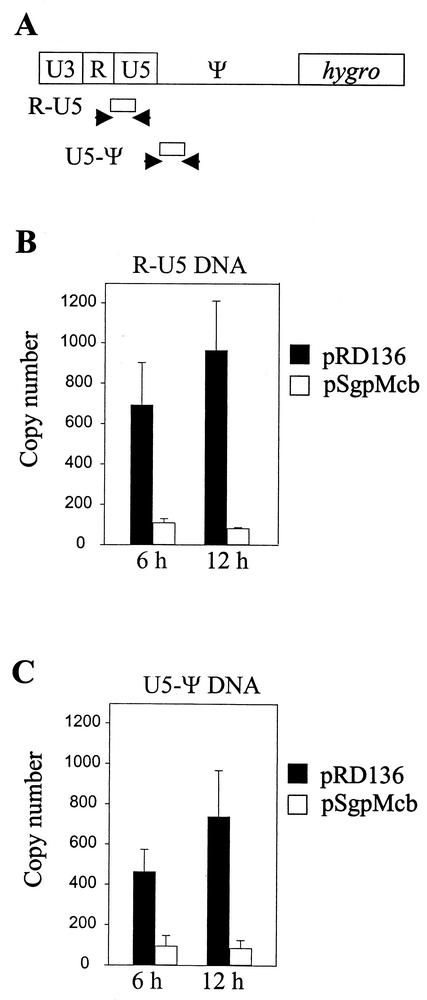
NC is also thought to play an important role during the reverse transcription of the viral genome. In this study, it is possible that the chimeric NC affected the efficiency of reverse transcription. To examine this hypothesis, viruses generated from pSgpMcb- or pRD136-transfected E1 cells were used to infect target D17 cells and we isolated DNA samples 6 or 12 h postinfection. The amounts of viral vector DNA synthesized by similar amounts of wild-type or chimeric virions (standardized by RT activity) in infected cells were quantified by real-time PCR analyses. Two sets of primers and probes were used in this assay (Fig. 3A): one set measured R-U5 DNA, the first DNA synthesized during reverse transcription (early product), whereas the other set measured U5-Ψ DNA, the DNA detected after plus-strand DNA transfer (late product). The measurements from three independent sets of experiments are shown in Fig. 3B and C. At the 6-h time points, levels of DNA synthesized from chimeric NC virions were on average 18% (R-U5) or 21% (U5-Ψ) of the wild-type level. The differences in the DNA synthesized from wild-type versus chimeric NC-containing viruses increased at the 12-h time point, and the levels of reverse transcription products from chimeric NC virions for R-U5 and U5-Ψ were both 10% of the wild-type level. The fact that the amounts of R-U5 DNAs synthesized by the chimeric viruses were lower indicates a defect in or before the early stage of reverse transcription. As negative controls, supernatants from pBSpac-transfected cells were incubated with D17 cells and DNA samples were isolated 6 or 12 h later. Real-time PCR analyses were also performed on these DNAs, and fewer than five copies of DNA were detected in all negative-control samples (data not shown).
FIG. 3.
Quantitative real-time PCR analyses of viral DNA synthesized by virions containing wild-type SNV NC or chimeric NC. (A) Primers and probes used in the analyses. R-U5 DNA was amplified by primers RU5-Fn (5′ TCCCAATAAAGCCTCTTGCTG 3′) plus RU5-Rn (5′ AGGAGACCCTCCCAAGGAAC 3′) and detected with probe RU5-Pn (5′ FAMTTGCATCCGAATCGTGGTCTCGCTAMRA 3′). U5-Ψ DNA was amplified by primers U5Ψ-Fn (5′ GCCTCTTGCTGTTTGCATCC 3′) plus U5Ψ-Rn (5′ GTCTCCAAATCCCGGACGA 3′) and detected with probe U5ψ-Pn (5′ FAMATCGTGGTCTCGCTGTTCCTTGGGAGTAMRA 3′). (B and C) Summary of viral DNA measurements from three independent sets of experiments. R-U5 and U5-Ψ DNAs are shown in panels B and C, respectively. DNA copy numbers are shown on the y axes. Black bars and white bars represent DNA synthesized by pRD136- and pSgpMcb-derived viruses, respectively. Standard errors are also shown for all the measurements.
Taken together, these data indicate that, during virus infection, pSgpMcb-derived viruses generated 10-fold less viral DNA than wild-type viruses. Therefore, reduction in the virus titer from pSgpMcb-derived virions is caused mainly by the decreased levels of reverse transcription products. This finding also suggests that the chimeric NC can carry out its function during virus integration with an efficiency similar to that of the wild-type NC protein.
Placement of tRNA in the primer-binding site (PBS) of viral RNA in chimeric NC virions.
It has been shown that, in MLV, Gag places the tRNA primers onto the PBS of the viral RNA (22). It has also been demonstrated in HIV-1 that mutations in the flanking regions of NC can inhibit tRNA placement (35, 36). Because our reverse transcription studies indicated that pSgpMcb-derived viruses had a defect in the early stage of reverse transcription, we questioned whether the chimeric Gag was able to carry out tRNA placement at the same efficiency as the wild-type Gag. A defect in tRNA placement could contribute to the decreased DNA synthesized by the chimeric-virus-infected cells.
The placement of tRNA on both wild-type virus and chimeric NC virus was examined by the primer-tagging assay as previously described (22). Viral vector RNA samples were isolated from wild-type and pSgpMcb-derived virions, and the amounts of vector RNA were quantified by nondenaturing Northern analyses (data not shown). Similar amounts of vector RNA isolated from pRD136- or pSgpMcb-generated virions were then incubated with RT in the presence of [α-32P]dATP. Because the first 2 nucleotides synthesized by RT would both be A, the primer tRNA annealed to the PBS would be able to use the radioactive dATP to extend these 2 nucleotides (22). The amounts of labeled primers reflected the numbers of tRNA that were already placed on the PBS in the vector RNA. Similar amounts of tRNA primers placed on vector RNA isolated from wild-type and chimeric virions were detected in these assays (data not shown). This result indicates that tRNA placement was carried out at similar efficiencies in the virions produced by pRD136- and pSgpMcb-transfected cells. Therefore, the reduced level of viral genome reverse transcription was caused by defects other than those in the tRNA placement.
Regions of MLV and SNV NCs can functionally replace their counterparts.
In this report, we generated four chimeric constructs containing portions of the NC sequences from another virus. Although relatively large stretches of amino acids were replaced with divergent sequences and the numbers of charged residues were altered, all four of the chimeric proteins were able to replicate with relatively mild defects. This result is in sharp contrast with studies showing that virus assembly and/or RNA packaging were severely affected when two or more amino acids from the same regions were altered (5, 10, 13, 24, 34, 49). For example, it was found that substituting one or two basic residues with neutral residues led to the moderate or strong attenuation of viral titers (34). Furthermore, the mutant with three basic residues substituted by neutral residues had a drastic reduced infectivity with severe defects in RNA packaging and reverse transcription (34). The NC in pMgpScb lost four basic residues (Fig. 1C); however, viruses derived from pMgpScb had only a mild defect in virus production and did not have a significant loss of RNA packaging and virus infectivity (Tables 1 and 2). We therefore conclude that, despite the divergence in primary sequences, flanking regions of NCs in MLV and SNV can functionally replace each other. This conclusion also lends credence to the hypothesis that the NC flanking regions in MLV and SNV are functionally conserved.
Phenotypes of NC chimeras.
The four NC chimeras had four different phenotypes based on the efficiency of virion production and infectivity of the viruses. Proteins encoded by pMgpSnb had a similar virion assembly efficiency and infectivity compared with that of the wild type. Proteins encoded by pMgpScb had a mild assembly defect and an infectivity comparable to that of the wild type. Proteins encoded by pSgpMnb had a mild assembly defect and a slight decrease in infectivity compared with those in the wild type, whereas proteins encoded by pSgpMcb had an assembly efficiency comparable to that of the wild type but a 10-fold-decreased infectivity.
The chimeric nature of NCs can impact the functions of the Gag polyprotein or the cleaved NC protein. We questioned whether defects of Gag polyprotein and NC protein could be independent from each other. The mild assembly defects observed in some of the chimeras were most likely caused by the Gag polyprotein with hampered functions. Defects in either the Gag polyprotein or the cleaved NC protein can cause a decrease in virus infectivity; therefore, it may be difficult to distinguish between the contributions of Gag and NC when the virus infectivity is reduced. However, since the chimeric viruses had an infectivity comparable to that of the wild-type virus, we conclude that the cleaved NC proteins can carry out their functions at efficiencies similar to those of wild-type NC. From these four chimeras, it appeared that the defects in virus assembly did not correlate with the decreased infectivity of the released virions. This observation suggests that whether the Gag polyprotein has a defect caused by NC is not a predictor of whether the cleaved NC protein can carry out its function during the replication cycle.
Charge distribution of flanking regions and possible effect on viral assembly.
The NC domain of the Gag polyprotein plays an important role in the Gag-Gag and Gag-RNA interactions that are critical to virion assembly (57, 60). One present view on the mechanisms of retroviral assembly is that the NC domain of Gag binds to RNA, which serves as a scaffold for the aggregation of viral proteins to allow virus assembly (45). It has been suggested that the basic nature of the NC is critical for its interaction with nucleic acid and virus assembly, which is supported by mutation analyses that demonstrated that substituting basic residues with neutral residues caused defects in virus assembly (5, 10, 13, 24, 34, 49).
The isoelectric points of MLV and SNV NCs were 10.41 and 9.95, respectively (Fig. 1C) (Genetics Computer Group program, University of Wisconsin). The isoelectric point for pMgpSnb- and pSgpMcb-encoded NCs was 10.79, which was close to that of the wild-type MLV NC, whereas the isoelectric point for pMgpScb- and pSgpMnb-encoded NCs was 8.36, which was far lower than those of the wild-type MLV and SNV NC proteins. The distribution of the isoelectric points correlated with the assembly phenotypes of these chimeras: proteins encoded by pMgpSnb and pSgpMcb were assembled with an efficiency equivalent to that of the wild type, whereas proteins encoded by pMgpScb and pSgpMnb had a mild assembly defect. This observation is in agreement with the mutational studies and suggests that the basic nature of the NC is important to virus assembly.
Chimeric NC and reverse transcription.
Virions produced by pSgpMcb-transfected cells had a 10-fold-reduced infectivity. However, the level of DNA synthesized by the chimeric NC-containing virions was reduced approximately 10-fold compared with that from the wild-type SNV NC-containing virions. Quantitative real-time PCR analyses revealed that both the early and late products of reverse transcription were reduced at similar levels. This finding suggests that the chimeric NC did not cause a defect in the processes needed to extend the reverse transcription products, including minus-strand DNA transfer, RT elongation during minus-strand DNA synthesis, and plus-strand DNA transfer. It has been proposed that the chaperone activity of NC impacts on the efficiency of strand-transfer events essential for the completion of reverse transcription (18, 33, 38, 51, 53). Our results demonstrated that the chimeric NC encoded by pSgpMcb could perform both strand transfer events at efficiencies similar to those of the wild-type NC. Our analyses indicated that the reduction in reverse transcription in pSgpMcb-derived virions appeared to occur prior to or at the very early stage of reverse transcription. Because the efficiency of tRNA placement in pSgpMcb-derived virions was similar to that in wild-type virions, it is possible that the chimeric NC was not as efficient as wild-type NC in forming or maintaining the initiation complex of reverse transcription. Alternatively, it is also possible that the chimeric NC-containing viruses suffered from inefficient uncoating or formation of the reverse transcription complex.
Structures of NC proteins and flexibility of flanking regions.
The nuclear magnetic resonance structures of HIV-1 and MLV NC have been determined (1, 14, 16, 55). In both NC proteins, the Cys-His boxes form well-defined zinc-binding structures whereas the flanking regions remain flexible. It is possible that, because the flanking regions have more flexible structures, they can tolerate large changes in the primary sequences, as demonstrated by the functional chimeric NC proteins in this study. However, it is important to note that the flexibility of the structures and the primary sequences does not imply that there are no requirements for sequences or motifs residing in the flanking regions, because the impact of the flanking regions on viral replication has been well documented by numerous mutation analyses (5, 10, 13, 24, 34, 49). The functional replacement of the MLV and SNV flanking regions also does not imply that the flanking regions of all retroviral NCs can be exchanged. First, retroviral NCs have a varied number of Cys-His boxes; therefore, elements important for NC function are likely to be distributed differently among these NCs. Secondly, viruses evolve to have the best-fit sequences; such sequence exchanges are likely to affect the fitness of the viruses in some manner. In our single-round replication assay, we could detect replication defects in three of the four chimeras. Nevertheless, the approach of generating and examining chimeras in this study has revealed novel information and can be used to further dissect elements important for the function of NCs.
Acknowledgments
We thank Vinay K. Pathak for his kind gift of the pWZH30 plasmid, the AIDS Vaccine Program and Nancy Rice (NCI-Frederick) for antibodies, Douglas Powell (NCI-Frederick) for assistance with the statistical analyses, Sara Cheslock for assistance with the GCG program, Sarah Palmer for assistance with setting up the real-time PCR analyses, and Anne Arthur for expert editorial assistance with the manuscript. We also thank Vinay K. Pathak for his intellectual input, helpful discussions, and reading of the manuscript and Alan Rein for helpful discussions and reading of the manuscript. We thank Que Dang, Sook-Kyung Lee, and Dexter Poon for critical reading of the manuscript.
This work was supported by the HIV Drug Resistance Program, National Cancer Institute, National Institutes of Health.
REFERENCES
- 1.Amarasinghe, G. K., R. N. De Guzman, R. B. Turner, K. J. Chancellor, Z. R. Wu, and M. F. Summers. 2000. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J. Mol. Biol. 301:491-511. [DOI] [PubMed] [Google Scholar]
- 2.Berkowitz, R., J. Fisher, and S. P. Goff. 1996. RNA packaging. Curr. Top. Microbiol. Immunol. 214:177-218. [DOI] [PubMed] [Google Scholar]
- 3.Berkowitz, R. D., A. Ohagen, S. Höglund, and S. P. Goff. 1995. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J. Virol. 69:6445-6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berthoux, L., C. Péchoux, M. Ottmann, G. Morel, and J.-L. Darlix. 1997. Mutations in the N-terminal domain of human immunodeficiency virus type 1 nucleocapsid protein affect virion core structure and proviral DNA synthesis. J. Virol. 71:6973-6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowzard, J. B., R. P. Bennett, N. K. Krishna, S. M. Ernst, A. Rein, and J. W. Wills. 1998. Importance of basic residues in the nucleocapsid sequence for retrovirus Gag assembly and complementation rescue. J. Virol. 72:9034-9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carteau, S., S. C. Batson, L. Poljak, J.-F. Mouscadet, H. de Rocquigny, J.-L. Darlix, B. P. Roques, E. Käs, and C. Auclair. 1997. Human immunodeficiency virus type 1 nucleocapsid protein specifically stimulates Mg2+-dependent DNA integration in vitro. J. Virol. 71:6225-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carteau, S., R. J. Gorelick, and F. D. Bushman. 1999. Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein. J. Virol. 73:6670-6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Certo, J. L., T. O. Kabdulov, M. L. Paulson, J. A. Anderson, and W. S. Hu. 1999. The nucleocapsid domain is responsible for the ability of spleen necrosis virus (SNV) Gag polyprotein to package both SNV and murine leukemia virus RNA. J. Virol. 73:9170-9177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Certo, J. L., B. F. Shook, P. D. Yin, J. T. Snider, and W. S. Hu. 1998. Nonreciprocal pseudotyping: murine leukemia virus proteins cannot efficiently package spleen necrosis virus-based vector RNA. J. Virol. 72:5408-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cimarelli, A., S. Sandin, S. Höglund, and J. Luban. 2000. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J. Virol. 74:3046-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dang, Q., and W. S. Hu. 2001. Effects of homology length in the repeat region on minus-strand DNA transfer and retroviral replication. J. Virol. 75:809-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darlix, J. L., A. Vincent, C. Gabus, H. de Rocquigny, and B. Roques. 1993. Trans-activation of the 5′ to 3′ viral DNA strand transfer by nucleocapsid protein during reverse transcription of HIV1 RNA. C. R. Acad. Sci. Ser. III 316:763-771. [PubMed] [Google Scholar]
- 13.Dawson, L., and X. F. Yu. 1998. The role of nucleocapsid of HIV-1 in virus assembly. Virology 251:141-157. [DOI] [PubMed] [Google Scholar]
- 14.De Guzman, R. N., Z. R. Wu, C. C. Stalling, L. Pappalardo, P. N. Borer, and M. F. Summers. 1998. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science 279:384-388. [DOI] [PubMed] [Google Scholar]
- 15.de la Luna, S., I. Soria, D. Pulido, J. Ortin, and A. Jimenez. 1988. Efficient transformation of mammalian cells with constructs containing a puromycin-resistance marker. Gene 62:121-126. [DOI] [PubMed] [Google Scholar]
- 16.Demene, H., N. Jullian, N. Morellet, H. de Rocquigny, F. Cornille, B. Maigret, and B. P. Roques. 1994. Three-dimensional 1H NMR structure of the nucleocapsid protein NCp10 of Moloney murine leukemia virus. J. Biomol. NMR 4:153-170. [DOI] [PubMed] [Google Scholar]
- 17.Dougherty, J. P., R. Wisniewski, S. L. Yang, B. W. Rhode, and H. M. Temin. 1989. New retrovirus helper cells with almost no nucleotide sequence homology to retrovirus vectors. J. Virol. 63:3209-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Driscoll, M. D., and S. H. Hughes. 2000. Human immunodeficiency virus type 1 nucleocapsid protein can prevent self-priming of minus-strand strong stop DNA by promoting the annealing of short oligonucleotides to hairpin sequences. J. Virol. 74:8785-8792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Embretson, J. E., and H. M. Temin. 1987. Lack of competition results in efficient packaging of heterologous murine retroviral RNAs and reticuloendotheliosis virus encapsidation-minus RNAs by the reticuloendotheliosis virus helper cell line. J. Virol. 61:2675-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng, Y. X., T. D. Copeland, L. E. Henderson, R. J. Gorelick, W. J. Bosche, J. G. Levin, and A. Rein. 1996. HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro. Proc. Natl. Acad. Sci. USA 93:7577-7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu, W., R. J. Gorelick, and A. Rein. 1994. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J. Virol. 68:5013-5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu, W., B. A. Ortiz-Conde, R. J. Gorelick, S. H. Hughes, and A. Rein. 1997. Placement of tRNA primer on the primer-binding site requires pol gene expression in avian but not murine retroviruses. J. Virol. 71:6940-6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu, W., and A. Rein. 1993. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J. Virol. 67:5443-5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu, X. D., R. A. Katz, A. M. Skalka, and J. Leis. 1988. Site-directed mutagenesis of the avian retrovirus nucleocapsid protein, pp 12. Mutation which affects RNA binding in vitro blocks viral replication. J. Biol. Chem. 263:2140-2145. [PubMed] [Google Scholar]
- 25.Gonsky, J., E. Bacharach, and S. P. Goff. 2001. Identification of residues of the Moloney murine leukemia virus nucleocapsid critical for viral DNA synthesis in vivo. J. Virol. 75:2616-2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorelick, R. J., R. E. Benveniste, T. D. Gagliardi, T. A. Wiltrout, L. K. Busch, W. J. Bosche, L. V. Coren, J. D. Lifson, P. J. Bradley, L. E. Henderson, and L. O. Arthur. 1999. Nucleocapsid protein zinc-finger mutants of simian immunodeficiency virus strain mne produce virions that are replication defective in vitro and in vivo. Virology 253:259-270. [DOI] [PubMed] [Google Scholar]
- 27.Gorelick, R. J., D. J. Chabot, D. E. Ott, T. D. Gagliardi, A. Rein, L. E. Henderson, and L. O. Arthur. 1996. Genetic analysis of the zinc finger in the Moloney murine leukemia virus nucleocapsid domain: replacement of zinc-coordinating residues with other zinc-coordinating residues yields noninfectious particles containing genomic RNA. J. Virol. 70:2593-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gorelick, R. J., D. J. Chabot, A. Rein, L. E. Henderson, and L. O. Arthur. 1993. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. J. Virol. 67:4027-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorelick, R. J., W. Fu, T. D. Gagliardi, W. J. Bosche, A. Rein, L. E. Henderson, and L. O. Arthur. 1999. Characterization of the block in replication of nucleocapsid protein zinc finger mutants from Moloney murine leukemia virus. J. Virol. 73:8185-8195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorelick, R. J., T. D. Gagliardi, W. J. Bosche, T. A. Wiltrout, L. V. Coren, D. J. Chabot, J. D. Lifson, L. E. Henderson, and L. O. Arthur. 1999. Strict conservation of the retroviral nucleocapsid protein zinc finger is strongly influenced by its role in viral infection processes: characterization of HIV-1 particles containing mutant nucleocapsid zinc-coordinating sequences. Virology 256:92-104. [DOI] [PubMed] [Google Scholar]
- 31.Gorelick, R. J., L. E. Henderson, J. P. Hanser, and A. Rein. 1988. Point mutants of Moloney murine leukemia virus that fail to package viral RNA: evidence for specific RNA recognition by a “zinc finger-like”protein sequence. Proc. Natl. Acad. Sci. USA 85:8420-8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gorelick, R. J., S. M. Nigida, Jr., J. W. Bess, Jr., L. O. Arthur, L. E. Henderson, and A. Rein. 1990. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J. Virol. 64:3207-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo, J., L. E. Henderson, J. Bess, B. Kane, and J. G. Levin. 1997. Human immunodeficiency virus type 1 nucleocapsid protein promotes efficient strand transfer and specific viral DNA synthesis by inhibiting TAR-dependent self-priming from minus-strand strong-stop DNA. J. Virol. 71:5178-5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Housset, V., H. De Rocquigny, B. P. Roques, and J.-L. Darlix. 1993. Basic amino acids flanking the zinc finger of Moloney murine leukemia virus nucleocapsid protein NCp10 are critical for virus infectivity. J. Virol. 67:2537-2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang, Y., A. Khorchid, J. Gabor, J. Wang, X. Li, J.-L. Darlix, M. A. Wainberg, and L. Kleiman. 1998. The role of nucleocapsid and U5 stem/A-rich loop sequences in tRNA3Lys genomic placement and initiation of reverse transcription in human immunodeficiency virus type 1. J. Virol. 72:3907-3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang, Y., A. Khorchid, J. Wang, M. A. Parniak, J.-L. Darlix, M. A. Wainberg, and L. Kleiman. 1997. Effect of mutations in the nucleocapsid protein (NCp7) upon Pr160gag-pol and tRNALys incorporation into human immunodeficiency virus type 1. J. Virol. 71:4378-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ji, X., G. J. Klarmann, and B. D. Preston. 1996. Effect of human immunodeficiency virus type 1 (HIV-1) nucleocapsid protein on HIV-1 reverse transcriptase activity in vitro. Biochemistry 35:132-143. [DOI] [PubMed] [Google Scholar]
- 38.Lapadat-Tapolsky, M., C. Pernelle, C. Borie, and J. L. Darlix. 1995. Analysis of the nucleic acid annealing activities of nucleocapsid protein from HIV-1. Nucleic Acids Res. 23:2434-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Linial, M. L. 1999. Foamy viruses are unconventional retroviruses. J. Virol. 73:1747-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martinez, I., and R. Dornburg. 1995. Improved retroviral packaging lines derived from spleen necrosis virus. Virology 208:234-241. [DOI] [PubMed] [Google Scholar]
- 41.Méric, C., and S. P. Goff. 1989. Characterization of Moloney murine leukemia virus mutants with single-amino-acid substitutions in the Cys-His box of the nucleocapsid protein. J. Virol. 63:1558-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Méric, C., and P.-F. Spahr. 1986. Rous sarcoma virus nucleic acid-binding protein p12 is necessary for viral 70S RNA dimer formation and packaging. J. Virol. 60:450-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller, A. D., and C. Buttimore. 1986. Redesign of retrovirus packaging cell lines to avoid recombination leading to helper virus production. Mol. Cell. Biol. 6:2895-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller, A. D., J. V. Garcia, N. von Suhr, C. M. Lynch, C. Wilson, and M. V. Eiden. 1991. Construction and properties of retrovirus packaging cells based on gibbon ape leukemia virus. J. Virol. 65:2220-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muriaux, D., J. Mirro, D. Harvin, and A. Rein. 2001. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. USA 98:5246-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oertle, S., and P.-F. Spahr. 1990. Role of the gag polyprotein precursor in packaging and maturation of Rous sarcoma virus genomic RNA. J. Virol. 64:5757-5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petropoulos, C. 1997. Retroviral taxonomy, protein structures, sequences, and genetic maps. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 48.Poon, D. T., G. Li, and A. Aldovini. 1998. Nucleocapsid and matrix protein contributions to selective human immunodeficiency virus type 1 genomic RNA packaging. J. Virol. 72:1983-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poon, D. T., J. Wu, and A. Aldovini. 1996. Charged amino acid residues of human immunodeficiency virus type 1 nucleocapsid p7 protein involved in RNA packaging and infectivity. J. Virol. 70:6607-6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rein, A., D. P. Harvin, J. Mirro, S. M. Ernst, and R. J. Gorelick. 1994. Evidence that a central domain of nucleocapsid protein is required for RNA packaging in murine leukemia virus. J. Virol. 68:6124-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rein, A., L. E. Henderson, and J. G. Levin. 1998. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci. 23:297-301. [DOI] [PubMed] [Google Scholar]
- 52.Riggs, J. L., R. M. McAllister, and E. H. Lennette. 1974. Immunofluorescent studies of RD-114 virus replication in cell culture. J. Gen. Virol. 25:21-29. [DOI] [PubMed] [Google Scholar]
- 53.Rodriguez-Rodriguez, L., Z. Tsuchihashi, G. M. Fuentes, R. A. Bambara, and P. J. Fay. 1995. Influence of human immunodeficiency virus nucleocapsid protein on synthesis and strand transfer by the reverse transcriptase in vitro. J. Biol. Chem. 270:15005-15011. [DOI] [PubMed] [Google Scholar]
- 54.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 55.Schuler, W., C. Dong, K. Wecker, and B. P. Roques. 1999. NMR structure of the complex between the zinc finger protein NCp10 of Moloney murine leukemia virus and the single-stranded pentanucleotide d(ACGCC): comparison with HIV-NCp7 complexes. Biochemistry 38:12984-12994. [DOI] [PubMed] [Google Scholar]
- 56.Stoltzfus, C. M., and P. N. Snyder. 1975. Structure of B77 sarcoma virus RNA: stabilization of RNA after packaging. J. Virol. 16:1161-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 58.Temin, H. M., and S. Mizutani. 1970. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature 226:1211-1213. [DOI] [PubMed] [Google Scholar]
- 59.Tsuchihashi, Z., and P. O. Brown. 1994. DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 68:5863-5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vogt, V. M. 1997. Retroviral virions and genomes. Cold Spring Harbor Press, Cold Spring Harbor, N.Y. [PubMed]
- 61.Yin, P. D., V. K. Pathak, A. E. Rowan, R. J. Teufel II, and W. S. Hu. 1997. Utilization of nonhomologous minus-strand DNA transfer to generate recombinant retroviruses. J. Virol. 71:2487-2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yovandich, J. L., E. N. Chertova, B. P. Kane, T. D. Gagliardi, J. W. Bess, Jr., R. C. Sowder II, L. E. Henderson, and R. J. Gorelick. 2001. Alteration of zinc-binding residues of simian immunodeficiency virus p8NC results in subtle differences in Gag processing and virion maturation associated with degradative loss of mutant NC. J. Virol. 75:115-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang, Y., and E. Barklis. 1997. Effects of nucleocapsid mutations on human immunodeficiency virus assembly and RNA encapsidation. J. Virol. 71:6765-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, Y., H. Qian, Z. Love, and E. Barklis. 1998. Analysis of the assembly function of the human immunodeficiency virus type 1 Gag protein nucleocapsid domain. J. Virol. 72:1782-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]